Abstract
The energy landscape of a peptide [Ace-Lys-Gln-Cys-Arg-Glu-Arg-Ala-Nme] in explicit water was studied with a multicanonical molecular dynamics simulation, and the AMBER parm96 force field was used for the energy calculation. The peptide was taken from the recognition helix of the DNA-binding protein, c-Myb. A rugged energy landscape was obtained, in which the random-coil conformations were dominant at room temperature. The CD spectra of the synthesized peptide revealed that it is in the random state at room temperature. However, the 300 K canonical ensemble, Q(300K), contained α-helix, 310-helix, β-turn, and β-hairpin structures with small but notable probabilities of existence. The complete α-helix, imperfect α-helix, and random-coil conformations were separated from one another in the conformational space. This means that the peptide must overcome energy barriers to form the α-helix. The overcoming process may correspond to the hydrogen-bond rearrangements from peptide–water to peptide–peptide interactions. The β-turn, imperfect 310-helix, and β-hairpin structures, among which there are no energy barriers at 300 K, were embedded in the ensemble of the random-coil conformations. Two types of β-hairpin with different β-turn regions were observed in Q(300K). The two β-hairpin structures may have different mechanisms for the β-hairpin formation. The current study proposes a scheme that the random state of this peptide consists of both ordered and disordered conformations. In contrast, the energy landscape obtained from the parm94 force field was funnel like, in which the peptide formed the helical conformation at room temperature and random coil at high temperature.
Keywords: Folding, rugged surface, funnel, β-hairpin, α-helix, random state, multicanonical, force field
The thermodynamic stability and the folding process of a polypeptide chain are determined by the energy landscape of the system. One picture for the landscape is a funnel-like surface, which has the advantage of quickly folding the chain into a unique tertiary structure. The other picture is a rugged surface, in which a number of energy local minima are widely spread, and the chain thermally fluctuates among the minima at a given temperature.
The folding funnel has been studied with simplified models (Bryngelson et al. 1995; Chan and Dill 1998; Dill 1999; Istrail et al. 1999; Nakamura and Sasai 1999), in which the chain was designed to fold into a unique, stable structure at the ground state or at a low temperature. An advantage of the simplified model is that the thermodynamically important states can be counted relatively precisely, and thus the free energies of the states are evaluated. The simplified model is also used to specify the determinant factors to fold the chain into unique tertiary structures (Irback and Potthast 1995; Li et al. 1996; Tatsumi and Chikenji 1999). A β-sheet protein, β-lactoglobulin, is known to pass through α-helical intermediates in the refolding process (Hamada et al. 1996), which cannot be explained with the simple funnel-like landscape. A simplified model by Chikenji and Kikuchi (2000) suggested the possibility that this protein kinetically moves down the slope of the free-energy landscape from the random-coil state to the α-helical state in the early stage of folding, and the thermal fluctuation in the helical state plays the role of a driving factor to carry the chain to the β-structure. However, due to simplicity of and adjustable parameters included in the model, the model itself cannot produce realistic discussions without the support of the atomic-level simulations or experiments.
A molecular dynamics (MD) simulation is a technique to trace the folding or unfolding process of a protein at an atomic resolution, and can complement experiments (Wong et al. 2000). The MD simulations (Boczko and Brooks 1995; Lazarids and Karplus 1997) implied that the funnel-like landscape is assigned to proteins with well-defined tertiary structures. Unfolding simulations (Tsai et al. 1999; Wang et al. 1999a) demonstrated that β-hairpin regions in proteins were considerably stable after the disruption of the other regions, due to the high stability of the β-turn.
Recently, the denatured states of proteins have been studied with unfolding simulations (Bond et al. 1997; Kazmirski and Dagget 1998; Wong et al. 2000). The long unfolding-simulation trajectories demonstrated that the denatured states involve residual structures consisting of thermally fluctuating secondary-structure elements. The residual structures also have been experimentally detected, and are considered to be important for protein folding (Neri et al. 1992; Shortle 1993, Shortle 1996; Blanco and Serrano 1995; Frank et al. 1995; Searle et al. 1996; Gillespie and Shortle 1997a,b; Wang and Shortle 1997; Blanco et al. 1998; Kamatari et al. 1999; Sinclair and Shortle 1999). Some proteins or protein segments are intrinsically unstructured in the native state (Wright and Dyson 1999), and this property was considered to be biologically important because the unstructured segments may have the ability to bind to several different targets.
Although the conventional MD technique provided valuable information about protein refolding, a full description of the protein folding process is still difficult because of the limitation of computation time and the inaccuracy in the force-field parameters (Brooks 1998). Thus, the full description was first attempted for short peptides. The folding mechanism of the α-helix has been studied for many years (Zimm and Bragg 1959). With the recent development of computer power, folding simulations of peptides are now achievable. Takano et al. (1999) observed the helix-coil transitions with an MD simulation of a 15-residue polyalanine peptide in explicit water. Daura et al. (1999) observed the folding and unfolding of the helix of a β-heptapeptide in methanol. The weighted histogram analysis method was used to provide more statistically reliable data on the helix propagation for the hydrophobic peptides Ace-(Ala)n-Nme, in which n = 4-15 (Yong and Brooks 1996). Duan and Kollman (1998) performed a 1-μs folding simulation of a 36-residue helical peptide in explicit water. In contrast, the folding mechanism of β-structures has not been well characterized (Munoz et al. 1997). Recently, the formation of β-structures was studied experimentally (Ramirez-Alvarado et al. 1996; Munoz et al. 1997; Alba et al. 1999; Honda et al. 2000). The MD simulations of the β-hairpin, the minimal β-structure element, were done and possible folding mechanisms were discussed (Dinner et al. 1999; Roccatano et al. 1999; Wang et al. 1999b; Bonvin and van Gunsteren, 2000). Galzitskaya et al. (2000) preformed the simulated annealing of a 16-residue peptide, which was taken from the β-hairpin region of the src SH3 domain, in explicit water starting from random conformations, and observed the formation of stable β-hairpins, in which the β-turn was first formed. Unfolding simulations of the β-hairpin (Pande and Rokhser 1999) showed that the unfolding process consisted of four distinguishable states.
A drawback of the conventional MD method is the sampling inefficiency in the conformational space, in which high-energy barriers are complicatedly distributed. Even then it is still difficult to obtain data statistically reliable for deriving the energy landscape even for the short peptides. Thus, to develop an effective sampling method is a key to describe precisely the energy landscape. Caves et al. (1998) reported that the integration of 10 short (120 ps) conventional MD runs provides a better sampling efficiency than a long (5 ns) conventional MD run. This exemplifies the fact that the sampling efficiency strongly depends on the sampling algorithm, although the study with the conventional MD simulation focused only on the conformational fluctuations around the native structure at 300 K. Recently, some powerful sampling methods have been developed to enhance the sampling of dense systems; multicanonical Monte Carlo sampling (Berg and Neuhaus 1992), entropic sampling (Lee 1993), replica-exchange Monte Carlo sampling (Hukushima and Nemoto 1996), simulated tempering (Lyubartsev et al. 1992), the 1/k-ensemble method (Hesselbo and Stinchcombe 1995), and Tsallis statistics (Tsallis 1988). These methods were applied to biological systems (Hansmann and Okamoto 1993, Hansmann and Okamoto 1997a,b; Kidera 1995; Andricioaei and Straub 1996; Hansmann et al. 1996; Bartels and Karplus 1997, Bartels and Karplus 1998; Bartels et al. 1998, 1999; Schaefer et al. 1998). The multicanonical algorithm was expanded to the simplified protein model (Iba et al. 1998; Chikenji et al. 1999; Kikuchi et al. 2000). Sugita and Okamoto have coupled the replica-exchange algorithm with the MD method (Sugita and Okamoto 1999, Sugita and Okamoto 2000a,b; Sugita et al. 2000).
Recently, Nakajima et al. (1997a) developed a multicanonical MD method. This method was applied to the structural prediction of CDR loops in antibodies (Shirai et al. 1998; Kim et al. 1999), the flexible docking between the CH3 domain and a pro-rich peptide (Nakajima et al. 1997b), and the evaluation of the free-energy landscape of peptide dimers in explicit water (Nakajima et al. 2000). This method was extended by dividing the total energy into two or more terms (Higo et al. 1997; Nakajima 1998), in which the sampling enhancement was done differently. Furthermore, the method was coupled with the weighted histogram analysis method to accurately evaluate high-energy barrier regions (transition states) in the conformational space (Ono et al. 1999). The advantage of the multicanonical method is that the canonical ensemble at any temperature can be derived from the sampling. Thus, we used this method for the conformational sampling of a peptide in explicit water to obtain the energy landscape, and measured the CD spectra for the synthesized peptide.
Results
The peptide we studied is [1Ace-2Lys-3Gln-4Cys-5Arg-6Glu-7Arg-8Ala-9Nme], in which Ace and Nme are, respectively, the N-terminal acetyl and C-terminal N-methyl groups. This sequence is derived from the recognition helix (third α-helix; the 128'th to 134'th residues) of the DNA-binding protein c-Myb, as determined by Ogata et al. (1995). The residue 8Ala is Phe in the original pdb data. This residue was replaced to reduce the system size. The amino acid residues 2Lys, 5Arg, 7Arg, and 6Glu were treated as charged. Note that the carbonyl oxygen (1O) in 1Ace and the hydrogen (9HN) of the amide group in 9Nme can participate in hydrogen bond (H-bond) formation. The peptide was immersed in an explicit water sphere with a radius = 17 Å (see Materials and Methods). The number of atoms in the system was 1818 (135 peptide atoms and 561 water molecules).
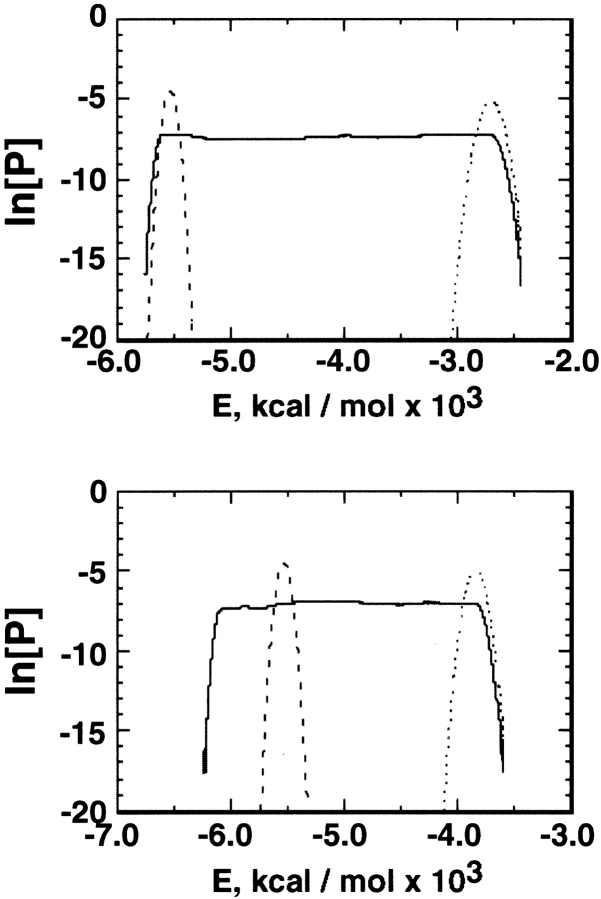
Two force fields, AMBER parm94 (Cornell et al. 1995) and parm96 (Kollman et al. 1997), were examined for the simulation (see Materials and Methods). The number of multicanonical iterative runs was 38 for the parm94 simulation, and 46 for the parm96 simulation. The number of MD steps in the last multicanonical run (i.e., sampling run) was 16.8 × 106 (16.8 ns) for parm94 and 50.4 × 106 (50.4) for parm96, and flat energy distributions (Fig. 1 ▶) were obtained. The parm96 simulation covered the range of 230–600 K, and the parm94 simulation covered 290–1000 K. As shown later, only the results from parm96 agreed with the experiment. Thus, we mainly describe the results from parm96.
Fig. 1.
Flat distribution (natural logarithm of probability P) of potential-energy (E) from the last multicanonical run. Top: parm94 simulation; bottom: parm96 simulation. In both, the dotted lines are the canonical distribution at T0 (see text), and the broken lines at 300 K.
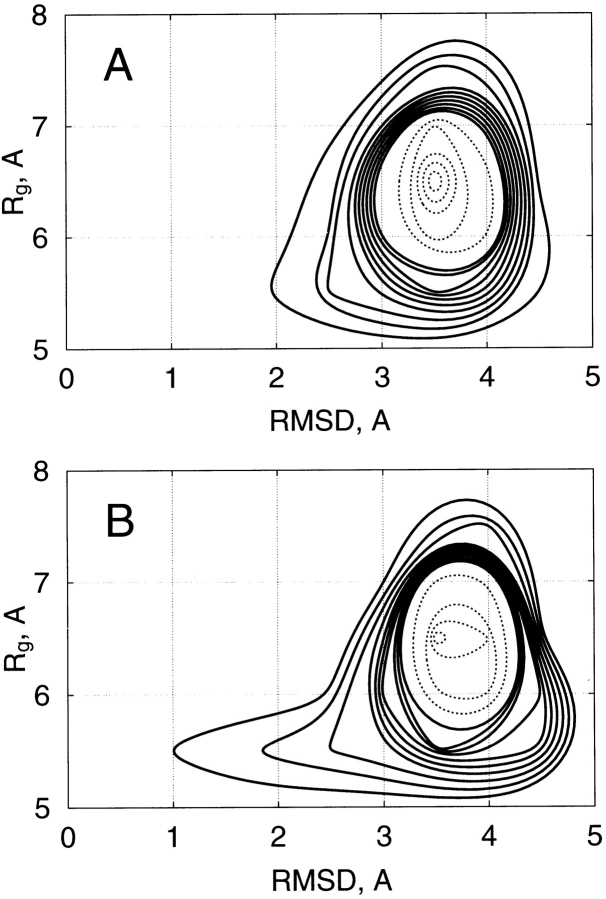
Figure 2A ▶ represents the relation between the potential energy (E) and the radius of gyration (Rg) of the peptide obtained from parm96: The Rg was widely distributed at any energy. Figure 2B ▶ is the relation between E and the main-chain root mean square deviation (rmsd), which was calculated between the sampled conformations from parm96 and the α-helix conformation (qcMyb) in the pdb data of c-Myb. The rmsd was distributed between 2.5 Å and 4.0 Å at any energy, and rarely approached 1 Å. Figure 3 ▶ is the conformational distribution from parm96 on the plane of rmsd and Rg at 300 and 500 K. The main conformational feature was the random-coil conformations at both temperatures, although the probability of the α-helix increased as temperature decreased.
Fig. 2.
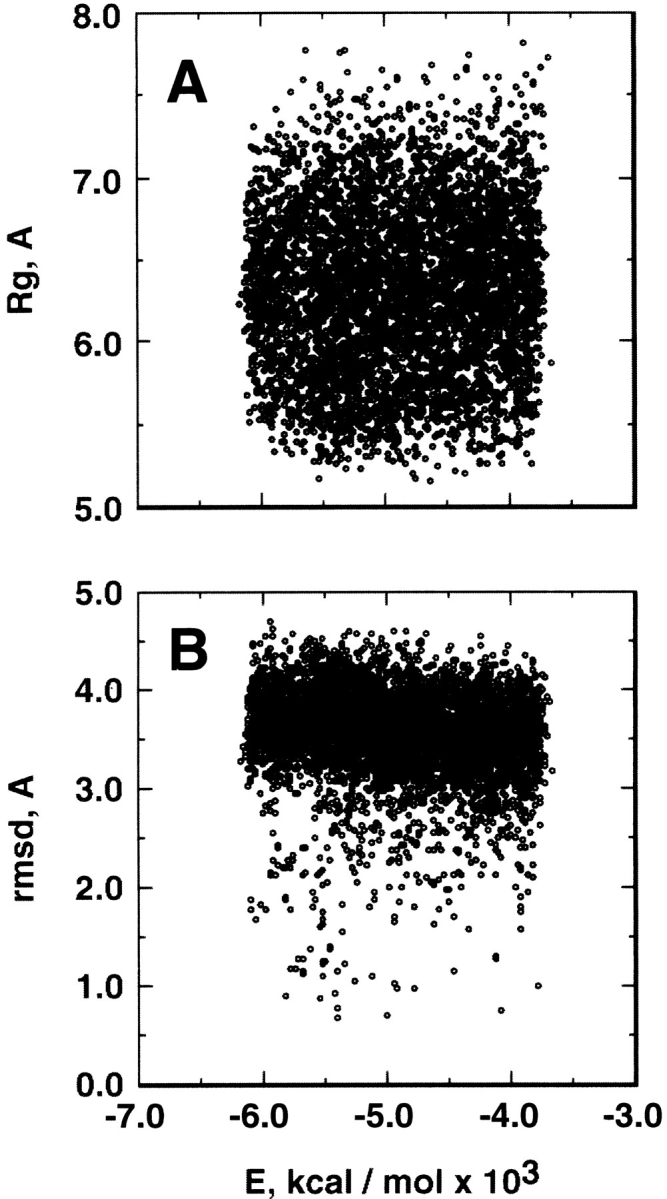
(A) Relation between potential energy, E, from parm96 and radius of gyration, Rg. (B) Relation between E from parm96 and the main-chain root mean square deviation, rmsd, between the sampled conformations and α-helix (qcMyb) in c-Myb.
Fig. 3.
Distribution of conformations from parm96 on plane of rmsd and Rg at 500 K (A) and 300 K (B). Solid contour lines represent probabilities from 0 to 0.2, with a spacing of 0.02, and broken lines from 0.2 to 1, with a spacing of 0.1.
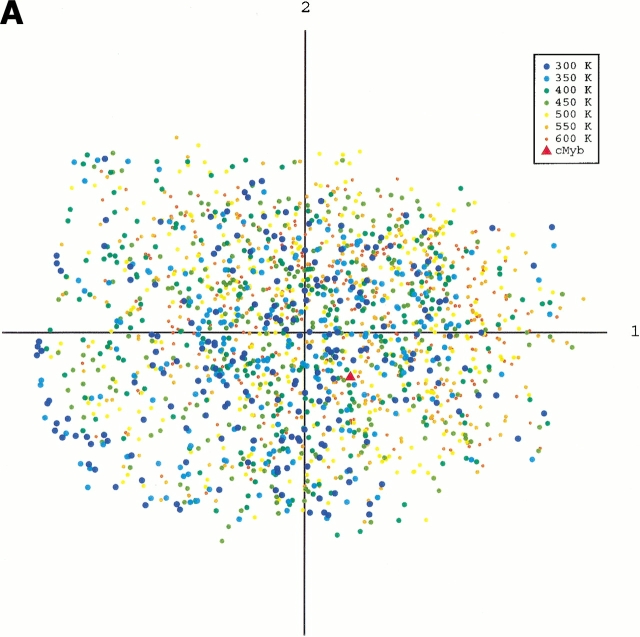
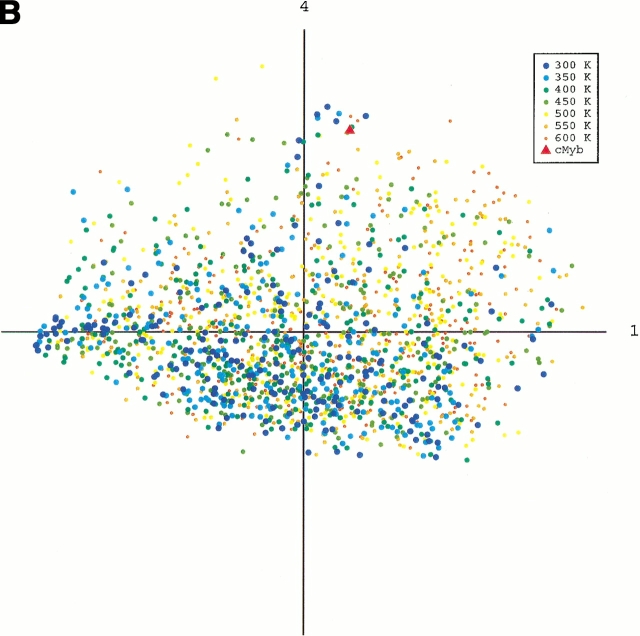
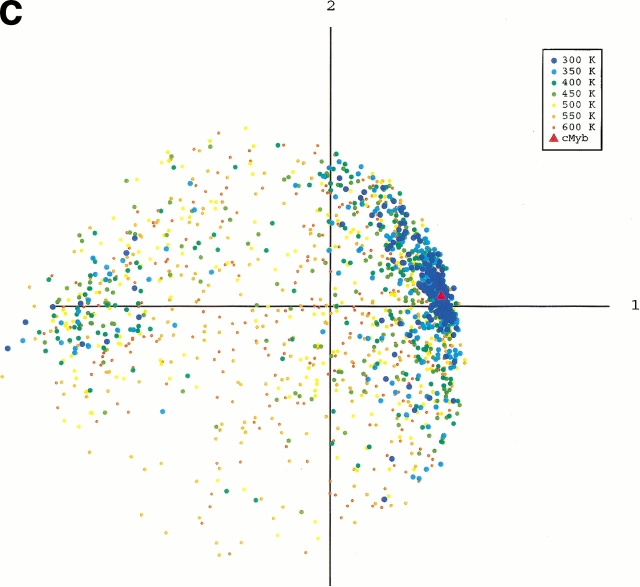
To obtain an image of the energy landscape from the sampled conformations, we did the following procedure: First, conformations were picked from the whole sampled conformations with the weight of the canonical energy distribution, P(E,T), at a temperature T. Note that the picked conformations generate a canonical ensemble Q(T), in which the probability of existence assigned to a conformation of energy E is P(E,T) in Q(T), and the energy distributions of Q(T0) and Q(300K) correspond to the dotted and broken lines, respectively, in Figure 1 ▶. Next, we combined the canonical ensembles at different temperatures and added qcMyb to them (i.e., Σ Q(T) + qcMyb). This combined ensemble covers a wide energy range. Last, we applied the principle component analysis (see Materials and Methods) to the ensemble. The eigen values were arranged in descending order. The conformation distribution on the principle axes gives the image of the potential energy landscape. The landscape from parm96 was rugged, as shown in Figure 4A ▶, in which the low-energy as well as the high-energy conformations were widely distributed on the plane of the first and second axes. The distribution (Fig. 4B ▶) on the plane of the first and fourth axes indicated the existence of a small α-helical cluster around qcMyb. This cluster is shown more clearly later. The quantity λiΣjλj represents the width of the distribution along the i'th principle axis, in which λi is the eigen value assigned to the i'th axis. The value was 22% for the first principle axis, 12% for the second, 10% for the third, 7% for the fourth, and so on. Afterward, we will see that not only the α-helix but also other secondary structures exist in the rugged landscape. In contrast, the landscape from parm94 was funnel like, and the α-helix was located at the bottom of the funnel (Fig. 4C ▶).
Fig. 4.
Potential-energy landscape of peptide on the first and second principle axes from parm96 (A), the first and fourth axes from parm96 (B), and the first and second axes from parm94 (C). Point labeled cMyb represents location of qcMyb. We did not use all of the conformations for the figures because the number (∼105) of conformations in Q(T) at each temperature is too large to show. Conformations were then picked randomly from Q(T). The numbers of the conformations shown in A and B are 207 (at 300K), 219 (350K), 253 (400K), 270 (450K), 302 (500K), 283 (550K), and 272 (600K), and those in C are 184 (at 300K), 199 (350K), 203 (400K), 191 (450K), 238 (500K), 263 (550K), and 259 (600K).
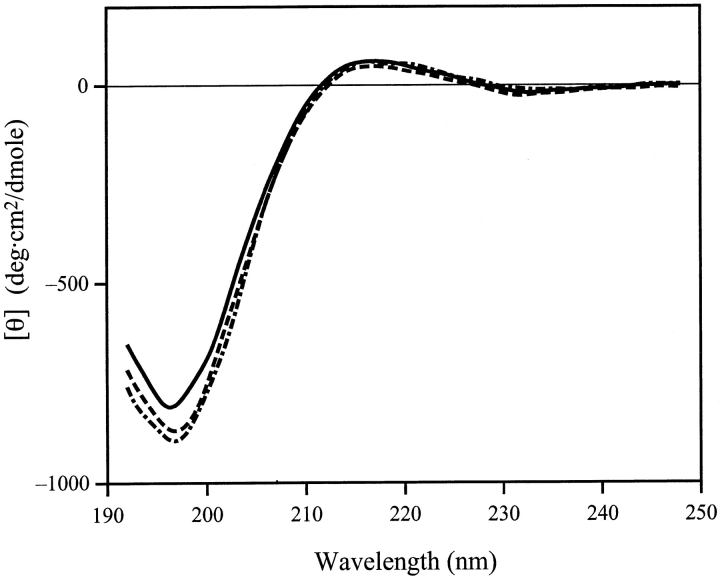
Figure 5 ▶ shows the CD spectra of the aqueous solution of this peptide at various pH values at 10°C. The large negative peak around 197 nm is typical of the disordered structure, and the peaks corresponding to α-helices or β-structures were absent. Neither raising the temperature to 25°C, nor the addition of salt (up to 0.5 M NaCl) changed the spectra significantly (data not shown). These results strongly suggest that the peptide does not adopt a rigid secondary structure under these conditions. Thus, parm96 is more plausible than parm94.
Fig. 5.
CD spectra of synthesized peptide at pH 6.0 (solid line), 7.0 (broken line), and 8.0 (dotted line). Temperature was 10°C and no salt was added.
Below, we describe the secondary structures included in Q(300 K) from parm96. The α-helix of the peptide is specified by the five intra-mainchain H-bonds; 1O-5HN (designated by HB1), 2O-6HN (HB2), 3O-7HN (HB3), 4O-8HN (HB4), and5O-9HN (HB5). Here, we designate a helical conformation with three bonds, HB2, HB3, and HB4, by HLX(2,3,4), for example. As shown later, the conformations HLX(2,4), HLX(2,3,4), HLX(1,2,3,4), and HLX(1,2,3,4,5) are close to qcMyb and generate a cluster. Thus, we call these conformations complete α-helix. In HLX(2,4), the helical conformation was well maintained by HB2 and HB4, although the central H-bond (HB3) was not formed. The other conformations (partly disordered helices) with more than two H-bonds, such as HLX(1,2,3), were located apart from the complete helix cluster, as also shown later. We call the partly disordered helices imperfect α-helices. In this study, an H bond was assigned to the pair of a donor and an acceptor when the distance between them was shorter than 2.5 A.
For β-hairpins, we considered two types, HPN1 and HPN2; the turn region of HPN1 is in residues 4–7, with the possible H-bonds of 2O-9HN, 3HN-8O, 3O-8HN, 4HN-7O, and 4O-7HN, and that of HPN2 is in residues 3–6 with 1O-8HN, 2HN-7O, 2O-7HN, 3HN-6O, and 3O-6HN. The H-bond (4O-7HN for HPN1 or 3O-6HN for HPN2) to form the β-turn is called a β-turn H-bond, and the other H-bonds are called inter-strand H-bonds. We defined a conformation as a hairpin when it contained both the β-turn H-bond and one or more inter-strand H-bonds. Although five H-bonds are possible for the hairpins, the maximum we observed was three (the β-turn and two inter-strand H-bonds) in HPN1 and two (the β-turn and one inter-strand H-bonds) in HPN2. We designate HPN1 with three H-bonds as HPN1(3), for example.
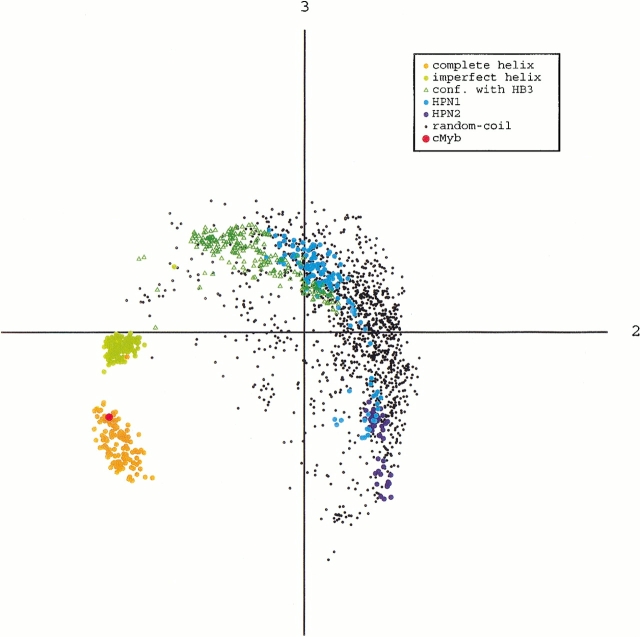
The principle component analysis was applied to Q(300K). The distributions of the α-helix, β-hairpin, random-coil conformations, as well as those only with HB3 (HB3 conformations), are shown in Figure 6 ▶. The complete helix, imperfect helix, and HB3 conformations were isolated from one another. This indicates that the energy barriers should be overcome for the transitions from the HB3 conformations to the imperfect helix, as well as from the imperfect helix to the complete helix. Note that the HB3 conformations were located in the random-coil ensemble. This suggests that the transitions from the random-coil to the HB3 conformations do not require jumps of energy barriers. Figure 6 ▶ also shows that the β-hairpin conformations were embedded in the ensemble of the random-coil conformations. We calculated the partial partition function Z at 300 K for each of the ordered structures and the disordered (i.e., random-coil) conformations, using the canonical distribution at 300 K, and estimated the partial free energy F = -RT ln[Z] of these structures, in which R is the gas constant and T = 300 K. Here, we define the partial free energy of each secondary structure relative to the random-coil conformations by ΔF = F(each ordered structure) - F(random-coil): ΔF(complete α-helix) = 2.617 kcal/mol [the probability of existence to Q(300K) = 1.18%], ΔF(imperfect α-helix) = 2.622 (1.17%), ΔF(HB3 conformations) = 1.67 (5.82%), ΔF(HPN1) = 2.58 (1.27%), ΔF(HPN2) = 3.09 (0.54%), and ΔF[HPN1(3)] = 3.89 (0.16%). The ΔF(complete α-helix) was smaller than ΔF[HPN1(3)], although it was slightly larger than that of ΔF(HPN1), in which HPN1 = HPN1(2) + HPN(3). In the estimation of ΔF, the random-coil conformations were defined by the conformations in Q(300K), except for the α-helices and β-hairpins. The probability of existence for the random coil was 95.84% of Q(300K) and that for the secondary structures was 4.16%.
Fig. 6.
Distributions of secondary structures and random-coil conformations in Q(300 K) from parm96, on the plane of the second and third principle axes. Point labeled cMyb represents location of qcMyb. Points labeled conf. with HB3 represent the conformations with only HB3. This plane most clearly showed the separation of the complete helix, imperfect helix, and random-coil conformations. Quantity λiΣj λj (see text) was 32% (i = 1), 21% (i = 2), and 14% (i = 3).
We calculated the partial internal energy ≤E> for each of the ordered structures and random conformations, and defined the relative internal energy as Δ≤E> = ≤E>(each ordered structure) - ≤E>(random-coil): Δ≤E>(complete α-helix) = -7.6 kcal/mol, Δ≤E>(imperfect α-helix) = -5.8, Δ≤E>(HB3 conformations) = -1.9, Δ≤E>(HPN1) = -2.6, and Δ≤E> (HPN2) = -4.2. The reference value, ≤E>(random-coil), was 5528.6 kcal/mol, and the internal energy ≤E>[Q(300K)] for the whole ensemble Q(300K) was −5528.7 kcal/mol. Note that the summation of ≤E> over the partial internal energies is not equal to ≤E>[Q(300K)]: ≤E>(complete α-helix) + ≤E>(imperfect α-helix) + ≤E>(HB3 conformations) + ≤E>(HPN1) + ≤E>(HPN2) ≠ ≤E>[Q(300K)] because the normalization of the partial internal energy for each ordered structure was done by the partial partition function. Remember that the probability of existence was nearly the same between the complete and imperfect α-helices. The result of ≤E> shows that the stabilizing factor is different between them, that is, ≤E>(complete α-helix) was lower than ≤E>(imperfect α-helix). Therefore, the complete α-helix is stabilized by the internal energy, and the imperfect α-helix is done by the entropy. This result is reasonable because the terminal region of the imperfect α-helix is more flexible than that of the complete α-helix.
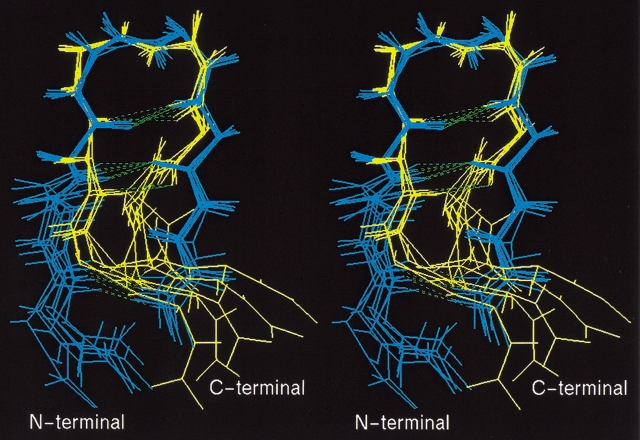
To investigate the structural differences between the complete and imperfect helices, snapshots of HLX(2,3,4) and HLX(1,2,3), picked up from Q(300K), are drawn in Figure 7A and B ▶, respectively, in which water molecules near the peptide atoms 4O or 8HN are shown together with the peptides. In HLX(1,2,3), 4O and 8HN were exposed to the solvent and formed H-bonds with water molecules (Fig. 7B ▶). In contrast, in HLX(2,3,4) 4O and 8HN formed an H-bond (HB5), and water molecules could not penetrate between 4O and 8HN (Fig. 7A ▶). A similar situation was found in the comparison of HLX(2,3,4) and HLX(3,4,5) (data not shown). This means that the imperfect helices, in which two sequential H-bonds (HB1 and HB2, or HB4 and HB5) at the N- or C-terminal were absent, were stabilized by the peptide-water H-bonds. The H-bond rearrangement from the peptide–water to peptide–peptide interactions may correspond to the process of overcoming the energy barriers. The absence of one H-bond (HB1 or HB5) at the N- or C-terminal did not largely affect the helical conformation, and water molecules could not penetrate into the packed peptide.
Fig. 7.
(A) Ten conformations randomly picked from HLX(2,3,4), and (B) those from HLX(1,2,3). In both A and B, water molecules close to 4O or 8HN with distances < 3.5 Å are shown. Bold bonds in the peptide are the covalent bonds 4C-4O and 8N-8HN. Broken lines represent hydrogen bonds.
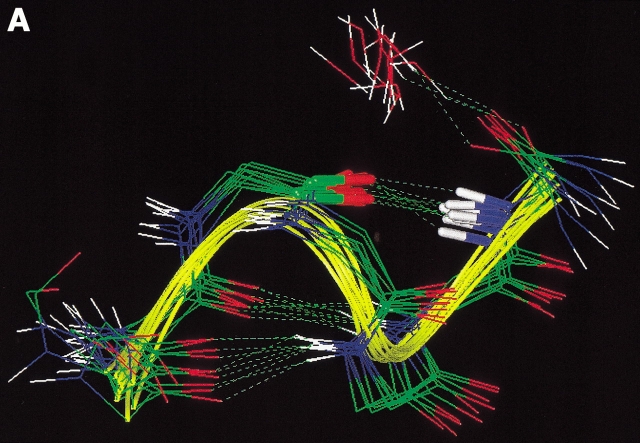
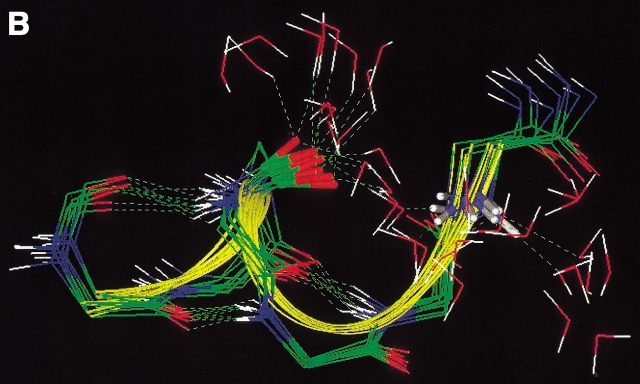
All of the conformations of HPN1(3) contained the same intra-mainchain H-bonds, for example, 2O-9HN, 4HN-7O, and 4O-7HN. However, they were clearly classified into two groups (Fig 8 ▶); one was characterized by the type I β-turn and the other by the type II' turn. Nakajima et al. (2000) recently performed the multicanonical MD simulation of a short peptide, and found that the energy barrier between the I and II' turns is relatively low. Another difference between the two groups was also found in the twisting of the strands (Fig. 8 ▶). Note that the peptide sequence is highly hydrophilic, so that a hydrophobic core cannot be formed. This property of the sequence may prevent the formation of the inter-strand H-bonds, 3HN-8O and 3O-8HN.
Fig. 8.
Stereo drawing of HPN1(3) picked up from Q(300K). Peptides in yellow are those characterized by type I β-turn and those in blue by type II'. Broken lines represent hydrogen bonds, 2O-9HN, 4HN-7O, and 4O-7HN. β-turn regions (residues 4–7) were superimposed among the conformations.
It is interesting to examine whether Q(300K) contained the 310-helix. By picking up conformations with H-bonds iO-i+3HN from Q(300K), we detected the imperfect 310-helix conformations with H-bonds 3O-6HN and 4O-7HN. The probability of existence for the 310-helix was small, 0.52% of Q(300K), which corresponded to ΔF = 3.09 kcal/mol. Note that the H-bond 3O-6HN is the β-turn H-bond of HPN2 and, 4O-7HN is that of HPN1.
We also detected conformations that have the inter-strand H-bonds but lack the β-turn H-bond. We call them closure conformations. The closure is generally classified as the random coil. The probability of existence in Q(300K) was 12.9% (ΔF = 1.19 kcal/mol): 7.3% (ΔF = 1.53) for HPN1, and 5.6% (ΔF = 1.68) for HPN2. Note that the free energy is not additive: ΔF(states A + B)≠ ΔF(A) + ΔF(B). The β-turn conformations, which include only the β-turn H-bond, had the probability of 6.64% (ΔF = 1.58); 1.93% (ΔF = 2.31) for HPN1 and 4.7% (ΔF = 1.78) for HPN2. The probability of the closure only with the H-bond 2O-9HN was 0.8% (ΔF = 2.87), and that only with 1O-8HN was 0.16% (ΔF = 3.81).
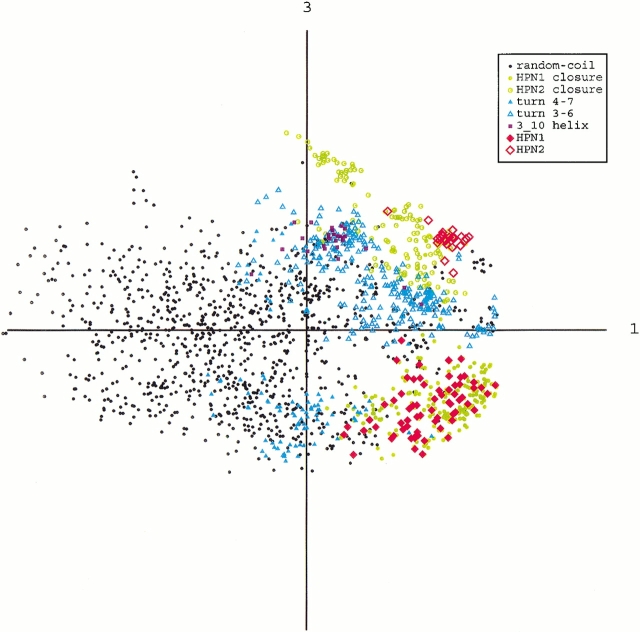
The distributions of the above structured conformations (i.e., β-hairpin, imperfect 310-helix, β-turn, and closure) may provide some scheme for the β-hairpin formation. Thus, we picked up these conformations from Q(300 K), and applied the principle component analysis to them, λiΣj λj = 34.3% (for i = 1), 18.6% (i = 2), 13.1% (i = 3), and so on. Figure 9 ▶ is the distribution on the plane of the first and third axes, in which the separation of HPN1 and HPN2 were most clearly shown. It is clear that the structured conformations did not form separated clusters from one another. This means that the transitions among them have no energy barriers. Note that HPN1 and the HPN1 closure are distributed in the same area, and the β-turn 4–7 conformations contact them. The HPN2 is distributed in a small region of the area of the HPN2 closure, and then the closure is spaced between the β-turn 3–6 conformations and HPN2. The imperfect 310-helix is located in a restricted region of the area of the β-turn 3–6 conformations, and has no contact with the β-hairpins.
Fig. 9.
Distribution of structured conformations and the other disordered conformations in Q(300K) on the plane of the first and third principle axes. Conformations are designated as follows: β-hairpin conformations are designated by HPN1 and HPN2, imperfect 310-helix by 3_10 helix, β-turn conformations by turn 4–7 and turn 3–6, which are the conformations with β-turn H-bonds only in residues 4–7 and 3–6, respectively, and closure conformations by HPN1 closure and HPN2 closure.
Discussion
The canonical ensemble Q(300K) from parm96 was subjected to the random state, which inherently consists not only of disordered conformations but also of several kinds of ordered ones. Accordingly, the potential-energy landscape was rugged, covering the wide variety of conformations. This result is expected, because the thermal fluctuations in the random state are larger than those in a state characterized by a well-defined structure. The current study may be useful to investigate the residual structures observed experimentally in the denatured state of proteins (Neri et al. 1992; Shortle 1993, Shortle 1996; Blanco and Serrano 1995; Frank et al. 1995; Searle et al. 1996; Gillespie and Shortle 1997a,b; Wang and Shortle 1997; Blanco et al. 1998; Kamatari et al. 1999; Sinclair and Shortle 1999), or help to interpret the unstructured segments in proteins in the native state (Wright and Dyson 1999). Remember that the current fragment is helical in the protein cMyb, but the actual conformation in water is random coil for the synthesized peptide fragment. The small cluster for α-helix in Q(300K) indicates that the peptide has a slight tendency to fold into the helix. Our study shows that the peptide can adopt different conformations, depending on the environment. This work supports the scheme proposed by Wright and Dyson (1999) that unstructured fragments are structured when functioning.
Although the conventional MD technique is becoming a powerful tool to study biomolecules, it is difficult to obtain statistically reliable data to view the energy landscape. Our multicanonical study provides a successful example that the simulation technique complements insufficient experimental data: The sampling covered a variety of secondary structures, which are not detectable experimentally because of the small probabilities of existence.
Because the multicanonical simulation provides an equilibrated picture, it is difficult to discuss precisely the kinetic process of chain folding. However, the potential-energy landscape yielded the information about energy barriers between the conformations. Our study showed that the propagation of helical content is achieved by the jumps between energy barriers from the HB3 conformations to the imperfect helix, and from the imperfect helix to the complete helix (Fig. 6 ▶). On the other hand, the HB3 conformations were accessible from the random-coil conformations without requiring jumps. The analysis of the snapshots suggested that the jumping over the barriers corresponds to the H-bond rearrangement from the peptide–water to the peptide–peptide interaction.
For β-hairpin formation, two mechanisms have been proposed as follows: (mechanism 1) the β-turn is formed first (Munoz et al. 1997; Alba et al. 1999; Wang et al. 1999b; Bonvin and van Gunsteren 2000; Galzitskaya et al. 2000), or (mechanism 2) the hydrophobic-core formation is formed first (Dinner et al. 1999). Because the current sequence is highly hydrophilic, a hydrophobic core cannot be formed. Thus, in the current case, the mechanism 2 should be replaced by a mechanism in which the closure conformations are formed first. Interestingly, Figure 9 ▶ implies that the formation process is different between HPN1 and HPN2. The HPN1 is accessible from either the β-turn conformations or the closure conformations, which suggests that both mechanisms 1 and 2 are possible. The HPN2 is accessible only from the closure conformations, which suggests mechanism 2. However, as the current peptide does not fold into a stable β-hairpin, due to the lack of hydrophobic-core residues, the folding mechanism proposed here does not provide enough information to develop a mechanism for the formation of stable hairpins. Studies should be done on other peptides that fold into thermally stable β-hairpins.
We observed an imperfect 310 helix in Q(300K). One may imagine that breaking the H-bond iO-i+3HN initiates the formation of the β-hairpin. Figure 9 ▶ shows that the 310 helix is located in a restricted area in the β-turn conformations, and is isolated from the β-hairpin ensemble. This means that the 310 helix did not directly contribute to the growth of the hairpins.
The multicanonical simulations (Ono et al. 2000) of the short hydrophobic peptides Ace-(Ala)n-Nme (n = 2 or 3), showed that parm94 had a high propensity to form the H-bonds iO-i+3HN and iO-i+4HN, and that parm96 was likely to form extended conformations. The peptide studied here was longer than those studied by Ono et al. (2000), and was more hydrophilic. Our parm96 results showed that the peptide has the ability to fold the chain into helix, hairpin, and random-coil conformations. The result on parm94, on the other hand, showed that the propensity of H-bond (iO-i+4HN) formation leads the chain into the α-helix, which is located at the bottom of the funnel-like potential-energy landscape. The refolding study with the conventional canonical MD simulation of the 36-residue peptide (Duan and Kollman 1998) also showed that parm94 has a high ability to fold the peptide into helical conformations. Our multicanonical simulation revealed that parm96 is more appropriate than parm94. Of course, it does not mean that parm96 is the best force field, because force fields other than AMBER have not yet been examined.
The current study may raise the question of whether MD studies performed with an inappropriate force field are valid. However, we know that the various MD trajectories of a protein provide a similar pattern of atomic-positional fluctuations that agree well with the X-ray B-factors, when the study is focused on the thermal fluctuations around the well-defined native conformation. Using a simple protein model, Higo et al. (1997) have shown that the folded pattern of the protein main chain and the tight atom packing in the protein interior are important factors to determine the overall conformational fluctuations of the main chain. Thus, the serious force-field problem is caused when the chain folding is studied. In this sense, care should be taken to simulate the unfolded state in the free-energy perturbation method.
The thermodynamic stability of a peptide chain is governed by the free energy differences between the native and denatured states. Given a flexible fragment in a protein or a short flexible peptide, there should exist a number of different stable conformations characterized by similar free-energy values. Such systems can be analyzed only by the rugged energy landscape, to accurately compare the computations with the experiments. In addition, the rates and the paths of the conformational changes among the different stable and meta-stable states can be evaluated by analyzing the energy barriers among them. When the reaction path is correctly assumed, the umbrella sampling technique helps to estimate precisely the heights of the free-energy barriers. An example is the cis-trans imide isomerization, in which our new method, called multicanonical WHAM (Nakamura et al. 1999; Ono et al. 1999), revealed the free-energy barrier between the cis and trans isomers of Ace-Ala-Pro-Nme. Thus, when the conformational sampling is highly enhanced and the force field is sufficiently reliable, a precise analysis of the free-energy landscape for a realistic molecular model can yield a more complete understanding of the dynamic conformations and energetics of a peptide chain. Very recently, Higo et al. (2001) succeeded in reconstructing the precise canonical ensemble of an 8-residue peptide, which was designed for a β-hairpin (Ramirez-Alvarado et al. 1996), in explicit water with parm96 force field, from the multicanonical ensemble covering the wide temperature range between 290 and 700 K. They found that the β-hairpin conformations are predominant in the canonical ensemble at 300 K. Our simulation studies together with experiments are now underway, with longer peptides and proteins.
Materials and methods
We used the multicanonical MD method (Nakajima et al. 1997a) to sample the peptide conformations in explicit water. Here, we briefly summarize the methodological advantage. This sampling is enhanced by introducing a modified energy function Emod =E + RT0 ln[P(E,T0)], in which E is the potential energy of the system, R is the gas constant, T0 is the temperature for the multicanonical MD simulation, and P(E,T0) is the canonical energy distribution at T0. The forces acting on the atoms are derived by -grad Emod. The temperature T0 is usually set high enough to overcome energy barriers in the potential energy surface during the prerun, as explained later. If P(E,T0) is known in a wide energy range, then a flat energy distribution is obtained in the range through the multicanonical simulation. A benefit of the method is that the canonical ensemble, P(E,T), at any temperature is derived using a reweighting formula (Shirai et al. 1998).
The distribution P(E,T0) is not known a priori, but is obtained through an iteration of the multicanonical MD runs. Prior to the multicanonical runs, a preparative canonical MD simulation (prerun) is done at T0, and the distribution P0(E,T0) obtained from the prerun accurately covers only the high-energy range around T0. Then, P0(E,T0) is extrapolated to the lower-energy direction, and the modified energy Emod is reset by use of the extrapolated P0(E,T0). The first multicanonical run generates a flat energy distribution covering a wider energy range than the prerun. Then, P1(E,T0), derived from the flat distribution using the reweighting formula, is again extrapolated to the lower-energy direction, and Emod is reset for the second run. This procedure is repeated until the sampling reaches the room-temperature region. Finally, the conformations sampled in the last multicanonical run are used for the analysis.
The peptide was immersed in a water sphere (radius = 17 Å) of the flexible TIP3P water model (Jorgensen et al. 1987). The sphere center was placed on the geometrical center of the peptide. A harmonic potential was applied to the water-oxygen atoms, but only when the water molecules were moving to the outside of the sphere, to confine the molecules within the sphere. The momentum and the angular momentum of the peptide were constrained at zero during the simulation, to keep the peptide position around the center of the water sphere. If the peptide assumed an extended conformation, then the methyl groups of 1Ace and 9Nme may touch the spherical boundary, and this extended conformation may be stabilized because of the hydrophobic interaction with the vacuum (i.e., the outside of the sphere). Thus, a harmonic potential was applied to the atoms in 1Ace and 9Nme, but only when these atoms were moving outside of a sphere (radius = 14 Å), for which the center was set to the water-sphere center. Due to this procedure, the space for the peptide fluctuations became smaller. However, the space was wide enough for the transitions among the random-coil and secondary structures, as shown in the Results section.
The computer program PRESTO (Morikami et al. 1992) with a modification for multicanonical MD (Nakajima et al. 1997a) was used for the sampling. The electrostatic interaction was calculated by using the cell-multipole expansion (Ding et al. 1992). The SHAKE algorithm (Ryckaert et al. 1977) was used to constrain the covalent bonds between hydrogen and heavy atoms. The time step of the MD simulation was 1 fs. The temperature was controlled by the constant-temperature method (Evans and Morriss 1983). The initial conformation of the peptide for the first multicanonical run was a random conformation, and the initial one for the (+1)'th run was the last one of the i'th run. Therefore, the sampled conformations were not influenced by the α-helix conformation (qcMyb) in the original pdb data of c-Myb. The multicanonical simulation was examined with two force fields: AMBER parm94 (Cornell et al. 1995) and parm96 (Kollman et al. 1997). The temperature T0 was 1000 K for the parm94 simulation, and 600 K for the parm96 simulation. We verified that T0 is high enough to unfold qcMyb into random-coil conformations with conventional canonical MD simulations in advance.
We used the principle component analysis in the same way as done by Shirai et al. (1998) to study the sampled conformations. Given an ensemble of n conformations, the main-chain rmsd was calculated for all conformation pairs in the ensemble, generating a distance matrix of [n × (n + 1)/2] elements. Next, the distance matrix was diagonalized, and a set of eigen vectors and eigen values were obtained. The eigen vectors construct a conformational space, in which the positions of the n conformations are assigned. An eigen vector with a larger eigen value corresponds to the principle axis along that the conformational distribution is wider.
We measured the CD spectra of this sequence to investigate the stable conformation in solution. The peptide was obtained from Kurabo (Osaka, Japan), and its purity was higher than 90%. The spectra were taken by use of a Jasco J-720 spectropolarimeter and a cell with a 1-mm path length. The sample concentration was 10 mM and pH was maintained by either 10 mM potassium phosphate buffer (pH 7.0 or 8.0) or 10 mM MES buffer (pH 6.0). Thirty two scans were accumulated for each spectra.
Acknowledgments
H.N. and N.N. were supported by grant-in-aid for Scientific Research from the Japan Society for the Promotion of Science (12558083 and 12680657).
The publication costs of this article were defrayed in part by payment of page charges.This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at www.proteinscience.org/cgi/doi/10.1110/ps.44901.
References
- Alba, E.D., Rico, M., and Jimenez, A. 1999. The turn sequence directs β-strand alignment in designed β-hairpins. Protein Sci. 8 2234–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andricioaei, I. and Straub, J.E. 1996. Generalized simulated annealing algorithms using Tsallis statistics: Application to conformational optimization of a tetrapeptide. Phys. Rev. E. 53 R3055–R3058. [DOI] [PubMed] [Google Scholar]
- Bartels, C. and Karplus, M. 1997. Multidimensional adaptive umbrella sampling: Application to main chain and side chain peptide conformations. J. Compt. Chem. 18 1450–1462. [Google Scholar]
- ———. 1998. Probability distributions for complex systems: Adaptive umbrella sampling of the potential energy. J. Phys. Chem. B 102 865–880. [Google Scholar]
- Bartels, C., Stote, R.H., and Karplus, M. 1998. Characterization of flexible molecules in solution: The RGDW peptide. J. Mol. Biol. 284 1641–1660. [DOI] [PubMed] [Google Scholar]
- Bartels, C., Schaefer, M., and Karplus, M. 1999. Determination of equilibrium properties of biomolecular systems using multidimensional adaptive umbrella sampling. J. Chem. Phys. 111 8048–8067. [Google Scholar]
- Berg, B.A. and Neuhaus, T. 1992. Multicanonical ensemble: A new approach to simulate first order phase transitions. Phys. Rev. Lett. 68 9–12. [DOI] [PubMed] [Google Scholar]
- Blanco, F.J. and Serrano, L. 1995. Folding of protein G B1 domain studied by the conformational characterization of fragments comprising its secondary structure elements. Eur. J. Biochem. 230 634–649. [DOI] [PubMed] [Google Scholar]
- Blanco, F.J., Serrano L, and Forman-Kay, J.D. 1998. High populations of non-native structures in the denatured state are compatible with the formations of the native fold state. J. Mol. Biol. 284 1153–1164. [DOI] [PubMed] [Google Scholar]
- Boczko, E.M. and Brooks, III, C.L. 1995. First-principle calculation of the folding free energy of a three-helix bundle protein. Science 269 393–396. [DOI] [PubMed] [Google Scholar]
- Bond, C.J., Wong, K.-B., Clarke, J., Fersht, A.R., and Daggett, V. 1997. Characterization of residual structure in the thermally denatured state of barnase by simulation and experiment: Description of the folding pathway. Proc. Natl. Acad. Sci. 94 13409–13413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonvin, A.M.J.J. and van Gunsteren, W.F. 2000. β-hairpin stability and folding: Molecular dynamics studies of the first β-hairpin of tendamistat. J. Mol. Biol. 296 255–268. [DOI] [PubMed] [Google Scholar]
- Brooks, III, C.L. 1998. Simulations of protein folding and unfolding. Curr. Opin. Struc. Biol. 8 222–226. [DOI] [PubMed] [Google Scholar]
- Bryngelson, J.D., Onuchic, J.N., Socci, N.D., and Wolynes, P.G. 1995. Funnels, pathways, and the energy landscape of protein folding: A synthesis. Proteins 21 167–195. [DOI] [PubMed] [Google Scholar]
- Caves, L.S.D., Evanseck, J.D., and Karplus, M. 1998. Locally accessible conformations: Multiple molecular dynamics simulations of crambin. Protein Sci. 7 649–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, H.S, and Dill, K.A. 1998. Protein folding in landscape perspective: Chevron plots and non-Arrhenius kinetics. Proteins 30 2–33. [DOI] [PubMed] [Google Scholar]
- Chikenji, G. and Kikuchi, M. 2000. A lattice model study of α - β transitions in protein folding —the free-energy landscape analysis—. Prog. Theor. Physics Suppl. 138 406–407 [Google Scholar]
- Chikenji, G., Kikuchi, M., and Iba, Y. 1999. Multi-self-overlap ensemble for protein folding: Ground state search and thermodynamics. Phys. Rev. Lett. 83 1886–1889. [Google Scholar]
- Cornell, W.D., Cieplak, P., Bayly, C.I., Gould, I.R., Kenneth, J., Merz, M., Ferguson, D.M., Spellmeyer, D.C., Fox, T., Caldwell, J.W., et al. 1995. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 117 5179–5197. [Google Scholar]
- Daura, X., van Gunsteren, W.F., and Mark, A.E. 1999. Folding-unfolding thermodynamics of a β-heptapeptide from equiliblium simulations. Proteins 34 269–280. [DOI] [PubMed] [Google Scholar]
- Dill, K.A. 1999. Polymer principles and protein folding. Protein Sci. 8 1166–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, H-Q., Karasawa, N., and Goddard, III, W.A. 1992. Atomic level simulations on a million particles: The cell multipole method for Coulomb and London nonbond interactions. J. Chem. Phys. 97 4309–4315. [Google Scholar]
- Dinner, A.R., Lazaridis, T., and Karplus, M. 1999. Understanding β-hairpin formation. Proc. Natl. Acad. Sci. 96 9068–9073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan, Y. and Kollman, PA. 1998. Pathways to protein folding intermediate observed in a 1-microseond simulation in aqueous solution. Science 282 740–744. [DOI] [PubMed] [Google Scholar]
- Evans, D.J. and Morriss, G.P. 1983. The isothermal/isobaric molecular dynamics ensemble. Phys. Lett. A 98 433–436. [Google Scholar]
- Frank, M.K., Clore, G.M., and Gronenborn, A.M. 1995. Structural and dynamic characterization of the urea denatured state of the immunoglobulin binding domain of streptococcal protein G by multidimensional heteronuclear NMR spectroscopy. Protein Sci. 4 2605–2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galzitskaya, O.V., Higo, J., Kuroda, M., and Nakamura, H. 2000. β-hairpin folds by molecular dynamics simulations. Chem. Phys. Lett 325 421–429. [Google Scholar]
- Gillespie, J.R. and Shortle, D. 1997a. Characterization of long-range structure in the denatured state of staphylococcal nuclease. I. Paramagnetic relaxation enhancement by nitroxide spin labels.J. Mol. Biol. 268 158–169. [DOI] [PubMed] [Google Scholar]
- ———. 1997b. Characterization of long-range structure in the denatured state of staphylococcal nuclease. II. Distance restraints from paramagnetic relaxation and calculation of ensemble structures. J. Mol. Biol. 268 170–184. [DOI] [PubMed] [Google Scholar]
- Hamada, D., Segawa, S., and Goto, Y. 1996. Non-native α-helical intermediate in the refolding of β-lactoglobulin, a predominantly β-sheet protein. Nature Struct. Biol. 3 868–873. [DOI] [PubMed] [Google Scholar]
- Hansmann, U.H.E, and Okamoto, Y. 1993. Prediction of peptide conformation by multicanonical algorithm: New approach to the multi-minima problem. J. Compt. Chem. 14 1333–1338. [Google Scholar]
- ———. 1997a. Generalized-ensemble Monte Carlo method for systems with rough energy landscape. Phys. Rev. E 56 2228–2233. [Google Scholar]
- ———. 1997b. Numerical comparison of three recently proposed algorithms in the protein folding problem. J. Comp. Chem. 18 920–933. [Google Scholar]
- Hansmann, U.H.E,, Okamoto, Y., and Eisenmenger, F. 1996. Molecular dynamics, Langevin, and hybrid Monte Carlo simulations in multicanonical ensemble. Chem. Phys. Lett. 259 321–330. [Google Scholar]
- Hesselbo, B. and Stinchcombe, R.B. 1995. Monte Carlo simulation and global optimization without parameters. Phys. Rev. Lett. 74 2151–2155. [DOI] [PubMed] [Google Scholar]
- Higo, J. and Umeyama, H. 1997. Protein dynamics determined by backbone conformation and atom packing. Protein Eng. 10 373–380. [DOI] [PubMed] [Google Scholar]
- Higo, J., Nakajima, N., Shirai, H., Kidera, A., and Nakamura, H. 1997. Two-component multicanonical Monte Carlo method for effective conformation sampling. J. Comp. Chem. 18 2086–2092. [Google Scholar]
- Higo, J., Galzitskaya, O.V., Ono, S., and Nakamura, H. 2001. Energy landscape of a β-hairpin peptide in explicit water. Studied by multicanonical molecular dynamics. Chem. Phys. Lett. 337 169–175. [Google Scholar]
- Honda, S., Kobayashi, N., and Munekata, E. 2000. Thermodynamics of a β-hairpin structure: Evidence for coorperative formation of folding nucleus. J. Mol. Biol. 295 269–278. [DOI] [PubMed] [Google Scholar]
- Hukushima, K. and Nemoto, K. 1996. Exchange Monte Carlo method and application to spin glass simulations. J. Phys. Soc. Jpn. 65 1604–1608. [Google Scholar]
- Iba, Y., Chikenji, G., and Kikuchi, M. 1998. Simulation of lattice polymers with multi-self-overlap ensemble. J. Phys. Soc. Jpn. 67 3327–3330. [Google Scholar]
- Irback, A. and Potthast, F. 1995. Studies of an off-lattice model for protein folding: Sequence dependence and improved sampling at finite temperature. J. Chem. Phys. 103 10298–10305. [Google Scholar]
- Istrail, S., Schwartz, R., and King, J. 1999. Lattice simulation of aggregation funnels for protein folding. J. Comput. Biol. 6 143–162. [DOI] [PubMed] [Google Scholar]
- Jorgensen, W.L., Chandrasekhar, J., Madura, J.D., Impley, R.W., and Klein, M.L. 1987. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79 926–935. [Google Scholar]
- Kamatari, Y.O., Ohji, S., Konno, T., Seki, Y., Soda, K., Kataoka, M., and Akasaka, K. 1999. The compact and extended denatured conformations of apomyoglobin in the methanol-water solvent. Protein Sci. 8 873–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazmirski, S.L. and Daggett, V. 1998. Non-native interactions in protein folding intermediates: Molecular dynamics simulations of hen lysozyme. J. Mol. Biol. 284 793–806. [DOI] [PubMed] [Google Scholar]
- Kidera, A. 1995. Enhanced conformation sampling in Monte Carlo simulations of proteins: Application to constrained peptide. Proc. Natl. Acad. Sci. 92 9886–9889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi, M., Chikenji, G., and Iba, Y. 2000. Multi-selfoverlap-ensemble Monte Carlo method for lattice proteins and heteropolymears. Prog. Theor. Physics Supp. 138 404–405. [Google Scholar]
- Kim, S.T., Shirai, H., Nakajima, N., Higo, J., and Nakamura, H. 1999. Enhanced conformational diversity search of CDR-H3 in antibodies: Role of the first CRD-H3 residue. Proteins 37 683–696. [PubMed] [Google Scholar]
- Kollman, P.A., Dixon, R.W., Cornell, W.D., Chipot, C., and Pohorille, A. 1997. The development/application of a "minimalist" organic/biochemical molecular mechanic force field using a combination of ab initio calculations and experimental data. In: Computer simulations of biological systems. (ed. W.F. van Gunsteren), ESCOM, Dordrecht, The Netherlands.
- Lazarids, T. and Karplus, M. 1997. "New view" of protein folding reconciled with the old through multiple unfolding simulations. Science 278 1928–1931. [DOI] [PubMed] [Google Scholar]
- Lee, J. 1993. New Monte Carlo algorithm: Entropic sampling. Phys. Rev. Lett. 71 211–214. [DOI] [PubMed] [Google Scholar]
- Li, H., Helling, R., Tang, C., and Wingreen, N. 1996. Emergence of preferred structures in a simple model of protein folding. Science 273 666–669. [DOI] [PubMed] [Google Scholar]
- Lyubartsev, A.P., Martsinovski, A.A., Shevkunov, S.V., and Vorontsov-Velyaminov, P.N. 1992. New approach to Monte Carlo calculation of the free energy: method of expanded ensembles. J. Chem. Phys. 96 1776–1783. [Google Scholar]
- Morikami, K., Nakai, T., Kidera, A., Saito, M., and Nakamura, H. 1992. PRESTO: A vectorized molecular mechanics program for biopolymers. Comput. Chem. 16 243–248. [Google Scholar]
- Munoz, V., Thompson, P.A., Hofrichter, J., and Eaton, W.A. 1997. Folding dynamics and mechanism of β-hairpin formation. Nature 390 196–199. [DOI] [PubMed] [Google Scholar]
- Nakajima, N. 1998. A selectively enhanced multicanonical molecular dynamics method for conformational sampling of peptides in realistic water molecules. Chem. Phys. Lett. 288 319–326. [Google Scholar]
- Nakajima, N., Nakamura, H., and Kidera, A. 1997a. Multicanonical ensemble generated by molecular dynamics simulation for enhanced conformational sampling of peptides. J. Phys. Chem. B 101 817–824. [Google Scholar]
- Nakajima, N., Higo, J., Kidera, A., and Nakamura, H. 1997b. Flaxible docking of a ligand peptide to a receptor protein by multicanonical molecular dynamics simulation. Chem. Phys. Lett. 278 297–301. [Google Scholar]
- Nakajima, N., Higo, J., Kidera, A., and Nakamura, H. 2000. Free energy landscape of peptides by enhanced conformational sampling. J. Mol. Biol. 296 197–216. [DOI] [PubMed] [Google Scholar]
- Nakamura, H., Ono, S., and Higo, J. 1999. A general ab initio approach for free energy landscape of biological molecules around the transition states: Fusion of the classical molecular mechanics simulation and the quantum chemical calculation. Proc. Japan Acad. B75 291–294. [Google Scholar]
- Nakamura, H.K. and Sasai, M. 1999. Hierarchy and connectivity in the folding funnel. In: Old and new views of protein folding. (ed. K. Kuwajima and M. Arai), pp. 125–131. Elsevier, Amsterdam.
- Neri, D., Billeter, M., Wider, G., and Wuthrich, K. 1992. NMR determination of residual structure in a urea-denatured protein, 434-repressor. Science 257 1559–1563. [DOI] [PubMed] [Google Scholar]
- Ogata, K., Morikawa, S., Nakamura, H., Hojo, H., Yoshimura, S., Zhang, R., Aimoto, S., Ametani, Y., Hirata, Z., Sarai, A., et al. 1995. Comparison of the free DNA-complexed forms of the DNA-binding domain from c-Myb. Struct. Biol. 2 309–320. [DOI] [PubMed] [Google Scholar]
- Ono, S., Nakajima, N., Higo, J., and Nakamura, H. 1999. The multicanonical weighted histogram analysis method for the free-energy landscape along structural transition paths. Chem. Phys. Lett. 312 247–254. [Google Scholar]
- ———. 2000. Peptide free-energy profile is strongly dependent on the force field: Comparison of C96 and AMBER95. J. Comp. Chem. 21 748–762. [Google Scholar]
- Pande, V.S. and Rokhsar, A.S. 1999. Molecular dynamics simulations of unfolding and refolding of a β-hairpin fragment of protein G. Proc. Natl. Acad. Sci. 96 9062–9067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Alvarado, M., Blanco, F.J., and Serrano, L. 1996. De novo design and structural analysis of a model β-hairpin peptide system. Nature Struct. Biol. 3 604–611. [DOI] [PubMed] [Google Scholar]
- Roccatano, D., Amadei, A., Di Nora, A., and Berendsen, H.J.C. 1999. A molecular dynamics study of the 41–56 β-hairpin from B1 domain of protein G. Protein Sci. 8 2130–2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryckaert, J-P., Ciccotti, G., and Berendsen, H.J.C. 1977. Numerical integration of Cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J. Comp. Phys. 23 327–341. [Google Scholar]
- Schaefer, M., Bartels, C., and Kurplus, M. 1998. Solution conformations and thermodynamics of structured peptides: Molecular dynamics simulation with an implicit solvation model. J. Mol. Biol. 284 835–848. [DOI] [PubMed] [Google Scholar]
- Searle, M.S., Zerella, R., Williams, D.H., and Packman, L.C. 1996. Native-like β-hairpin structure in an isolated fragment from ferredoxin: NMR and CD studies of solvent effect on the N-teminal 20 residues. Protein Eng. 9 559–565. [DOI] [PubMed] [Google Scholar]
- Shirai, H., Nakajima, N., Higo, J., Kidera, A,, and Nakamura, H. 1998. Conformational sampling of CDR-H3 in antibodies by multicanonical molecular dynamics simulation. J. Mol. Biol. 278 481–496. [DOI] [PubMed] [Google Scholar]
- Shortle, D. 1993. Denatured states of proteins and their roles in folding and stability. Curr. Opin. Struct. Biol. 3 66–74. [Google Scholar]
- ———. 1996. Structural analysis of non-native states of proteins by NMR methods. Curr. Opin. Struct. Biol. 6 24–30. [DOI] [PubMed] [Google Scholar]
- Sinclair, J.F. and Shortle, D. 1999. Analysis of long-range interactions in a model denatured state of staphylococcal nuclease based on correlated changes in backbone dynamics. Protein Sci. 8 991–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugita, Y. and Okamoto, Y. 1999. Replica-exchange molecular dynamics method for protein folding. Chem. Phys. Lett. 314 141–151. [Google Scholar]
- ———. 2000a. An analysis on protein folding problem by replica-exchange method. Prog.Theor. Physics Supp. 138 402–403. [Google Scholar]
- ———. 2000b. Replica-exchange multicanonical algorithm and multicanonical replica-exchange method for simulating systems with rough energy landscape. Chem. Phys. Lett. 329 261–270. [Google Scholar]
- Sugita, Y., Kitao, A., and Okamoto, Y. 2000. Multidimensional replica-exchange method for free energy calculations J. Chem. Phys. 113 5065–5071. [Google Scholar]
- Takano, M., Yamato, T., Higo, J., Suyama, A., and Nagayama, K. 1999. Molecular dynamics of a 15-residue poly(L-alanine) in water: Helix formation and energetics. J. Am. Chem. Soc. 121 605–612. [Google Scholar]
- Tatsumi, R. and Chikenji, G. 1999. Origin of the designability of protein structues. Phys. Rev. E 60 4696–4700. [DOI] [PubMed] [Google Scholar]
- Tsai, J., Levitt, M., and Baker, D. 1999. Hierarchy of structure loss in MD simulations of src SH3 domain unfolding. J. Mol. Biol. 291 215–225. [DOI] [PubMed] [Google Scholar]
- Tsallis, C. 1988. Possible generalization of Boltzmann-Gibbs statistics. J. Stat. Phys. 52 479–487. [Google Scholar]
- Wang, H., Varady, J., Ng, L., and Sung, S-S. 1999b. Molecular dynamics simulations of β-hairpin folding. Proteins 37 325–333. [DOI] [PubMed] [Google Scholar]
- Wang, L., Duan, Y., Shortle, R., Imperiali, B., and Kollman, P.A. 1999a. Study of stability and unfolding mechanism of BBA1 by molecular dynamics simulations at different temperatures. Protein Sci. 8 1292–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. and Shortle, D. 1997. Residual helical and turn structure in the denatured state of staphylococcal nuclease: Analysis of peptide fragments. Folding & Design 2 93–100. [DOI] [PubMed] [Google Scholar]
- Wong, K-B., Clarke, J., Bond, C.J., Neira, J.L., Freund, S.M.V., Fersht, A.R., and Daggett, V. 2000. Towards a complete description of the structural and dynamic properties of the denatured state of barnase and the role of residual structure in folding. J. Mol. Biol. 296 1257–1282. [DOI] [PubMed] [Google Scholar]
- Wright, P.E. and Dyson, H.J. 1999. Intrinsically unstructured proteins: Re-assessing the protein structure-function paradigm. J. Mol. Biol. 293 321–331. [DOI] [PubMed] [Google Scholar]
- Yong, W.S. and Brooks, III, C.L. 1996. A microscopic view of helix propagation: N and C-terminal helix growth in alanine helices. J. Mol. Biol. 259 560–572. [DOI] [PubMed] [Google Scholar]
- Zimm, H.B. and Bragg, J.K. 1959. Theory of phase transition between helix and random coil in polypeptide chains. J. Chem. Phys. 31 526–535. [Google Scholar]