Abstract
In a study of 114 epidemiologically linked Zambian transmission pairs, we evaluated the impact of human leukocyte antigen class I (HLA-I)–associated amino acid polymorphisms, presumed to reflect cytotoxic T lymphocyte (CTL) escape in Gag and Nef of the virus transmitted from the chronically infected donor, on the plasma viral load (VL) in matched recipients 6 mo after infection. CTL escape mutations in Gag and Nef were seen in the donors, which were subsequently transmitted to recipients, largely unchanged soon after infection. We observed a significant correlation between the number of Gag escape mutations targeted by specific HLA-B allele–restricted CTLs and reduced VLs in the recipients. This negative correlation was most evident in newly infected individuals, whose HLA alleles were unable to effectively target Gag and select for CTL escape mutations in this gene. Nef mutations in the donor had no impact on VL in the recipient. Thus, broad Gag-specific CTL responses capable of driving virus escape in the donor may be of clinical benefit to both the donor and recipient. In addition to their direct implications for HIV-1 vaccine design, these data suggest that CTL-induced viral polymorphisms and their associated in vivo viral fitness costs could have a significant impact on HIV-1 pathogenesis.
Control of viral replication after infection has been attributed, at least in part, to CTL (CD8+) activity (1). Although all infected individuals can mount HIV-specific CTL responses, sustained, near-complete, viral control is only evident in a few who are recognized as aviremic controllers (2). Progression to AIDS occurs in the vast majority of untreated HIV infected patients, and this loss of immune control is partly caused by CTL escape mutations (3). Escape mutations modify a viral genome during infection for optimal replication in the absence of an immune response. Thus, CTLs targeting structurally or functionally conserved epitopes might benefit the host by diminishing the ability of the virus to develop escape mutations while at the same time retaining replicative fitness. Independent studies have demonstrated that CTLs directed against Gag, a relatively conserved protein with structurally critical domains, correlate with improved clinical markers of disease progression (4, 5). A recent study of >500 antiretroviral therapy naive HIV-1 patients in South Africa demonstrated that the best level of viral control occurred in patients who targeted Gag-specific CTL epitopes that are presented by specific HLA class I (HLA-I) alleles (6). In many cases, such targeting leads to the selection of specific HLA-I–associated amino acid polymorphisms that result in reduced epitope recognition (1). It remains unclear whether the clinical benefit observed in these individuals is caused by the quality of the immune responses, to viral fitness constraints resulting from CTL escape, or to both.
Evidence that it is a combination of these factors that results in reduced plasma viral load (VL) is supported by studies in individuals who carry certain HLA class I alleles and who control HIV well in the absence of antiretroviral therapy. HLA-B*57 and HLA-B*27, in particular, have been consistently associated with improved markers of clinical outcome and slower rates of progression to AIDS (7). HIV-specific CTLs restricted by these two HLA molecules dominate in individuals expressing these alleles (8), with Gag being the major CTL target. Distinct qualitative differences have been demonstrated in CTLs restricted by HLA-B*57 (9) or HLA-B*27 (10). Recent studies demonstrate reversion of CTL escape mutations upon transmission into a new host in the absence of HLA-mediated selection pressure (11, 12). These in vivo findings have been supported by in vitro studies showing a reduced replication capacity of viruses with certain CTL escape mutations (13, 14). Nevertheless, none of these mutations have been shown to negatively affect VL or modify other biological markers of clinical outcome in chronic infection. One explanation may be that CTL escape mutations, which reduce the immunological suppression of virus replication (3), can simultaneously reduce the replication capacity of the virus (11, 13, 14). In other words, although a CTL escape mutation may result in a reduced viral replication capacity, it is not likely that this will result in a lower VL in the chronically infected host, where this mutation was induced, because the virus is gaining a fitness advantage by avoiding CTLs. Such fitness effects may therefore only be evident upon transmission to a recipient lacking the same restricting HLA-I molecule.
In this report, we took advantage of the HLA-I–associated HIV-1 polymorphisms that had been defined in Gag and Nef from studies of a large South African cohort with chronic clade C infection (unpublished data). We used this information to study—in a cohort of epidemiologically linked, clade C–infected Zambian HIV-1 transmission pairs—the effects of mutations induced in the transmitted virus by donor CTLs on an early VL set point in the newly infected recipient. Our findings suggest that HLA-B–restricted mutations in Gag, but not Nef, result in a reduced VL upon transmission, and directly demonstrate for the first time in vivo that a clinically significant viral fitness cost is induced when the virus escapes immune recognition by donor CTLs.
RESULTS AND DISCUSSION
To identify HLA-I–associated HIV-1 subtype C polymorphisms, we performed HLA-I typing on 701 untreated South African patients with chronic HIV-1 infection and sequenced the gag and nef genes of their circulating virus (unpublished data) (12, 15). Associations were corrected for viral phylogeny, as this approach was shown to significantly improve the specificity of HLA-I associations (16, 17). We limited our initial analysis to HLA-I viral polymorphisms that occurred within or flanked (within five amino acids of the N or C termini of the epitope) known HLA-I–restricted CTL epitopes. Such HIV-1 polymorphisms are more likely to represent CTL escape and less likely to represent confounding secondary compensatory mutations (11, 13, 14). These analyses resulted in the identification of 20 mutations within Gag (18 within p24) and 11 within Nef (Table I). The majority of the mutations within Gag (17 out of 20; 85%) and Nef (8 out of 11; 73%) were HLA-B restricted. Of those mutations that have been fully evaluated, all (five in Gag and two in Nef) were formally shown to be CTL escape variants (12, 15, 18–20).
Table I.
HLA class I allele–associated HIV-1 polymorphisms occurring within or flanking known CTL epitopes in chronically clade C–infected South Africans
| HIV-1 subregion | Common mutationsa | Defined epitopesb | Specific HLA-I associationc |
|---|---|---|---|
| Gag-p17 | EKIRLRPGGKKHYML | RY10 (Gag 20–29) | B*4201, P = 1.1 × 10−4; q = 0.07 |
| EKIRLRPGGKKHYML | RY10 (Gag 20–29) | B*4201, P = 1.7 × 10−4; q = 0.17 | |
| Gag-p24 | HQAISPRTLNAWVKV | IW9 (Gag 147–155) | B*5703, P = 2.5 × 10−14; q = 0 |
| HQAISPRTLNAWVKV | IW9 (Gag 147–155) | B*5703, P = 1.6 × 10−8; q = 0 | |
| QAISPRTLNAWVKVI | SV9 (Gag 148–156) | B*8101, P = 2.8 × 10−3; q = 0.15 | |
| IEEKAFSPEVIPMFT | KF11 (Gag 162–171) | B*5703, P = 1.4 × 10−15; q = 0 | |
| IEEKAFSPEVIPMFT | KF11 (Gag 162–171) | B*5703, P = 2.9 × 10−4; q = 0.04 | |
| EGATPQDLNTMLNTV | TL9 (Gag 180–188) | B*8101, P = 4.3 × 10−7; q = 0 | |
| EGATPQDLNTMLNTV | TL9 (Gag 180–188) | B*8101, P = 3.6 × 10−5; q = 0.01 | |
| EGATPQDLNTMLNTV | TL9 (Gag 180–188) | B*8101, P = 1.9 × 10−3; q = 0.11 | |
| AGTTSTLQEQIAWMT | TW10 (Gag 240–249) | B*57/5801, P = 1.9 × 10−28; q = 0 | |
| AGTTSTLQEQIAWMT | TW10 (Gag 240–249) | B*5703, P = 1.7 × 10−5; q = 0 | |
| TSNPPIPVGDIYKRW | PY9 (Gag 254–262) | B*35, P = 1.1 × 10−7; q = 0 | |
| DYVDRFFKTLRAEQA | DA9 (Gag 298–306) | B*1401, P = 2.5 × 10−11; q = 0 | |
| FRDYVDRFFKTLRAE | YL9 (Gag 296–304) | Cw*0304, P = 3 × 10−4; q = 0.04 | |
| RAEQATQDVKNWMTD | QW9 (Gag 308–316) | B*5801, P = 3.7 × 10−7; q = 0 | |
| RAEQATQDVKNWMTD | AW11 (Gag 306–316) | B*4403, P = 3 × 10−9; q = 0.03 | |
| TILRALGPGATLEEM | RL9 (Gag 335–343) | Cw*08, P = 9.3 × 10−4; q = 0.09 | |
| TILRALGPGATLEEM | RL9 (Gag 335–343) | Cw*08, P = 8.5 × 10−4; q = 0.09 | |
| GVGGPGHKARVLAEA | GL9 (Gag 355–363) | B*0702/05, P = 6.7 × 10−10; q = 0.03 | |
| Nef | EEEEEVGFPVRPQVP | EV11 (Nef 65–75) | B*4501, P = 8.3 × 10−9; q = 0 |
| FPVRPQVPLRPMTYK | RM9 (Nef 72–80) | B*0702, P = 10−9; q = 0 | |
| FPVRPQVPLRPMTYK | RM9 (Nef 72–80) | B*8101, P = 1.8 × 10−10; q = 0 | |
| RPQVPLRPMTYKAAF | VY8 (Nef 74–81) | B*35, P = 1.1 × 10−7; q = 0 | |
| MTYKAAFDLSFFLKE | KF9 (Nef 82–90) | B*57/5801, P = 7.3 × 10−6; q = 0.07 | |
| FFLKEKGGLEGLIYS | KL9 (Nef 91–100) | B*4403, P = 1.9 × 10−8; q = 0 | |
| YSKKRQEILDLWVYH | KY11 (Nef 105–115) | Cw*0701, P = 7.9 × 10−7; q = 0 | |
| YSKKRQEILDLWVYH | KY11 (Nef 105–115) | B*18, P = 1.4 × 10−9, q = 0 | |
| GVRYPLTFGWCFKLV | YF9 (Nef 135–143) | B*35, P = 6.7 × 10−6; q = 0 | |
| PGVRYPLTFGWCFKL | RW8 (Nef 134–141) | A*2402, P = 10−4; q = 0.20 | |
| PGVRYPLTFGWCFKL | RW8 (Nef 134–141) | A*2301, P = 6.8 × 10−12; q = 0 |
These data are taken from 59 HLA associations (q < 0.2) defined in a clade C South African cohort (unpublished data), in which analysis of gag sequences was performed using data from 672 study subjects and analysis of nef sequences was performed using data from 443 study subjects. The Los Alamos National Laboratory HIV Molecular Immunology Database (available at http://www.hiv.lanl.gov/content/index) was used to define known CTL epitopes.
HLA-associated mutations (bolded) shown in the context of the known CTL epitopes (underlined).
Defined CTL epitopes with amino acid residues based on the HXB2 sequences in parentheses.
Methods for calculating p- and q-values have been previously described (reference 16).
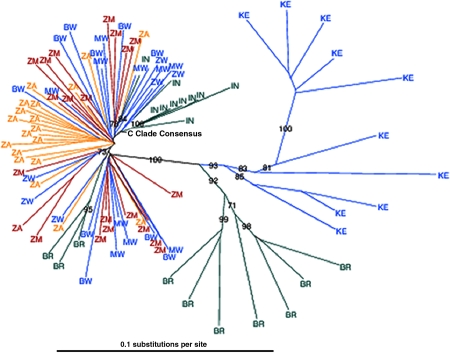
We then applied these HLA-I–associated polymorphisms, found in chronic infection, to the analyses of early HIV-1 transmission within a cohort of 114 epidemiologically linked Zambian transmission pairs. These couples were initially identified as HIV-1 discordant (i.e., one partner was HIV-1 infected and their spouse was uninfected). Despite counseling and condom provision, transmission from the chronically infected partner to their spouse continues, but at a significantly reduced rate (∼8% per year) (21). Epidemiological linkage of the transmission pairs was established through sequence analysis of the regions encoding gp41 and Gag (22). Population-based sequencing of gag and nef genes was performed on DNA RT-PCR amplified from plasma virus RNA obtained ∼6 mo after the estimated date of infection. A phylogenetic tree, constructed to determine the relationships of HIV-1 gag gene sequences between the two cohorts studied in this paper and other clade C sequences derived from the Los Alamos National Laboratory HIV Molecular Immunology Database (Fig. 1), demonstrated that Southern African sequences tend to cluster together, distinct from those prevalent in representative countries in East Africa, South America, and Asia. These data suggest that sequence analyses derived from one country may be closely applicable to viruses found in neighboring countries in this region. This is supported by previous experiments that have reported the identical escape mutations selected in geographically distinct but otherwise similar clade B–infected cohorts (12).
Figure 1.
Southern African clade C Gag sequences tend to cluster together. Neighbor-joining phylogenetic tree of Gag (p17 and p24) clade C taxa is shown. These taxa (20 from each cohort) were randomly selected from the total pool of sequencing data available from the South African (ZA, yellow) and Zambian (ZM, red) cohorts. We used an HIV database (Table I) to obtain clade C consensus and sequences from other countries affected by the clade C epidemic. Other African countries are shown (blue), with 10 sequences each from Botswana (BW), Kenya (KE), and Malawi (MW), and 6 sequences from Zimbabwe (ZW). Non-African taxa (green) are represented by 10 sequences each from India (IN) and Brazil (BW). Bootstrap values >70% are shown.
After HLA-I typing of both partners from each of the 114 epidemiologically linked Zambian transmission pairs, plasma VL was determined from the same samples that viral gag and nef gene sequences were derived. We examined the relationship of amino acid polymorphisms in Gag and Nef sequences to early VL in the newly infected partners of this Zambian transmission pair cohort. We reasoned that early VL (after seroconversion) would be more reflective of the characteristics of the virus transmitted from the donor than VL seen later during chronic infection, when reversion and compensatory mutations might occur. Indeed, the majority of HIV-1 polymorphisms (579 out of 610; 94.9%) were identical in the donor and recipient at 6 mo after transmission. Gag mutations were more likely to be unchanged in the recipient when compared with Nef mutations, because the latter had a greater tendency to exist as the clade C consensus amino acid in the recipient (97.1 vs. 91.3%, respectively; P = 0.002).
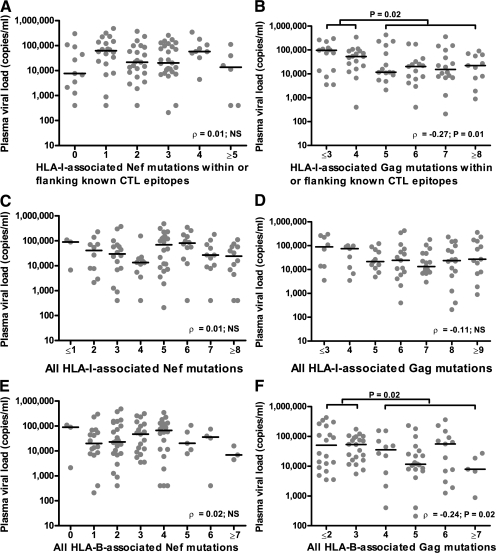
Next, we analyzed the relationship between the numbers of transmitted HLA-I–linked Gag and Nef mutations within or flanking known CTL epitopes (Table I) to early VL in recipient partners. Accumulation of HLA-I–associated polymorphisms in Nef was not linked to either a higher or lower VL in recipients (ρ = 0.01; NS; Fig. 2 A). In contrast, a higher number of HLA-I–associated mutations within Gag was significantly associated with lower VL (ρ = −0.27; P = 0.01; Fig. 2 B).
Figure 2.
HLA-B–associated mutations in donor Gag affect HIV-1 plasma VL in linked recipients. (A and B) In linked recipients in the Zambian cohort (n = 114), the association between the numbers of mutations within or flanking known CTL epitopes (Table I) in HIV-1 Nef (A) and Gag (B) with VL is shown. (C and D) All HLA-I–restricted polymorphisms associated with Nef and Gag mutations, respectively, were included (P < 0.001 and q < 0.2), regardless of whether they were within or flanking known CTL epitopes (Table I and Table S1). (E and F) all HLA-B–restricted mutations (P < 0.001 and q < 0.2) for Nef and Gag, respectively. The horizontal bars represent median VL values. The Mann-Whitney U test was used to compare median VL between groups, and the nonparametric Spearman rank correlation test was used to determine whether a correlation exists between the number of mutations and VL.
Although limiting the HLA-I–associated HIV-1 polymorphisms to those occurring within or flanking known CTL epitopes strengthens the case that these changes represent CTL escape mutations, it potentially introduces selection bias into our dataset. We therefore included all HLA-I–associated polymorphisms (P < 0.001; q < 0.2) whether or not they were coupled with known CTL epitopes (Table I; and Table S1, available at http://www.jem.org/cgi/content/full/jem.20072457/DC1). This provided an additional 16 mutations in Gag and 12 mutations in Nef. Unlike what was observed for mutations associated with known CTL epitopes, we saw a more even distribution of the HLA-I restriction of these mutations for both Gag (seven HLA-A, four HLA-B, and four HLA-C restricted) and Nef (four restricted by each HLA-I allele). No associations were seen when comparing the numbers of Nef mutations with VL (ρ = 0.01; NS; Fig. 2 C), consistent with the previous analysis, and there was no longer a significant association with the total number of Gag mutations and VL (ρ = −0.11; NS; Fig. 2 D). In contrast, when we focused the analysis on HLA-B–associated Gag polymorphisms, these remained significantly associated with diminished VL (ρ = −0.24; P = 0.02; Fig. 2 F). HLA-A– and HLA-C–associated Gag polymorphisms were not associated with a lower recipient VL (ρ = 0.08; NS; unpublished data). Similarly, HLA-B–associated Nef mutations were not associated with either positive or negative VL changes in the recipients (ρ = 0.02; NS; Fig. 2 E).
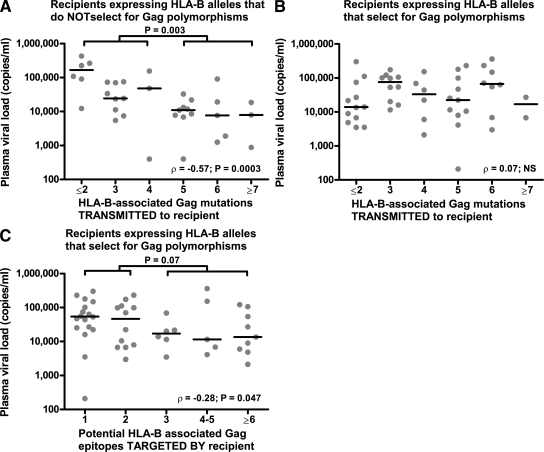
These results (Fig. 2 F) support previous studies demonstrating that the HIV-1–specific CTL responses with the greatest impact on VL are HLA-B restricted (23) and are directed against Gag epitopes associated with CTL escape mutations (6), perhaps reflecting the selection pressure imposed by these effective responses. Thus, a confounding factor in this analysis of the in vivo fitness cost of escape mutations may be the new HIV-1–specific CTL responses, certainly present a 6 mo after infection, which could by themselves reduce VL. This would imply that newly infected individuals lacking the HLA-B molecules that drive CTL escape mutations in Gag (Table I) would benefit most from the receipt of virus with detrimental escape mutations in Gag. When only these latter recipients were included in the analysis, the association between the increasing number of Gag mutations and reduced VL was much stronger (ρ = −0.57; P = 0.0003; Fig. 3 A).
Figure 3.
Recipients lacking HLA-B alleles associated with Gag mutations benefit most from these mutations. (A and B) The relationship between VL and HLA-B–associated polymorphisms in Gag is shown for recipients who lack (A) or carry (B) the HLA-B alleles associated with these mutations (Table I). (C) The relationship between VL and the number of Gag epitopes that are potentially targeted in the same recipients as in B. Potential epitopes are defined as the number of Gag polymorphisms that are associated with the HLA-B alleles of each recipient. Panel A represents 35 recipients, whereas B and C represent 50 recipients each. The horizontal bars represent median VL values. Statistics were performed as described in Fig. 2.
This finding also suggests that the recipients who are able to effectively target Gag themselves would derive less absolute benefit from CTL-induced Gag mutations because of the equipoise between CTL-derived virus suppression and the fitness costs associated with escape mutations. Indeed, we did not see any association between the number of Gag mutations and VL in recipient carriers of HLA-B alleles that select for the Gag CTL escape mutations described in Table I (ρ = 0.07; NS; Fig. 3 B). To determine whether newly induced Gag-specific CTL responses could be contributing to VL control in this latter recipient group, the number of HLA-B–restricted Gag epitopes that could potentially be targeted in each individual was quantified and compared with VL. In line with previous findings (4, 6), the greater the number of Gag CTL epitopes potentially targeted by these recipients, the greater the decrease in VL (ρ = −0.28; P = 0.047; Fig. 3 C). This latter finding in recipients with Gag-specific CTLs capable of inducing mutations is also similar to a recently published paper in which an inverse relationship was demonstrated between VL and HLA-I–associated polymorphisms in Gag, Pol, and Nef in chronically infected hosts (24). Our analysis expands on this study because CTL escape mutations induced by the recipient's immune system were almost nonexistent at the time point analyzed (unpublished data). Therefore, it is likely that the potency of CTLs associated with mutations in Gag plays an important role in early viral control, even if these mutations do not occur.
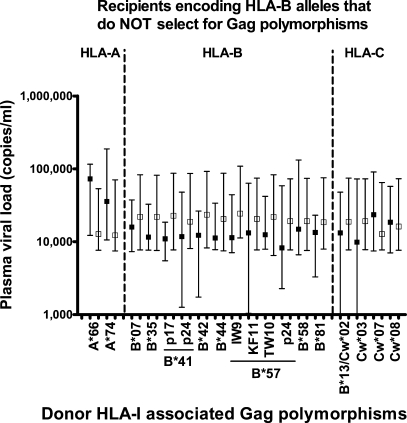
Because certain HLA-B alleles are associated with improved disease outcomes (7, 23), it was important to consider the possibility that the significant associations with VL were driven by Gag mutations restricted only by those HLA-I alleles previously associated with a lack of disease progression (e.g., HLA-B*57 or -B*5801). We therefore stratified the data according to which HLA-I–associated Gag mutation was present in recipients who lacked HLA-B alleles that drive escape (Fig. 4). Although none of the VL associations with any individual HLA-I allele were statistically significant, certain patterns could be seen. All 12 HLA-B–associated Gag mutations were linked to lower VLs in HLA-mismatched recipients (compared with only one out of five HLA-A– or HLA-C–associated mutations; P = 0.002). Of note, this trend was not unique to HLA-B alleles associated with good clinical markers, but instead was also observed with HLA-B*07, -B*35, -B*41, and -B*44. These data suggest that it is number of CTL epitopes targeted by any individual HLA allele (as assessed by the frequency of Gag mutations induced) that drives its association with lower VL. Notably, 13 out of 20 (65%) of the mutations in Gag (Table I) were restricted by HLA molecules (HLA-B*42, -B*57, -B*5801, and -B*8101) previously associated with lower VLs and a better clinical outcome (7, 23).
Figure 4.
Lower VL in recipients is not restricted to a select class of donor HLA-B alleles. VLs based on transmission of viruses with (closed squares) or without (open squares) Gag mutations restricted by individual HLA-I alleles are shown. The data represent only recipients (n = 36) who do not express HLA-B molecules associated with Gag polymorphisms, as shown in Table I. Only mutations present in at least two of the recipients are shown. The Gag mutation restricted by either HLA-B*13 or -Cw*02 could not be resolved to either individual allele. Squares represent median values, and error bars represent the interquartile range of the data.
Previous studies have examined VL differences based on CTL escape that occurs during the course of chronic infection. These analyses are complicated by the fact that the occurrence of a CTL escape mutation in an individual abolishes immunological suppression of viral replication in that individual. Indeed, escape from an HLA-B*27–restricted Gag response results in a higher VL (3) despite the fact that it is associated with reduced replicative capacity in vitro (14). It is therefore difficult to assess whether any particular escape mutation results in a viral fitness cost in chronically infected patients. This current study is unique in that the identity of the transmitted virus was known in each of the individuals early after infection. This allowed us to observe the impact of HLA-I–induced mutations on virus replication after it entered a new immunogenetic environment where the fitness cost of the mutations would be most pronounced. Importantly, we demonstrate two different mechanisms of viral control in newly infected recipients. Similar to previous experiments (6), those recipients expressing certain HLA-B alleles likely control VL by effective CTL Gag targeting. In contrast, newly infected recipients who lack HLA-B alleles associated with Gag polymorphisms can derive significant benefit from the transmission of viruses containing Gag mutations induced in partners with potent HLA-B–restricted responses.
One possible explanation for the finding that mutations in Gag, but not Nef, affect viral fitness is that the former protein (and in particular p24) must make multiple interactions with other Gag molecules during the assembly of both immature viral capsids and mature viral cores, as well as with cellular components such as cyclophilin A (25). This relative structural inflexibility may allow the immune system, through its targeting of Gag, to significantly affect viral replication, and would provide a strong mechanistic rationale for the repeated observation that Gag-specific CTLs correlate with biological markers of improved clinical outcome (4–6).
It is not clear why HLA-B–restricted responses directed against HIV-1 play such an important role in immunopathogenesis, although several groups have demonstrated this association (6, 8, 23). It is possible the HLA-B alleles just happen to target important regions in p24 with a higher binding affinity (26). Furthermore HLA-A, -B, and -C are differentially expressed depending on the cell type (27), and it is plausible that certain antigen-presenting cells preferentially express HLA-B molecules. In addition to CTL function, HLA-B Bw4 interacts with NK cells, and this was recently demonstrated to play an important role in HIV-1 control (28).
The ∼10-fold VL reduction associated with increasing numbers of escape mutations (fewer than two versus more than six Gag mutations; Fig. 3 A) observed in this report may be clinically significant, because even a 2.5-fold lower VL among recent HIV-1 seroconverters was associated with a benefit in AIDS-free survival (29). Compensatory mutations (11, 13) or reversions (30) occurring after our analyzed time point could dampen the impact of CTL escape on viral fitness constraints. Follow-up VL data were available for 39 recipients at ∼1 yr after seroconversion or 6 mo after the first evaluation. Although the VL significantly increased at the second time point compared with the first (70,928 vs. 21,961 copies/ml, respectively; P < 0.0001 using the Wilcoxon signed-rank test), it was higher in all patients irrespective of the number of Gag polymorphisms or whether or not the patients carried the “protective” HLA-I alleles. Therefore, it is difficult to know whether the increase in plasma VL seen after the first year of infection is caused by the development of compensatory mutations, the loss of immune control, or other factors. Nevertheless, these HLA-I–induced mutations are certainly present at the earliest point of infection and would be expected to also influence peak VL. Because much of the CD4 depletion occurs before seroconversion (31), these viral fitness mutations may contribute to long-lasting clinical benefit even if their biological impact is lost at some point after seroconversion. It would have been interesting to evaluate the impact of viral fitness mutations on CD4 T cells; however, this clinical test was not available at the time of the study.
Although these findings need to be replicated in other cohorts, they strengthen the argument for CTL targeting of Gag antigens as an important mechanism for sustained viral control. Moreover, if a virus encoding escape mutations in gag were to be transmitted to a new host in the absence of fully compensatory mutations (as observed in this report), these new recipients would likely benefit as well. These data imply that for CTL-based HIV vaccines to effectively control VL, they must simultaneously target multiple Gag epitopes, thereby ensuring that fitness constraints prevent the virus from easily mutating. Thus, efforts to identify other CTL-induced HIV-1 mutations in structurally important regions of Gag (or other proteins), and to define mechanisms by which these mutations influence VL set point and disease progression will ultimately benefit the design of an AIDS vaccine.
MATERIALS AND METHODS
Patients.
The South African cohort consisted of 680 patients with chronic HIV-1 infection from Durban, South Africa (6, 23). The Zambian participants included 114 transmission pairs, shown to have epidemiologically linked HIV-1, from the Zambia-Emory HIV Research Project (21, 22). All patients in both cohorts were antiretroviral therapy naive. Zambian linked recipients were identified at 5.6 mo (mean; median = 3.1 mo) after the estimated time of infection, at which time plasma samples were obtained from both the donor and the recipient. All research protocols were approved by the ethics committees in Durban, South Africa and Lusaka, Zambia, and by the Emory University and Oxford University Institutional Review Boards.
HIV-1 sequencing.
Viral RNA from Zambian samples was extracted from 200 μl of plasma using a robotic system with the MagNA Pure LC Total Nucleic Acid Isolation Kit (Roche). 10 and 1 μl RNA were amplified using a one-step RT-PCR high fidelity kit (Invitrogen) for the RT and first-round amplification steps using gene-specific primers. Gag primers included (forward) 5′-ATTTGACTAGCGGAGGCTAGAA-3′ and (reverse) 5′-GACAGGTGTAGGTCCTACTAATACTGTACC-3′. Nef primers included (forward) 5′-AATAGAGTTAGGCAGGGATAC-3′ (32) and (reverse) 5′-GCACTCAAGGCAAGCTTTATTGAGGCTTA-3′. For nested PCR reactions, primer sequences for gag amplification were (inner forward) 5′-TTTGACTAGCGGAGGCTAGAAGGA-3′ and (inner reverse) 5′-GTATCATCTGCTCCTGTGTCTAAGAGAGC-3′. The primer sequences for nef gene amplification were (inner forward) 5′-GAGAGACTTCATATTGGTTGCAGCG-3′ (a gift from S. Mallal, Murdoch University, Murdoch, Australia) and (inner reverse) 5′-AAAGCAGCTGCTTATATGCAGCATCT-3′. PCR products were purified using Montage MultiScreen PCR96 filtration plates (Millipore), and amplicons were sequenced by Macrogen. Sequences were analyzed using Sequencher 4.7 software (Gene Codes Corp.). Multiple peaks were recorded using International Union of Pure and Applied Chemistry codes whenever the lower peak level exceeded one third of the level of the maximum peak, and nucleotide sequences were then converted to amino acid sequences using the universal code. We also performed gag sequencing of multiple single genomes from both the donor and the recipient (mean = 20 clones in each group) in a subset of 11 transmission pairs. The consensus sequencing method mirrored the major polymporphisms transmitted from the donor to the recipient (unpublished data). HIV-1 sequencing of South African plasma samples was performed in a similar fashion (unpublished data).
Plasma VL.
Plasma VL was determined at the Emory Center for AIDS Research Virology Core Laboratory using the Amplicor HIV-1 Monitor Test (version 1.5; Roche).
HLA class I typing.
Using genomic DNA prepared from whole blood or buffy coats (QIAamp blood kit; QIAGEN), HLA class I genotyping relied on a combination of PCR-based techniques, including PCR with sequence-specific primers (Invitrogen) and reference-strand conformation analysis (Invitrogen), as described previously (33). Ambiguities were resolved by sequencing-based typing using kits (Abbott Molecules, Inc.) designed for capillary electrophoresis and the ABI 3130xl DNA Analyzer (Applied Biosystems).
Calculation of HLA-I–associated HIV polymorphisms.
We used a method similar to those previously described (16, 34) to identify HLA-I–associated HIV polymorphisms. This approach corrected for the phylogenetic structure of the sequences. Additionally, we included multiple HLA predictors, allowing for the elimination of spurious associations caused by HLA linkage disequilibrium. In brief, for both Gag and Nef, a maximum likelihood phylogenetic tree was constructed from the corresponding sequences. For every HLA allele, amino acid position, and amino acid at that position, two generative—or directed graphical—models of the observed presence or absence of the amino acid in each sequence were created: one represented the null hypothesis that the observations are generated by the phylogenetic tree alone, and the other represented the alternative hypothesis that additional escape or reversion takes place because of HLA pressure in the subjects for which the sequences are observed. The likelihood of the observations was then maximized over the parameters of both models using an expectation-maximization algorithm, and a p-value was computed using a likelihood ratio test based on those likelihoods. To increase power, the tests were binarized such the presence or absence of a given HLA allele was correlated with the presence or absence of a given amino acid. HLA polymorphism pairs were also analyzed only when the actual or expected count in every cell of the corresponding two-by-two contingency table was greater than or equal to three. For every amino acid at each position, the HLA allele with the strongest association (and its corresponding p-value) was added to the list of identified associations. The analysis was repeated after removing individuals having or possibly having this HLA allele. This procedure was iterated until no HLA allele yielded an association with P < 0.05. A q-value statistic, estimating the proportion of false positives among the associations identified, was computed for each association by repeating this analysis on null data. These q-value statistics were estimated separately for Gag and Nef associations using one million null tests per protein. Associations with q-values less than or equal to 0.2 were deemed significant.
Analysis of HLA-I–associated HIV-1 polymorphisms in linked Zambian transmission pairs.
HIV-1 gag and nef sequences were analyzed in 88 and 96 out of 114 Zambian transmission pairs, respectively. HIV-1 polymorphisms were defined based on associations calculated in the South African cohort. Only predicted CTL escapes and attractions were analyzed. HLA-I associations that were either not present in the Zambian cohort or were associated with less than or equal to one mutation were not included in the analysis. In cases where more than one HLA-I was associated with a specific polymorphism, the linked amino acid also associated with a known CTL epitope was used. If no CTL association was present, then the HLA-I allele demonstrating the greatest association with the HIV-1 polymorphism was used. The donor sequence was treated as the transmitted sequence, except in cases where the donor encoded an escape mutation and the recipient encoded the clade C consensus amino acid (69 out of 1,004, or 6.9%, of the mutations).
Statistics.
Comparisons between VL and the number of mutations were performed by the nonparametric Spearman rank correlation and Mann-Whitney U tests.
Online supplemental material.
Table S1 lists HLA-I allele–associated HIV-1 polymorphisms not occurring within or near known CTL epitopes. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20072457/DC1.
Supplementary Material
Acknowledgments
We thank the Zambia-Emory HIV Research Project participants and those in Durban who made possible the collection of blood samples for this study.
This work was supported by National Institutes of Health (NIH) grants AI-64060 (E. Hunter, P.A. Goepfert, and R.A. Kaslow) and AI-46995 (P.J.R. Goulder), the International AIDS Vaccine Initiative (S. Allen and E. Hunter), the Wellcome Trust (P.J.R. Goulder), the Medical Research Fund UK (P. Matthews and A. Prendergast), Microsoft Research (D. Heckerman and J.M. Carlson), and the Emory Center for AIDS Research Virology Core through a NIH center grant (P30 AI050409). E. Hunter is a Georgia Research Alliance Eminent Scholar, and P.J.R. Goulder is an Elizabeth Glaser Pediatric AIDS Foundation scientist.
The authors have no conflicting financial interests.
References
- 1.Goulder, P.J., and D.I. Watkins. 2004. HIV and SIV CTL escape: implications for vaccine design. Nat. Rev. Immunol. 4:630–640. [DOI] [PubMed] [Google Scholar]
- 2.Bailey, J.R., T.M. Williams, R.F. Siliciano, and J.N. Blankson. 2006. Maintenance of viral suppression in HIV-1–infected HLA-B*57+ elite suppressors despite CTL escape mutations. J. Exp. Med. 203:1357–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goulder, P.J., R.E. Phillips, R.A. Colbert, S. McAdam, G. Ogg, M.A. Nowak, P. Giangrande, G. Luzzi, B. Morgan, A. Edwards, et al. 1997. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat. Med. 3:212–217. [DOI] [PubMed] [Google Scholar]
- 4.Edwards, B.H., A. Bansal, S. Sabbaj, J. Bakari, M.J. Mulligan, and P.A. Goepfert. 2002. Magnitude of functional CD8+ T-cell responses to the gag protein of human immunodeficiency virus type 1 correlates inversely with viral load in plasma. J. Virol. 76:2298–2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramduth, D., P. Chetty, N.C. Mngquandaniso, N. Nene, J.D. Harlow, I. Honeyborne, N. Ntumba, S. Gappoo, C. Henry, P. Jeena, et al. 2005. Differential immunogenicity of HIV-1 clade C proteins in eliciting CD8+ and CD4+ cell responses. J. Infect. Dis. 192:1588–1596. [DOI] [PubMed] [Google Scholar]
- 6.Kiepiela, P., K. Ngumbela, C. Thobakgale, D. Ramduth, I. Honeyborne, E. Moodley, S. Reddy, C. de Pierres, Z. Mncube, N. Mkhwanazi, et al. 2007. CD8(+) T-cell responses to different HIV proteins have discordant associations with viral load. Nat. Med. 13:46–53. [DOI] [PubMed] [Google Scholar]
- 7.Kaslow, R.A., M. Carrington, R. Apple, L. Park, A. Munoz, A.J. Saah, J.J. Goedert, C. Winkler, S.J. O'Brien, C. Rinaldo, et al. 1996. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat. Med. 2:405–411. [DOI] [PubMed] [Google Scholar]
- 8.Altfeld, M., E.T. Kalife, Y. Qi, H. Streeck, M. Lichterfeld, M.N. Johnston, N. Burgett, M.E. Swartz, A. Yang, G. Alter, et al. 2006. HLA Alleles Associated with Delayed Progression to AIDS Contribute Strongly to the Initial CD8(+) T Cell Response against HIV-1. PLoS Med. 3:e403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Migueles, S.A., A.C. Laborico, W.L. Shupert, M.S. Sabbaghian, R. Rabin, C.W. Hallahan, D. Van Baarle, S. Kostense, F. Miedema, M. McLaughlin, et al. 2002. HIV-specific CD8+ T cell proliferation is coupled to perforin expression and is maintained in nonprogressors. Nat. Immunol. 3:1061–1068. [DOI] [PubMed] [Google Scholar]
- 10.Almeida, J.R., D.A. Price, L. Papagno, Z.A. Arkoub, D. Sauce, E. Bornstein, T.E. Asher, A. Samri, A. Schnuriger, I. Theodorou, et al. 2007. Superior control of HIV-1 replication by CD8+ T cells is reflected by their avidity, polyfunctionality, and clonal turnover. J. Exp. Med. 204:2473–2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crawford, H., J.G. Prado, A. Leslie, S. Hue, I. Honeyborne, S. Reddy, M. van der Stok, Z. Mncube, C. Brander, C. Rousseau, et al. 2007. Compensatory mutation partially restores fitness and delays reversion of escape mutation within the immunodominant HLA-B*5703-restricted Gag epitope in chronic human immunodeficiency virus type 1 infection. J. Virol. 81:8346–8351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leslie, A.J., K.J. Pfafferott, P. Chetty, R. Draenert, M.M. Addo, M. Feeney, Y. Tang, E.C. Holmes, T. Allen, J.G. Prado, et al. 2004. HIV evolution: CTL escape mutation and reversion after transmission. Nat. Med. 10:282–289. [DOI] [PubMed] [Google Scholar]
- 13.Brockman, M.A., A. Schneidewind, M. Lahaie, A. Schmidt, T. Miura, I. Desouza, F. Ryvkin, C.A. Derdeyn, S. Allen, E. Hunter, et al. 2007. Escape and compensation from early HLA-B57-mediated cytotoxic T-lymphocyte pressure on human immunodeficiency virus type 1 Gag alter capsid interactions with cyclophilin A. J. Virol. 81:12608–12618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schneidewind, A., M.A. Brockman, R. Yang, R.I. Adam, B. Li, S. Le Gall, C.R. Rinaldo, S.L. Craggs, R.L. Allgaier, K.A. Power, et al. 2007. Escape from the dominant HLA-B27-restricted cytotoxic T-lymphocyte response in Gag is associated with a dramatic reduction in human immunodeficiency virus type 1 replication. J. Virol. 81:12382–12393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leslie, A., D. Kavanagh, I. Honeyborne, K. Pfafferott, C. Edwards, T. Pillay, L. Hilton, C. Thobakgale, D. Ramduth, R. Draenert, et al. 2005. Transmission and accumulation of CTL escape variants drive negative associations between HIV polymorphisms and HLA. J. Exp. Med. 201:891–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhattacharya, T., M. Daniels, D. Heckerman, B. Foley, N. Frahm, C. Kadie, J. Carlson, K. Yusim, B. McMahon, B. Gaschen, et al. 2007. Founder effects in the assessment of HIV polymorphisms and HLA allele associations. Science. 315:1583–1586. [DOI] [PubMed] [Google Scholar]
- 17.Brumme, Z.L., C.J. Brumme, D. Heckerman, B.T. Korber, M. Daniels, J. Carlson, C. Kadie, T. Bhattacharya, C. Chui, J. Szinger, et al. 2007. Evidence of Differential HLA Class I-Mediated Viral Evolution in Functional and Accessory/Regulatory Genes of HIV-1. PLoS Pathog. 3:e94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bansal, A., E. Gough, S. Sabbaj, D. Ritter, K. Yusim, G. Sfakianos, G. Aldrovandi, R.A. Kaslow, C.M. Wilson, M.J. Mulligan, et al. 2005. CD8 T-cell responses in early HIV-1 infection are skewed towards high entropy peptides. AIDS. 19:241–250. [PubMed] [Google Scholar]
- 19.Draenert, R., S. Le Gall, K.J. Pfafferott, A.J. Leslie, P. Chetty, C. Brander, E.C. Holmes, S.C. Chang, M.E. Feeney, M.M. Addo, et al. 2004. Immune selection for altered antigen processing leads to cytotoxic T lymphocyte escape in chronic HIV-1 infection. J. Exp. Med. 199:905–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leslie, A., D.A. Price, P. Mkhize, K. Bishop, A. Rathod, C. Day, H. Crawford, I. Honeyborne, T.E. Asher, G. Luzzi, et al. 2006. Differential selection pressure exerted on HIV by CTL targeting identical epitopes but restricted by distinct HLA alleles from the same HLA supertype. J. Immunol. 177:4699–4708. [DOI] [PubMed] [Google Scholar]
- 21.Fideli, U.S., S.A. Allen, R. Musonda, S. Trask, B.H. Hahn, H. Weiss, J. Mulenga, F. Kasolo, S.H. Vermund, and G.M. Aldrovandi. 2001. Virologic and immunologic determinants of heterosexual transmission of human immunodeficiency virus type 1 in Africa. AIDS Res. Hum. Retroviruses. 17:901–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trask, S.A., C.A. Derdeyn, U. Fideli, Y. Chen, S. Meleth, F. Kasolo, R. Musonda, E. Hunter, F. Gao, S. Allen, and B.H. Hahn. 2002. Molecular epidemiology of human immunodeficiency virus type 1 transmission in a heterosexual cohort of discordant couples in Zambia. J. Virol. 76:397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kiepiela, P., A.J. Leslie, I. Honeyborne, D. Ramduth, C. Thobakgale, S. Chetty, P. Rathnavalu, C. Moore, K.J. Pfafferott, L. Hilton, et al. 2004. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature. 432:769–775. [DOI] [PubMed] [Google Scholar]
- 24.Frater, A.J., H. Brown, A. Oxenius, H.F. Gunthard, B. Hirschel, N. Robinson, A.J. Leslie, R. Payne, H. Crawford, A. Prendergast, et al. 2007. Effective T-cell responses select human immunodeficiency virus mutants and slow disease progression. J. Virol. 81:6742–6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lanman, J., T.T. Lam, M.R. Emmett, A.G. Marshall, M. Sakalian, and P.E. Prevelige Jr. 2004. Key interactions in HIV-1 maturation identified by hydrogen-deuterium exchange. Nat. Struct. Mol. Biol. 11:676–677. [DOI] [PubMed] [Google Scholar]
- 26.Borghans, J.A., A. Molgaard, R.J. de Boer, and C. Kesmir. 2007. HLA alleles associated with slow progression to AIDS truly prefer to present HIV-1 p24. PLoS ONE. 2:e920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson, D.R. 2000. Differential expression of human major histocompatibility class I loci: HLA-A, -B, and -C. Hum. Immunol. 61:389–396. [DOI] [PubMed] [Google Scholar]
- 28.Alter, G., M.P. Martin, N. Teigen, W.H. Carr, T.J. Suscovich, A. Schneidewind, H. Streeck, M. Waring, A. Meier, C. Brander, et al. 2007. Differential natural killer cell–mediated inhibition of HIV-1 replication based on distinct KIR/HLA subtypes. J. Exp. Med. 204:3027–3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lyles, R.H., A. Munoz, T.E. Yamashita, H. Bazmi, R. Detels, C.R. Rinaldo, J.B. Margolick, J.P. Phair, and J.W. Mellors. 2000. Natural history of human immunodeficiency virus type 1 viremia after seroconversion and proximal to AIDS in a large cohort of homosexual men. Multicenter AIDS Cohort Study. J. Infect. Dis. 181:872–880. [DOI] [PubMed] [Google Scholar]
- 30.Kuntzen, T., J. Timm, A. Berical, L.L. Lewis-Ximenez, A. Jones, B. Nolan, J. Schulze zur Wiesch, B. Li, A. Schneidewind, A.Y. Kim, et al. 2007. Viral sequence evolution in acute HCV infection. J. Virol. 81:11658–11668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brenchley, J.M., T.W. Schacker, L.E. Ruff, D.A. Price, J.H. Taylor, G.J. Beilman, P.L. Nguyen, A. Khoruts, M. Larson, A.T. Haase, and D.C. Douek. 2004. CD4+ T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract. J. Exp. Med. 200:749–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shugars, D.C., M.S. Smith, D.H. Glueck, P.V. Nantermet, F. Seillier-Moiseiwitsch, and R. Swanstrom. 1993. Analysis of human immunodeficiency virus type 1 nef gene sequences present in vivo. J. Virol. 67:4639–4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tang, J., S. Tang, E. Lobashevsky, A.D. Myracle, U. Fideli, G. Aldrovandi, S. Allen, R. Musonda, and R.A. Kaslow. 2002. Favorable and unfavorable HLA class I alleles and haplotypes in Zambians predominantly infected with clade C human immunodeficiency virus type 1. J. Virol. 76:8276–8284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carlson, J., C. Kadie, S. Mallal, and D. Heckerman. 2007. Leveraging hierarchical population structure in discrete association studies. PLoS ONE. 2:e591. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.