Abstract
In this paper we address the question of whether the burial of polar and nonpolar groups in the protein locale is indeed accompanied by the heat capacity changes, ΔCp, that have an opposite sign, negative for nonpolar groups and positive for polar groups. To accomplish this, we introduced amino acid substitutions at four fully buried positions of the ubiquitin molecule (Val5, Val17, Leu67, and Gln41). We substituted Val at positions 5 and 17 and Leu at position 67 with a polar residue, Asn. As a control, Ala was introduced at the same three positions. We also replaced the buried polar Gln41 with Val and Leu, nonpolar residues that have similar size and shape as Gln. As a control, Asn was introduced at Gln41 as well. The effects of these amino acid substitutions on the stability, and in particular, on the heat capacity change upon unfolding were measured using differential scanning calorimetry. The effect of the amino acid substitutions on the structure was also evaluated by comparing the 1H-15N HSQC spectra of the ubiquitin variants. It was found that the Ala substitutions did not have a considerable effect on the heat capacity change upon unfolding. However, the substitutions of aliphatic side chains (Val or Leu) with a polar residue (Asn) lead to a significant (> 30%) decrease in the heat capacity change upon unfolding. The decrease in heat capacity changes does not appear to be the result of significant structural perturbations as seen from the HSQC spectra of the variants. The substitution of a buried polar residue (Gln41) to a nonpolar residue (Leu or Val) leads to a significant (> 25%) increase in heat capacity change upon unfolding. These results indicate that indeed the heat capacity change of burial of polar and nonpolar groups has an opposite sign. However, the observed changes in ΔCp are several times larger than those predicted, based on the changes in water accessible surface area upon substitution.
Keywords: Heat capacity changes, hydration, amino acid substitutions, differential scanning calorimetry, NMR
One of the major factors that define the stability of the native protein structure is interaction with solvent water. Thermodynamic parameters that describe interactions of different protein groups with water have been the subject of numerous experimental and theoretical studies (Baldwin 1986; Ben-Naim 1990; Ben-Naim et al. 1990; Dill et al. 1989; Kauzmann 1959; Makhatadze 1998a; Makhatadze et al. 1995; Makhatadze and Privalov 1990; Makhatadze and Privalov 1995; Murphy and Gill 1991; Myers et al. 1995; Privalov and Gill 1988; Sharp and Madan 1996; Spolar et al. 1992; Sturtevant 1977; Woolfson et al. 1993; Yang et al. 1992). The thermodynamic parameter that is of particular importance for characterizing interactions with water is the heat capacity change upon transfer of a solute into an aqueous solution. It is long established that the transfer of aliphatic compounds into water is accompanied by an increase in heat capacity (Edsall 1935). Further, the unfolding of proteins is also accompanied by an increase in heat capacity (Kauzmann 1959). These two observations led to a hypothesis that the heat capacity changes upon protein unfolding are defined by the nonpolar groups of the protein that are buried in the native state and become solvent exposed in the unfolded state (Privalov and Khechinashvili 1974). It was proposed that the transfer of a nonpolar compound from a nonpolar environment to water could be correlated with the parameter derived from the structure (Baldwin 1986; Privalov and Gill 1988; Spolar et al. 1989; Sturtevant 1977). This parameter, called water accessible surface area (ASA), can be used to extrapolate the experimental thermodynamic data on the transfer of model compounds to describe protein unfolding. Several years later it was noted that in the native protein structure, not only nonpolar, but also polar groups are buried. Thus the exposure of the polar groups to water upon protein unfolding should contribute to the heat capacity changes. The model compound studies indicated that the heat capacity change upon transfer of polar groups into water should have negative heat capacity changes, that is, a sign opposite to that for the transfer of nonpolar groups (Makhatadze and Privalov 1990). Computer simulations using Monte Carlo and random network models of water provide a qualitative explanation for the physical basis of such opposite effects (Gallagher and Sharp 1998; Madan and Sharp 1996; Madan and Sharp 1997; Sharp and Madan 1996). It appeared that the use of the exposure of both polar and nonpolar ASA can describe adequately the heat capacity changes upon unfolding of a large number of proteins that differ in size, structure, and amino acid composition (Makhatadze and Privalov 1990; Murphy and Freire 1992; Robertson and Murphy 1997; Spolar et al. 1992). However, there is an underlying caution on the correctness of direct transfer of the results obtained on the model compounds, which are small organic molecules, to the proteins, which are substantially larger molecules.
To address this, we decided to explore the possibility of measuring the effects of polar to nonpolar substitutions on the heat capacity change upon unfolding at sites completely buried in the protein interior. We used the ubiquitin molecule for these experiments. Ubiquitin is an excellent model for biophysical studies because it is a small globular protein of 76 amino acid residues that is soluble and relatively stable (Makhatadze et al. 1998; Wintrode et al. 1994; Woolfson et al. 1993). The latter is an important factor because the amino acid substitutions we planned to make were expected to have a significant impact on the stability of the variants. Three sites (positions 5, 17, and 67) were chosen for nonpolar to polar substitutions and one site (position 41) was used for polar to nonpolar substitutions. The variants of ubiquitin were purified and the thermodynamics of their unfolding was measured using differential scanning calorimetry. We found that indeed the nonpolar to polar substitutions lead to a decrease in the heat capacity changes, whereas the polar to nonpolar substitutions lead to an increase in heat capacity change upon protein unfolding.
Results
Amino acid substitution sites
All mutations were performed in the background of the yeast ubiquitin variant carrying the Lys63Arg substitution (Makhatadze et al. 1998; Thomas and Makhatadze 2000). In addition, a 6xHis-tag was added to the C-terminus of the molecule to facilitate protein purification. The amino acid substitutions performed in this background are designated by U6H63. Five amino acid substitutions were generated in the background of U6H63: Gln41Val, Gln41Leu, Gln41Asn, Leu67Ala, and Leu67Asn. Several amino acid substitutions were strongly destabilizing so they were introduced in the background of an ubiquitin variant carrying an additional substitution R42E that significantly increases protein stability, and is designated as U6H63/42 (Loladze et al. 1999). Four substitutions were generated in the background of U6H63/42: Val5Ala, Val5Asn, Val17Ala, and Val17Asn.
At all four substitution sites, the side chains of Val5, Val17, Gln41, and Leu67 are completely buried (calculated solvent accessibility for all four side chains in the native state is zero). Val5 is located in the β-strand 1; Val17 is located in the loop connecting β-strand 2 and helix 1; Gln41 is located in β-strand 3; Leu67 is located in β-strand 4. The side chains of Val5, Val17, and Leu67 are only surrounded by aliphatic side chains and are part of the hydrophobic core of the ubiquitin molecule (Lazar et al. 1997). Inspection of the three-dimensional structure of ubiquitin shows that the buried side chain of Gln41 can make two hydrogen bonds with the backbone oxygen atoms of residues Lys27 (3.02 Å) and Leu36 (2.85 Å). Thus we can expect that the Gln41Val or Gln41Leu substitutions will not only replace a buried polar group with a nonpolar but will also eliminate two side-chain/main-chain hydrogen bonds. It is important to note that the environment for side chains of residues at all four sites is largely nonpolar. Table 1 shows the number of heavy atoms (C, O, N) located within 7 Å of the side chain. The total number of the heavy atoms is similar, indicating that the environment around all four sites has similar packing. Nonpolar atoms from the side chains are more than 60% of the total number of atoms within 7 Å distance. Of the total 213 atoms located within 7 Å from all four sites, 130 are nonpolar and only six are from polar side-chain atoms, and the remainder are mostly backbone atoms (C, O, N).
Table 1.
Number of heavy atoms within 7 Å distance from the Cγ of the side chaina
| Side-chain: polar atoms | Side-chain: nonpolar atoms | Backbone atoms | Total number of atoms | |
| Val5 | 0 | 33 | 22 | 55 |
| Val17 | 5 | 32 | 15 | 52 |
| Gln41 | 1 | 31 | 23 | 55 |
| Leu67 | 0 | 34 | 17 | 51 |
a Calculations were done using the Protein Data Bank entry 1UBQ. For comparison, the solvent-exposed residues will have on average ∼25 total heavy atoms within 7 Å distance, and of those on average only ∼35% will belong to the nonpolar side-chain atom class.
Effect of amino acid substitutions on the thermostability of the ubiquitin variants
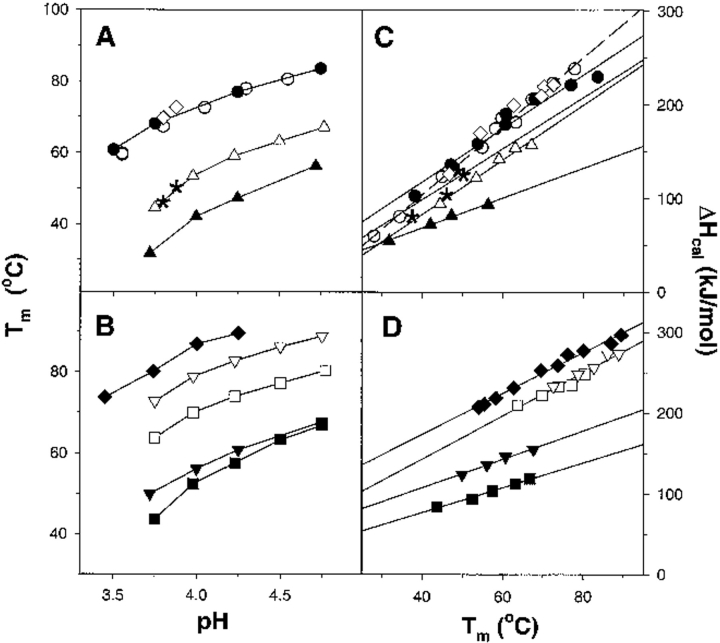
Figure 1 ▶ shows the dependence of the transition temperature of different ubiquitin variants on the pH of the solution. For positions Val5, Val17, and Leu67, substitutions to both Ala and Asn decrease significantly the transition temperature. However, the Val5Ala, Val17Ala, and Leu67Ala substitutions show less destabilization than the variants that contain Asn (Val5Asn, Val17Asn, Leu67Asn). The decrease in Tm for the Val17Ala variant was 8°C, and for Val5Ala and Leu67Ala variants, it was 17°C. For comparison, the decrease in thermostability for Val5Asn, Val17Asn, and Leu67Asn variants is almost twice that of Ala variants: 30°C. In contrast, the substitution of a polar residue at position 41 to nonpolar residues, Gln41Val or Gln41Leu, did not produce a significant change in Tm (Fig. 1 ▶), whereas the substitution of Gln41 to a smaller Asn led to a decrease in the stability by 15°C. The fact that even Val→Ala or Leu→Ala substitutions produce such dramatic changes in thermostability reconfirms our previous assessment that the hydrophobic core of ubiquitin was optimized for stability (Thomas and Makhatadze 2000).
Fig. 1.
Thermodynamic parameters of unfolding of the ubiquitin variants measured by DSC. (A,B) The dependence of the transition temperature (Tm) of unfolding of the ubiquitin variants on the pH of the solution. (C,D) The dependence of the calorimetric enthalpy of unfolding (ΔHcal) of the ubiquitin variants on the transition temperature (Tm). Symbols represent the following proteins: U6H63 (•), U6H63-Gln41Leu (○), U6H63-Gln41Val (⋄), U6H63-Gln41Asn (asterisk), U6H63-Leu67Ala (▵), U6H63-Leu67Asn (▴), U6H63/42 (♦), U6H63/42-Val5Ala (□), U6H63/42-Val5Asn (▪) U6H63/42-Val17Ala (∇), U6H63/42-Val17Asn (|ap).
Effect of amino acid substitutions on the enthalpy of unfolding of the ubiquitin variants
In addition to the decrease in the transition temperature, the variants of ubiquitin also have lower enthalpy of unfolding. The amino acid substitutions of a large nonpolar residue to a smaller one have a moderate effect on the enthalpy of unfolding (Fig. 1 ▶ and Table 2). The Val to Ala substitutions at positions 5 and 17 decrease the enthalpy of unfolding of ubiquitin by approximately 10%. This decrease in enthalpy is certainly on the order of the experimental error of enthalpy determination estimated to be 5%. This uncertainty in enthalpy determination is due entirely to the inaccuracy in the concentration measurement. The Leu67 to Ala substitution leads to a 20% decrease in the enthalpy of unfolding (Fig. 1 ▶). Decreases in the enthalpy of unfolding upon substitution of Ile to Val, Leu, or Phe at two hydrophobic positions in the ubiquitin molecule have been reported before (Thomas and Makhatadze 2000). In the case of these 15 ubiquitin variants, the magnitude in enthalpy decrease (up to 30%) is comparable with the instances reported here (Thomas and Makhatadze 2000). This decrease in the enthalpy of unfolding was attributed to two factors: the changes in the packing of hydrophobic residues upon substitutions and the difference in the hydration enthalpies of different residues. These two enthalpies are expected to have an opposite sign (Makhatadze and Privalov 1995; Thomas and Makhatadze 2000). Substitution of a large nonpolar residue, buried in the hydrophobic core of a protein, to a smaller residue is expected to lower the enthalpy of unfolding because of the potential decrease in the packing interactions. On the other hand, the change in the enthalpy of hydration upon larger to smaller nonpolar residue substitutions is expected to increase the enthalpy of unfolding. The final effect of these two factors depends on the site of the substitution but seems to always decrease the enthalpy when a large nonpolar residue is replaced with a smaller residue.
Table 2.
Comparison of the experimental and calculated values for the heat capacity changes upon unfolding of ubiquitin variants
| Experimental | Calculated | ||||||
| ΔVa | ΔCp | ΔΔCp | ΔΔCp(MP)b | ΔΔCp(MF)c | ΔΔCp(SLR)d | ΔΔCp(MPS)e | |
| U6H63 | 2.8 ± 0.2 | — | |||||
| U6H63-Leu67Ala | −75 | 2.9 ± 0.2 | 0.1 | −0.15 | −0.13 | −0.09 | −0.08 |
| U6H63-Leu67Asn | −40 | 1.6 ± 0.1 | −1.2 | −0.26 | −0.25 | −0.16 | −0.13 |
| U6H63-Gln41Asn | −24 | 2.7 ± 0.3 | −0.1 | −0.11 | −0.11 | −0.07 | −0.06 |
| U6H63-Gln41Val | −11 | 3.6 ± 0.2 | 0.8 | 0.10 | 0.11 | 0.07 | 0.05 |
| U6H63-Gln41Leu | 15 | 3.7 ± 0.2 | 0.9 | 0.15 | 0.14 | 0.09 | 0.07 |
| U6H63/42 | 2.9 ± 0.3 | — | |||||
| U6H63/42-Val5Ala | −49 | 2.7 ± 0.3 | −0.2 | −0.11 | −0.09 | −0.07 | −0.06 |
| U6H63/42-Val17Ala | −49 | 2.7 ± 0.3 | 0.0 | −0.11 | −0.09 | −0.07 | −0.06 |
| U6H63/42-Val5Asn | −13 | 1.5 ± 0.1 | −1.4 | −0.22 | −0.21 | −0.14 | −0.11 |
| U6H63/42-Val17Asn | −13 | 1.8 ± 0.2 | −1.1 | −0.22 | −0.21 | −0.14 | −0.11 |
a Volume data from Tsai et al. (1999) and has units of Å3. b Calculated according to equation 1 (Makhatadze and Privalov 1995). c Calculated according to equation 2 (Murphy and Freire 1992). d Calculated according to equation 3 (Spolar et al. 1992). e Calculated according to equation 4 (Myers et al. 1995). All heat capacity data ΔCp are in kJ • mol−1 • K−1.
The situation is even less certain when a buried nonpolar residue is substituted with a polar residue. It seems that one can expect the decrease in the enthalpy of unfolding because of the fact that the hydration of polar groups is characterized by a much more negative enthalpy than the enthalpy of hydration of nonpolar groups (Makhatadze and Privalov 1995). The estimates show that Val or Leu substitutions to Asn are expected to decrease the enthalpy of unfolding because of the changes in hydration by ∼60 kJ•mol−1. This is somewhat less than the experimentally observed decrease (Fig. 1 ▶). In the case of the ubiquitin molecule, substitution of nonpolar residues Val or Leu at positions 5, 17, and 67 to a polar Asn leads to a dramatic decrease in the enthalpy of unfolding. This decrease is as high as 80–110 kJ•mol−1 or 40% to 50% of the enthalpy of the corresponding reference protein at 60°C. Thus the effect of the changes in hydration upon nonpolar to polar substitutions appears to be further enhanced by the decrease in the enthalpy of internal interactions.
Interestingly, the substitution of the buried polar residue at position 41 to a nonpolar residue of similar size and shape did not dramatically affect the enthalpy of unfolding. Actually, the enthalpies of unfolding of Gln41Val and Gln41Leu and variants of ubiquitin at 60°C are practically identical to the enthalpy of unfolding of the reference wild-type protein. This can be rationalized by the fact that the side chain of Gln41 forms two hydrogen bonds in the native state. The enthalpy of hydration of the Gln side chain is estimated to be on the order of −60 kJ•mol−1 at 60°C. The energy of a hydrogen bond in a vacuum is also estimated to be large. According to different estimates, it can vary between 30–70 kJ•mol−1, depending on the geometry and the nature of the groups involved in the hydrogen bond formation (Makhatadze and Privalov 1995). Although the interior of a protein has a low dielectric constant, it is not similar to the dielectric constant of a vacuum or nonpolar liquid. According to some recent experimental measurements, it can be as high as 12 (Dwyer et al. 2000; Stites et al. 1991). A higher dielectric constant for the protein interior means that lower estimates for the enthalpy of the internal hydrogen bond are more realistic. This certainly agrees with the 15 kJ/mol decrease in enthalpy of unfolding for the Gln41Asn variant. Thus the unfavorable enthalpy of hydration upon the burial of Gln41 is probably largely compensated by the enthalpy of formation of the hydrogen bond in the low dielectric environment of the protein interior. As a result, substitution of a hydrogen-bonded polar residue with the nonpolar residue of the same size has very small effect on the enthalpy of protein unfolding. This conclusion is in accord with previous empirical calculations (Makhatadze and Privalov 1995).
NMR spectroscopy
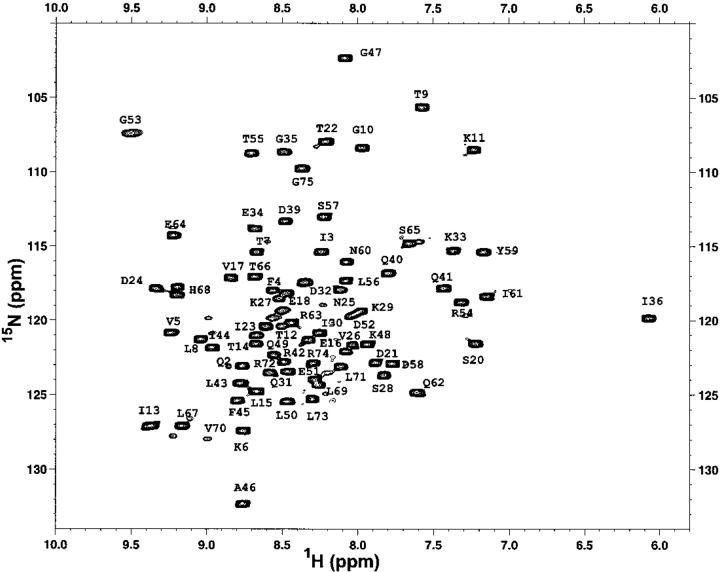
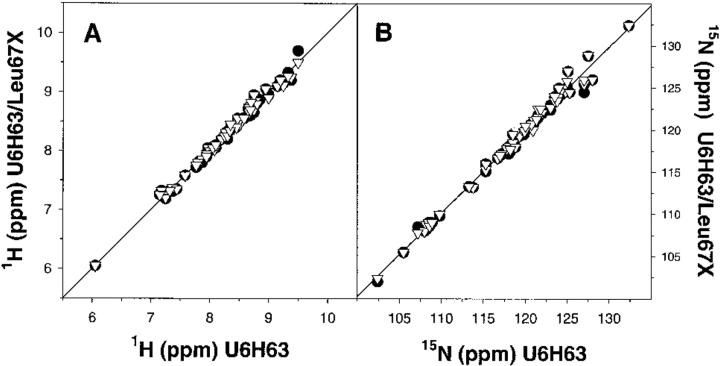
The changes in ΔCp upon protein unfolding are thought to arise primarily from the hydration of the surface area that is exposed upon unfolding of the protein. Thus, structural changes away from the substitution site leading to the changes in the buried area might have an effect on the ΔCp. The effect of amino acid substitutions on the structure of the ubiquitin variants was evaluated using nuclear magnetic resonance (NMR) spectroscopy. Figure 2 ▶ shows the 1H-15N correlation spectrum of the U6H63 variant of ubiquitin. The spectrum shows intense cross peaks and good resonance displacement indicative of a native protein. The chemical shifts are in general agreement with the chemical shifts assignment reported earlier for human ubiquitin (Schneider et al. 1992; Weber et al. 1987). Furthermore, comparison between the U6H63 protein and ubiquitin variants U6H63-Leu67Ala and U6H63-Leu67Asn shows very small chemical shift differences for both 1H and 15N protons (Fig. 3 ▶). From this we conclude that there are only very small rearrangements in the structures of these three proteins and that the differences between the three structures can, at least in a first approximation, be considered insignificant.
Fig. 2.
1H-15N chemical shift correlation spectrum of the U6H63 variant of yeast ubiquitin with the partial assignments of the backbone 15N amide protons.
Fig. 3.
Comparison of 1H (A) and 15N (B) chemical shifts for U6H63 with its two variants containing the substitutions Leu67Ala (•) and Leu67Asn (∇). In all four cases, the correlation coefficients are greater than 0.989.
Discussion
Heat capacity change upon unfolding, ΔCp
The decrease in the enthalpy of unfolding of ubiquitin variants in some cases also leads to the changes in the temperature dependence of this parameter. The primary goal of this study was to estimate the effect of changes in the polarity of a buried residue on the temperature dependence of the enthalpy of unfolding.
The amino acid substitutions generated at positions 5, 17, 41, and 67 of the ubiquitin molecule produced very different effects on the temperature dependence of the enthalpy of unfolding. Figure 1 ▶ and Table 2 show that there is a large variety in the slopes of ΔH versus Tm plots. This slope represents the heat capacity change upon protein unfolding, ΔCp. When nonpolar residues were substituted with polar (Val5Asn, Val17Asn, Leu67Asn), the decrease in the slope from 2.9 ± 0.3 kJ•mol−1•K−1 to an average of 1.7±0.3 kJ•mol−1•K−1 is observed (Table 2). This 1.2 kJ•mol−1•K−1, or a 40% decrease in the ΔCp values upon a nonpolar to polar substitution, is significantly higher than the error in estimating this parameter. The decrease in ΔCp is independent of which buried position of ubiquitin the substitution of nonpolar residue to polar residue is made. Furthermore, substitutions to Ala residues at the same positions do not have any appreciable effect on the ΔCp values. This reassures that the effects observed in the case of nonpolar to polar substitutions are not consequences of particular properties at the substitution sites but are consequences of the type of substitution. This conclusion is also supported by the fact that the HSQC spectra corresponding to the variants with substitutions at position 67 indicate significant structural similarity between the wild-type, Leu67Ala, and Leu67Asn proteins (Fig. 3 ▶). This fact is very important because it is believed that the heat capacity changes upon protein unfolding are largely defined by the exposure to water of polar and nonpolar groups that were buried in the native state (Makhatadze et al. 1995; Makhatadze and Privalov 1995; Murphy and Freire 1992; Murphy and Gill 1991; Spolar et al. 1992). Furthermore, it is believed that the heat capacity changes are directly proportional to the amount of the water-accessible surface area, ASA, a geometrical parameter that provides a measure of the extent of contacts between residues in a protein and the solvent water. If there is no significant change in the structure of the protein, the decrease in the values of ΔCp means that exposure of buried polar groups will lead to a lower increment of heat capacity than the exposure of the buried nonpolar group. It must be mentioned that the changes in ASA upon amino acid substitutions in many cases deviate from what one would predict just based on the difference in ASA of the amino acid being replaced, but these deviations are usually small (see e.g., Kim et al. 1993; Matthews 1993; Matthews 1995; Matthews 1996; Takano et al. 1997; Takano et al. 1999; Yamagata et al. 1998).
Further support that exposure of buried polar groups will lead to a lower increment of heat capacity comes from the analysis of the ΔCp values for the amino acid substitution that reverses the polarity of a buried residue in the other direction, that is, from the polar to nonpolar substitution, Gln41Val and Gln41Leu. Based on the results discussed above, one would expect that a polar to nonpolar substitution would lead to an increase in the ΔCp values. This is exactly what is observed (Table 2). The slopes of ΔH versus Tm for the Gln41Val and Gln41Leu ubiquitin variants are 3.6 and 3.7 ± 0.2 kJ•mol−1•K−1, respectively, or ∼25% higher than the wild-type protein. For comparison, substitution with the smaller polar residue in Gln41Asn did not have a noticeable effect on ΔCp (Table 2). This undoubtedly shows that the exposure of a polar group has a much smaller effect on heat capacity than the exposure of the nonpolar group of a similar size.
There are three major reasons why these estimates of ΔCp seem to be reliable. First, the change in ΔCp does not depend on the site at which the substitutions are made. The decrease in ΔCp upon a nonpolar to polar substitution is very similar at positions 5, 17, and 67, which are separated by 9 to 15 Å. Second, the change of a polar to a nonpolar residue produces an effect on ΔCp very similar in magnitude but opposite in sign to the effect of a nonpolar to a polar substitution. Third, there do not seem to be significant structural changes in the ubiquitin molecule upon these substitutions.
Based on these results, we can conclude that exposure to the solvent of a polar group upon protein unfolding produces a far lower change in ΔCp than the exposure of a nonpolar group. Similar conclusions have been proposed by several groups and were based on the analysis of the thermodynamics of transfer of different model compounds into water (Makhatadze and Privalov 1993; Murphy and Gill 1991; Spolar et al. 1992). It was shown that the heat capacity changes upon transfer of polar and nonpolar surfaces to water are of opposite sign (Makhatadze and Privalov 1995): The transfer of nonpolar groups is accompanied by a positive ΔCp and the transfer of polar groups is accompanied by negative ΔCp values.
Comparison of experimental and empirically calculated ΔΔCp
Several empirical equations for the calculations of the heat capacity changes upon protein unfolding based on the changes in ASA as a result of unfolding have been proposed. Equation 1 was proposed by Makhatadze and Privalov (1995), equation 2 by Murphy and Freire (1992), and equation 3 by Spolar et al. (1992). These equations are based on the experimental heat capacity change upon transfer for different types of model compounds. Equation 4 by Myers et al. (1995) was a result of the fit of the experimental ΔCp data for a number of proteins.
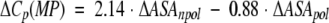 |
1 |
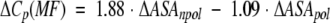 |
2 |
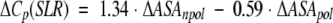 |
3 |
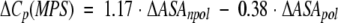 |
4 |
The surface areas for use in these equations were taken from Miller et al. (1987): 67, 117, 137, 44, and 91 Å2 for the nonpolar ΔASAnpol of the side chains of Ala, Val, Leu, Asn, and Gln, respectively, and 69 and 53 Å2 for the polar ΔASApol for the parts of the Asn and Gln side chains. Calculations performed by either of the four equations predict that substitutions of either Val or Leu with Ala will produce very small changes in ΔCp, something on the order of 0.1 kJ•mol−1•K−1 (Table 2). Changes of this magnitude are within the experimental error of measurements of the ΔCp (Pace et al. 1999). The experimental data follow this prediction and show that Leu/Val to Ala substitutions have an undetectable effect on the heat capacity changes upon unfolding (Fig. 1 ▶; Table 2). Calculations according to equations 1 to 4 for the amino acid substitutions from a polar to a nonpolar amino acid residue or from nonpolar to a polar, predict larger changes in ΔCp, up to 0.26 kJ•mol−1•K−1 (Table 2). However, the experimental values are 3 to 4 times larger (Table 2). What causes this discrepancy? The first possible explanation is that the calculations for the changes in ΔCp using equations 1 to 4 are not applicable to the systems that undergo small structural perturbations. It seems that when used for the unfolding of a protein, during which the changes in the ASA are large (≫: 5000 Å2), the averaging produces a reasonable prediction of the ΔCp (Robertson and Murphy 1997). Yet, when the reaction involves much smaller changes in the ASA, such as from protein-protein or protein-ligand interactions, the predictions were not always found to be valid (see e.g., Henriques et al. 2000) However, other studies were in excellent agreement between experimental and calculated values (e.g., Brokx et al. 2001). The calculations also neglect the heat capacity changes associated with the internal interactions and assume that the effect is entirely because of the hydration. Several groups have used different approaches to examine closely this assumption. It was shown that the internal interactions contribute < 5% to 10% to the observed ΔCp of globular proteins (Makhatadze 1998a; Makhatadze and Privalov 1990; Privalov and Makhatadze 1992; Robertson and Murphy 1997; Sturtevant 1977). The second possible explanation of the observed discrepancies between the experimental and predicted changes in ΔΔCp on nonpolar to polar and polar to nonpolar substitutions is water penetration. It has been proposed from both experimental and theoretical studies that transient and permanent water molecules can be found in the protein interior (Yu et al. 1999; Dwyer et al. 2000; Garcia and Hummer 2000). One of the most convincing examples is the appearance of new water molecules seen in the X-ray structure of staphylococcal nuclease variant that buries an acidic Glu in place of the nonpolar Val66 (Dwyer et al. 2000). More importantly, these buried water molecules appear to be positionally ordered. Thus, one would expect that release of buried water will increase the apparent polar ASA exposed upon unfolding, and correspondingly, to lower ΔCp values. However, this intuitive conclusion disagrees with the reported estimates of the heat capacity change upon release of buried water. For example, Morton and Ladbury (1996) estimated the ΔCp of burying a water molecule to be between −25 and −50 J•K−1• (mol of water)−1. These estimates indicate that burial of water will produce higher ΔCp values. The third possible explanation of the observed discrepancies between the experimental and predicted changes in ΔΔCp involves the effects of nonpolar to polar and polar to nonpolar substitutions on the structure of the unfolded state ensemble. One can imagine that the amount of apparent ASA exposed in the unfolded state is dependent on the amino acid substitution (see e.g., Wrabl and Shortle 1999). However, there are three facts that argue against it: (1) The difference between experimental and calculated ΔΔCp is observed only when the substitutions change residues from nonpolar to polar or vice versa (Table 2). There is no obvious difference between experimental and calculated ΔΔCp (Table 2) when a large nonpolar residue is replaced with a smaller one or when a larger polar residue is replaced with a smaller polar residue; (2) the observed differences between experimental and calculated ΔΔCp seem to be independent of the position of the substitution site in the sequence of ubiquitin, that is, is the same for nonpolar to polar substitutions at all three positions 5, 17, and 67; (3) the absolute values ΔΔCp do not depend on whether the substitution is from a nonpolar to a polar residue (positions 5, 17, 67) or from a polar to a nonpolar residue (position 41). Thus, at present time there is no logical explanation for the discrepancy between calculated and experimentally observed ΔΔCp that can be proposed.
Convergence temperature and ΔCp
It was believed for quite some time that there is an intimate relationship between the enthalpy of protein unfolding and the heat capacity change upon unfolding, such that the ΔH functions for different proteins converge to a common value at ∼110°C (Privalov and Khechinashvili 1974). Later, Murphy and Gill (1991), using a larger set of globular proteins, showed the convergence to a single value of the ΔH(T) functions at high temperature. Numerous explanations for the meaning of this convergence have been proposed, mostly suggesting that the interactions of certain protein groups with the solvent are zero at the convergence temperature. Later, more detailed analysis of the denaturation of a larger set of proteins (Makhatadze and Privalov 1995; Robertson and Murphy 1997) showed that the convergence temperature is not as obvious as it seemed from the analysis of the initial set of globular proteins. The results presented above clearly show that the convergence temperature for the unfolding of globular proteins is probably purely superficial: Proteins can be properly folded into a three-dimensional structure and yet the enthalpy (and for the same reason) entropy functions will extrapolate to a different value at high temperature.
We must note, however, that the convergence behavior of enthalpy and entropy functions was observed for the naturally occurring proteins and there is a possibility that they evolved according to certain rules. The benefit (if any) to have both thermodynamic functions converge to the same value at high temperatures remains to be uncovered.
Concluding remarks
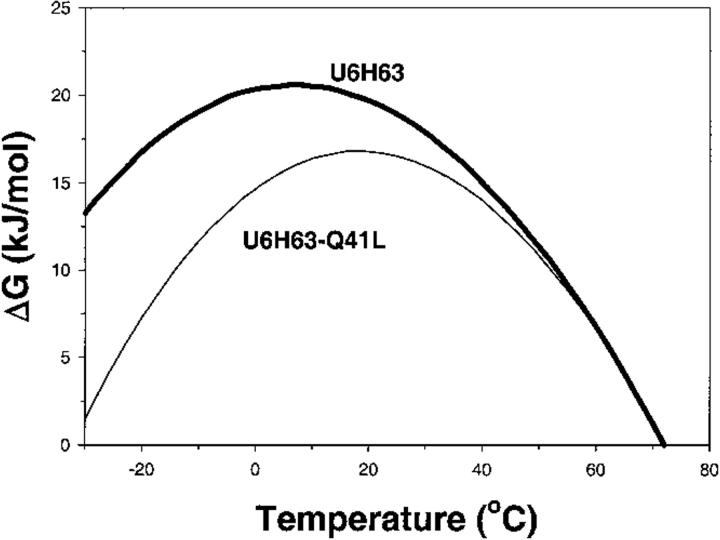
We have shown that it is possible to modulate ΔCp of a protein in a rational way. This fact is very important in the rational engineering of proteins for biotechnological needs because ΔCp defines the temperature dependence of the stability curve as measured by the Gibbs energy, ΔG. In many cases it is far more important to have an enzyme with high thermostability (high Tm) than with high thermodynamic stability (high ΔG) (Sanchez-Ruiz and Makhatadze 2001). Based on the results presented in this paper, one can imagine a situation in which amino acid substitution produces protein with a lower stability at 25°C but with a higher transition temperature. A good example of such behavior was recently shown by Willis et al., (2000). The opposite situation, in which two proteins are generated that have similar thermostability but different thermodynamic stability at 25°C, can also be achieved. In fact, this is the case for the U6H63 and U6H63-Gln41Leu variant proteins (Fig. 4 ▶). The knowledge of just one parameter, Tm or ΔG at single temperature, is not enough to describe adequately the thermodynamic properties of a protein, and ΔCp is thus a parameter of paramount importance.
Fig. 4.
Comparison of the temperature dependences of the stability (ΔG) functions for U6H63 (thick line) and U6H63-Gln41Leu (thin line). The temperature dependence of ΔG was calculated assuming temperature-independent ΔCp as 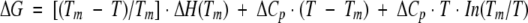 using the data from Figure 1. ▶
using the data from Figure 1. ▶
Materials and methods
Cloning and site-directed mutagenesis
His-tagged ubiquitin was generated using the following primers: Usph-f (5′GATATACGCATGCAGATCTTCGTC) and UecoH-r (5′GCCGAATTCGGTCATCAGTGATGGTGATGGTGATGAC CACCTCTTAGTCTTAAG) and the yeast ubiquitin gene as a template (Makhatadze et al. 1998; Thomas and Makhatadze 2000). SphI and EcoRI restriction enzymes were used for directional cloning of the His-tagged ubiquitin gene under the control of the T7 promoter. Mutations in the ubiquitin gene were generated using the QuikChange Site-Directed Mutagenesis Kit and the His-tagged ubiquitin gene as a template. The incorporation of mutations was confirmed by sequencing the entire ubiquitin gene using the dideoxy chain termination sequencing method on an ABI PRISM 377 DNA sequencer.
Overexpression and purification of ubiquitin variants
A plasmid carrying an His-tagged gene for ubiquitin variant was transformed into Escherichia coli BL21(DE3) cells and grown at 37°C in 2YT medium containing 100μg/mL ampicillin. Ubiquitin expression was induced with 1 mM isopropyl-β-D-thiogalactoside (IPTG) when the optical density of cells was approximately 0.8 o.u. at 600 nm. Cells were harvested by centrifugation after 5 h and were resuspended in buffer A (100 mM NaH2PO4 10 mM Tris, 6M Urea at pH 8.0; Novagen, His Binding Kit). Lysis was achieved by passing the cell suspension through the French pressure cell twice. The cell debris was removed by centrifugation at 25,000g for 1 h at 4°C. The supernatant was loaded onto a Ni-NTA His-Binding Resin (Novagen). His-tagged protein was eluted from the column according to the protocol from the manufacturer. Fractions containing ubiquitin were combined and applied onto Sephadex G-50 gel-filtration column (2.5 × 120 cm) equilibrated with 50 mM Tris, 100 mM KCl at pH 7.5. Ubiquitin containing fractions were dialyzed against water, lyophilized, and stored at −20°C.
Overexpression and purification of the 15N-labeled ubiquitin variants
The BL21 (DE3) strain of E. coli, containing the plasmid for the ubiquitin variant, was grown in MOPS-based minimal media (Neidhardt et al. 1974), with 0.2% glucose as the carbon source and 0.2% ammonium chloride (NH4Cl) as the nitrogen source. The media was also supplemented with 2 mg/mL thiamine and 100 μg/mL ampicillin. Uniformally enriched 15N samples were obtained by growing on 15NH4Cl as the sole nitrogen source. Stable isotopically labeled ammonium chloride (15NH4Cl) was purchased from Cambridge Isotope Laboratories. The ubiquitin production was induced when the cell density reached an absorbance of 1 o.u. at 600 nm by adding 1 mM IPTG. The cells were harvested by centrifugation and washed once in the MOPS medium without the addition of any supplements or nutrients. Further purification of the overexpressed ubiquitin variants was done according to the protocol described above for the unlabeled proteins.
Differential scanning calorimetry
The calorimetric experiments were performed on a VP-DSC calorimeter (MicroCal) as described before (Makhatadze 1998b). Protein concentration was 1.8 to 2.5 mg/mL, and experiments were performed in 20 mM glycine (pH 2.0–3.5) or 20 mM sodium acetate buffers (pH 3.5–4.75). The concentration of the His-tagged ubiquitin and its variants was measured spectrophotometrically at 280 nm using an extinction coefficient of 0.136 o.u. for a 0.1% solution. All experiments were performed at a heat rate of 1.5 deg/min. All ubiquitin variants were routinely checked for reversibility by recording the second scan. In all cases, the reversibility was better than 90%. Calorimetric profiles were analyzed according to a two-state transition model as described (Thomas and Makhatadze 2000).
NMR spectroscopy
1H-15N HSQC spectra were acquired on a Bruker spectrometer (DRX-600) in 95% H2O/5% 2H2O as described (Falzone et al. 1994; Falzone et al. 1996). Quadrature detection was achieved in the indirect 1H dimension by using TPPI (Drobny et al. 1979). Thirty-two scans for each spectrum were collected in 256 experiments. Water was suppressed using a WATERGATE sequence (Piotto et al. 1992). Processing was performed with Xwinnmr software. The solutions were approximately 1 mM in protein and 5 mM in acetate buffer at pH 5.0 with added 2H2O to yield a 90%H2O/10%2H2O solution.
Acknowledgments
We thank Chris Falzone for the NMR experiments and Marimar Lopez and Greg Bertenshaw for their discussions of the manuscript. This work was supported by the grant from the NIH (GM54537 to G.I.M).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at www.proteinscience.org/cgi/doi/10.1110/
References
- Baldwin, R.L. 1986. Temperature dependence of the hydrophobic interaction in protein folding. Proc. Natl. Acad. Sci. 83 8069–8072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Naim, A. 1990. Solvent effects on protein association and protein folding. Biopolymers 29 567–596. [DOI] [PubMed] [Google Scholar]
- Ben-Naim, A., Ting, K.L., and Jernigan, R.L. 1990. Solvent effect on binding thermodynamics of biopolymers. Biopolymers 29 901–919. [DOI] [PubMed] [Google Scholar]
- Brokx, R.D., Lopez, M.M., Vogel, H.J., and Makhatadze, G.I. 2001. Energetics of target peptide binding by calmodulin reveals different modes of binding. J. Biol. Chem. 276 14083–14091. [DOI] [PubMed] [Google Scholar]
- Dill, K.A., Alonso, D.O., and Hutchinson, K. 1989. Thermal stabilities of globular proteins. Biochemistry 28 5439–5449. [DOI] [PubMed] [Google Scholar]
- Drobny, G., Pines, A.S., Weitekamp, D.P., and Wen, D. 1979. Symp. Faraday Soc. 13 49–55. [Google Scholar]
- Dwyer, J.J., Gittis, A.G., Karp, D.A., Lattman, E.E., Spencer, D.S., Stites, W.E., and Garcia-Moreno, E.B. 2000. High apparent dielectric constants in the interior of a protein reflect water penetration. Biophys. J. 79 1610–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edsall, J.T. 1935. Apparent molal heat capacities of amino acids and other organic compounds. J. Am. Chem. Soc. 57 1506–1507. [Google Scholar]
- Falzone, C.J., Kao, Y.H., Zhao, J., MacLaughlin, K.L., Bryant, D.A., and Lecomte, J.T. 1994. 1H and 15N NMR assignments of PsaE, a photosystem I subunit from the cyanobacterium Synechococcus sp. strain PCC 7002. Biochemistry 33 6043–6051. [DOI] [PubMed] [Google Scholar]
- Falzone, C.J., Mayer, M.R., Whiteman, E.L., Moore, C.D., and Lecomte, J.T. 1996. Design challenges for hemoproteins: The solution structure of apocytochrome b5. Biochemistry 35 6519–6526. [DOI] [PubMed] [Google Scholar]
- Gallagher, K. and Sharp, K. 1998. Electrostatic contributions to heat capacity changes of DNA-ligand binding. Biophys. J. 75 769–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia, A.E. and Hummer, G. 2000. Water penetration and escape in proteins. Proteins 38 261–272. [PubMed] [Google Scholar]
- Henriques, D.A., Ladbury, J.E., and Jackson, R.M. 2000. Comparison of binding energies of SrcSH2-phosphotyrosyl peptides with structure-based prediction using surface area based empirical parameterization. Protein Sci. 9 1975–1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauzmann, W. 1959. Some factors in the interpretation of protein denaturation. Adv. Protein Chem. 14 1–63. [DOI] [PubMed] [Google Scholar]
- Kim, K.S., Tao, F., Fuchs, J., Danishefsky, A.T., Housset, D., Wlodawer, A., and Woodward, C. 1993. Crevice-forming mutants of bovine pancreatic trypsin inhibitor: Stability changes and new hydrophobic surface. Protein Sci. 2 588–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar, G.A., Desjarlais, J.R., and Handel, T.M. 1997. De novo design of the hydrophobic core of ubiquitin. Protein Sci. 6 1167–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loladze, V.V., Ibarra-Molero, B., Sanchez-Ruiz, J.M., and Makhatadze, G.I. 1999. Engineering a thermostable protein via optimization of charge-charge interactions on the protein surface. Biochemistry 38 16419–16423. [DOI] [PubMed] [Google Scholar]
- Madan, B. and Sharp, K. 1996. Heat capacity changes accompanying hydrophobic and ionic solvation: A Monte Carlo and random network model study. J. Phys. Chem. B 100 7713–7721. [Google Scholar]
- ———. 1997. Molecular origin of hydration heat capacity changes of hydrophobic solutes: Perturbation of water structure around alkanes. J. Phys. Chem. B 101 11237–11242. [Google Scholar]
- Makhatadze, G.I. 1998a. Heat capacities of amino acids, peptides and proteins. Biophys. Chem. 71 133–156. [DOI] [PubMed] [Google Scholar]
- ———. 1998b. Measuring protein thermostability by differential scanning calorimetry. Trans. J. Wiley. Current Protocols in Protein Chemistry, 2, John Wiley & Sons, New York. [DOI] [PubMed]
- Makhatadze, G.I. and Privalov, P.L. 1990. Heat capacity of proteins. I. Partial molar heat capacity of individual amino acid residues in aqueous solution: Hydration effect. J. Mol. Biol. 213 375–384. [DOI] [PubMed] [Google Scholar]
- ———. 1993. Contribution of hydration to protein folding thermodynamics. I. The enthalpy of hydration. J. Mol. Biol. 232 639–659. [DOI] [PubMed] [Google Scholar]
- ———. 1995. Energetics of protein structure. Adv. Protein Chem. 47 307–425. [DOI] [PubMed] [Google Scholar]
- Makhatadze, G.I., Clore, G.M., and Gronenborn, A.M. 1995. Solvent isotope effect and protein stability. Nat. Struct. Biol. 2 852–855. [DOI] [PubMed] [Google Scholar]
- Makhatadze, G.I., Lopez, M.M., Richardson, 3rd, J.M., and Thomas S.T. 1998. Anion binding to the ubiquitin molecule. Protein Sci. 7 689–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews, B.W. 1993. Structural and genetic analysis of protein stability. Annu. Rev. Biochem. 62 139–160. [DOI] [PubMed] [Google Scholar]
- ———. 1995. Studies on protein stability with T4 lysozyme. Adv. Protein Chem. 46 249–278. [DOI] [PubMed] [Google Scholar]
- ———. 1996. Structural and genetic analysis of the folding and function of T4 lysozyme. FASEB J. 10 35–41. [DOI] [PubMed] [Google Scholar]
- Miller, S., Janin, J., Lesk, A.M., and Chothia, C. 1987. Interior and surface of monomeric proteins. J. Mol. Biol. 196 641–656. [DOI] [PubMed] [Google Scholar]
- Morton, C.J. and Ladbury, J.E. 1996. Water-mediated protein-DNA interactions: The relationship of thermodynamics to structural detail. Protein Sci. 5 2115–2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, K.P. and Freire, E. 1992. Thermodynamics of structural stability and cooperative folding behavior in proteins. Adv. Protein Chem. 43 313–361. [DOI] [PubMed] [Google Scholar]
- Murphy, K.P. and Gill, S.J. 1991. Solid model compounds and the thermodynamics of protein unfolding. J. Mol. Biol. 222 699–709. [DOI] [PubMed] [Google Scholar]
- Myers, J.K., Pace, C.N., and Scholtz, J.M. 1995. Denaturant m values and heat capacity changes: Relation to changes in accessible surface areas of protein unfolding. Protein Sci. 4 2138–2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidhardt, F.C., Bloch, P.L., and Smith, D.F. 1974. Culture medium for enterobacteria. J. Bacteriol. 119 736–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace, C.N., Grimsley, G.R., Thomas, S.T., and Makhatadze, G.I. 1999. Heat capacity change for ribonuclease A folding. Protein Sci. 8 1500–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piotto, M., Saudek, V., and Sklenar, V. 1992. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR 2 661–665. [DOI] [PubMed] [Google Scholar]
- Privalov, P.L. and Gill, S.J. 1988. Stability of protein structure and hydrophobic interaction. Adv. Protein Chem. 39 191–234. [DOI] [PubMed] [Google Scholar]
- Privalov, P.L. and Khechinashvili, N.N. 1974. A thermodynamic approach to the problem of stabilization of globular protein structure: A calorimetric study. J. Mol. Biol. 86 665–684. [DOI] [PubMed] [Google Scholar]
- Privalov, P.L. and Makhatadze, G.I. 1992. Contribution of hydration and non-covalent interactions to the heat capacity effect on protein unfolding. J. Mol. Biol. 224 715–723. [DOI] [PubMed] [Google Scholar]
- Robertson, A.D. and Murphy, K.P. 1997. Protein structure and the energetics of protein stability. Chem. Rev. 97 1251–1267. [DOI] [PubMed] [Google Scholar]
- Sanchez-Ruiz, J.M. and Makhatadze, G.I. 2001. To charge or not to charge? Trends Biotechnol. 19 132–135. [DOI] [PubMed] [Google Scholar]
- Schneider, D.M., Dellwo, M.J., and Wand, A.J. 1992. Fast internal main-chain dynamics of human ubiquitin. Biochemistry 31 3645–3652. [DOI] [PubMed] [Google Scholar]
- Spolar, R.S., Ha, J.H., and Record, Jr., M.T. 1989. Hydrophobic effect in protein folding and other noncovalent processes involving proteins. Proc. Natl. Acad. Sci. 86 8382–8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spolar, R.S., Livingstone, J.R., and Record, Jr., M.T. 1992. Use of liquid hydrocarbon and amide transfer data to estimate contributions to thermodynamic functions of protein folding from the removal of nonpolar and polar surface from water. Biochemistry 31 3947–3955. [DOI] [PubMed] [Google Scholar]
- Stites, W.E., Gittis, A.G., Lattman, E.E., and Shortle, D. 1991. In a staphylococcal nuclease mutant the side-chain of a lysine replacing valine 66 is fully buried in the hydrophobic core. J. Mol. Biol. 221 7–14. [DOI] [PubMed] [Google Scholar]
- Sturtevant, J.M. 1977. Heat capacity and entropy changes in processes involving proteins. Proc. Natl. Acad. Sci. 74 2236–2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano, K., Yamagata, Y., Fujii, S., and Yutani, K. 1997. Contribution of the hydrophobic effect to the stability of human lysozyme: Calorimetric studies and X-ray structural analyses of the nine valine to alanine mutants. Biochemistry 36 688–698. [DOI] [PubMed] [Google Scholar]
- Takano, K., Yamagata, Y., Funahashi, J., Hioki, Y., Kuramitsu, S., and Yutani, K. 1999. Contribution of intra- and intermolecular hydrogen bonds to the conformational stability of human lysozyme(,). Biochemistry 38 12698–12708. [DOI] [PubMed] [Google Scholar]
- Thomas, S.T. and Makhatadze, G.I. 2000. Contribution of the 30/36 hydrophobic contact at the C-terminus of the alpha-helix to the stability of the ubiquitin molecule. Biochemistry 39 10275–10283. [DOI] [PubMed] [Google Scholar]
- Tsai, J., Taylor, R., Chothia, C., and Gerstein, M. 1999. The packing density in proteins: Standard radii and volumes. J. Mol. Biol. 290 253–266. [DOI] [PubMed] [Google Scholar]
- Weber, P.L., Brown, S.C., and Mueller, L. 1987. Sequential 1H NMR assignments and secondary structure identification of human ubiquitin. Biochemistry 26 7282–7290. [DOI] [PubMed] [Google Scholar]
- Willis, M.A., Bishop, B., Regan, L., and Brunger, A.T. 2000. Dramatic structural and thermodynamic consequences of repacking a protein hydrophobic core. Structure 8 1319–1328. [DOI] [PubMed] [Google Scholar]
- Wintrode, P.L., Makhatadze, G.I., and Privalov, P.L. 1994. Thermodynamics of ubiquitin unfolding. Proteins 18 246–253. [DOI] [PubMed] [Google Scholar]
- Woolfson, D.N., Cooper, A., Harding, M.M., Williams, D.H., and Evans, P.A. 1993. Protein folding in the absence of the solvent ordering contribution to the hydrophobic interaction. J. Mol. Biol. 229 502–511. [DOI] [PubMed] [Google Scholar]
- Wrabl, J. and Shortle, D. 1999. A model of the changes in denatured state structure underlying m value effects in staphylococcal nuclease. Nat. Struct. Biol. 6 876–883. [DOI] [PubMed] [Google Scholar]
- Yamagata, Y., Kubota, M., Sumikawa, Y., Funahashi, J., Takano, K., Fujii, S., and Yutani, K. 1998. Contribution of hydrogen bonds to the conformational stability of human lysozyme: Calorimetry and X-ray analysis of six tyrosine → phenylalanine mutants. Biochemistry 37 9355–9362. [DOI] [PubMed] [Google Scholar]
- Yang, A.S., Sharp, K.A., and Honig, B. 1992. Analysis of the heat capacity dependence of protein folding. J. Mol. Biol. 227 889–900. [DOI] [PubMed] [Google Scholar]
- Yu, B., Blaber, M., Gronenborn, A.M., Clore, G.M., Caspar, D.L. 1999. Disordered water within a hydrophobic protein cavity visualized by X-ray crystallography. Proc. Natl. Acad. Sci. 96 103–108. [DOI] [PMC free article] [PubMed] [Google Scholar]