Abstract
The nature of the dynamical coupling between a protein and its surrounding solvent is an important, yet open issue. Here we used temperature-dependent protein crystallography to study structural alterations that arise in the enzyme acetylcholinesterase upon X-ray irradiation at two temperatures: below and above the glass transition of the crystal solvent. A buried disulfide bond, a buried cysteine, and solvent exposed methionine residues show drastically increased radiation damage at 155 K, in comparison to 100 K. Additionally, the irradiation-induced unit cell volume increase is linear at 100 K, but not at 155 K, which is attributed to the increased solvent mobility at 155 K. Most importantly, we observed conformational changes in the catalytic triad at the active site at 155 K but not at 100 K. These changes lead to an inactive catalytic triad conformation and represent, therefore, the observation of radiation-inactivation of an enzyme at the atomic level. Our results show that at 155 K, the protein has acquired—at least locally—sufficient conformational flexibility to adapt to irradiation-induced alterations in the conformational energy landscape. The increased protein flexibility may be a direct consequence of the solvent glass transition, which expresses as dynamical changes in the enzyme's environment. Our results reveal the importance of protein and solvent dynamics in specific radiation damage to biological macromolecules, which in turn can serve as a tool to study protein flexibility and its relation to changes in a protein's environment.
Keywords: Temperature-dependent protein crystallography, dynamical transition in proteins, solvent glass transition, radiation damage, disulfide, acetylcholinesterase, enzyme radiation-inactivation
Conformational flexibility is a key component in protein function. A global dynamical transition in proteins at temperatures between 180 and 230 K has been revealed by various techniques, including Mössbauer (Parak et al. 1982) and neutron spectroscopy (Doster et al. 1989; Ferrand et al. 1993). The dynamical transition marks the onset of unharmonic large-scale protein motions, which can be essential for substrate binding to enzymes (Rasmussen et al. 1992) and for ligand escape in myoglobin (Ostermann et al. 2000), as shown by X-ray crystallography. However, evidence has been presented that enzymatic activity can persist at temperatures below the global dynamical transition (Daniel et al. 1998). In the concept of the protein energy landscape developed by Frauenfelder, a protein molecule can be found in one of a number of quasi-isoenergetic conformational substates (Frauenfelder et al. 1991). Transitions between these substates are possible only at temperatures above a dynamical transition. The protein energy landscape is hierarchically organized in tiers, and flexibility at each tier might be gained at discrete dynamical transition temperatures not necessarily identical to the dynamical transition temperature at 180–230 K characterizing the global protein dynamics.
It has been proposed that the state of the solvent surrounding a protein affects its dynamical behaviour (Réat et al. 2000; Vitkup et al. 2000; Walser and Van Gunsteren 2001). Flash-cooling a protein crystal to cryotemperatures—typically 100 K—leads to vitrification of the solvent if its viscosity is such that crystallization is prevented at the cooling rate employed (Garman and Schneider 1997). The viscosity of the solvent is influenced by the presence of salts and precipitants and by the fact that it is confined within the protein crystal. The structure and density of the amorphous solvent at cryotemperatures are similar to those of the liquid form at room temperature, yet it is trapped in a thermodynamically metastable state owing to its quasi-infinite viscosity. The fate of the amorphous solvent upon heating depends on its composition and confinement. Solvent in the trigonal crystals of the enzyme Torpedo californica acetylcholinesterase (TcAChE, Sussman et al. 1991) used in the present study is found within large channels (∼65 Å in diameter) that run straight through the crystal. Warming these flash-cooled crystals to temperatures >155 K leads to solvent crystallization within the channels, causing a drastic increase in unit cell volume, which eventually destroys the protein crystal (Weik et al. 2001). The analogy with the behavior of pure water is helpful in this context. Upon heating, amorphous solid water undergoes a reversible glass transition at 130–140 K which manifests itself as a drastic drop in viscosity (McMillan and Los 1965; Johari et al. 1987). Further heating leads to irreversible crystallization into cubic ice at 150–155 K (for a review, see Mishima and Stanley 1998). Prior to crystallization, amorphous solid water has been shown to exhibit liquid-like properties (Smith et al. 1997). The analogy with the behavior of pure water thus provided circumstantial evidence that the solvent in the trigonal TcAChE crystals undergoes a glass transition prior to crystallization (Weik et al. 2001), that is, at or below 155 K, yet above 130 K, which is the glass transition temperature for pure water. The change in viscosity of the solvent at its glass transition offers the opportunity to study possible changes in the flexibility of a protein accompanying dynamical changes in its environment.
The experimental concept of the present study exploits the solvent glass transition in TcAChE crystals. Two temperatures were chosen at which the solvent has different viscosities. At 100 K, it is in an amorphous, rigid state. At 155 K—after having undergone a glass transition—the solvent has the lowest possible viscosity at cryotemperatures but does not yet crystallize. In the present study, structural alterations reported to arise in a protein upon X-ray irradiation (Burmeister 2000; Ravelli and McSweeney 2000; Weik et al. 2000) were shown to be markers for protein dynamics. This allowed us to analyze the flexibility of the protein at the two temperatures. At 155 K, we observed that irradiation produces significant structural changes in the active site which involve the catalytic histidine. These changes do not occur at 100 K. In addition, the radiation sensitivities of a buried disulfide bond, a buried free cysteine, and solvent-exposed methionine residues are greatly increased at 155 K compared to 100 K.
Results and Discussion
Five complete data sets (A–E) were collected sequentially at 100 and 155 K, each on a single TcAChE crystal, on beamline ID14–EH2 of the European Synchrotron Radiation Facility (ESRF) at Grenoble, France.
Temperature-dependent damage of sulfur-containing groups
Sulfur-containing groups, in particular disulfide bonds, have been shown by protein crystallography to be among the most radiation-sensitive features of a protein (Burmeister 2000; Ravelli and McSweeney 2000; Weik et al. 2000). However, not every sulfur-containing group in a given protein is equally radiation-sensitive. TcAChE possesses three intramolecular disulfide bonds, one of which, Cys254-Cys265, is significantly more radiation-sensitive at 100 K than the other two, Cys402-Cys521 and Cys67-Cys94 (Weik et al. 2000). We found that the radiation sensitivities of these disulfide bonds also display differential temperature dependence. Figure 1 ▶ shows the temperature-dependent sensitivity to radiation of Cys254-Cys265 and Cys402-Cys521. The Cys254-Cys265 disulfide bond is equally radiation-sensitive at 100 and 155 K as assessed by featureless difference Fourier maps based on corresponding data sets collected at the two temperatures (Fig. 1a ▶). The Cys402-Cys521 disulfide bond, however, is significantly more affected by radiation at 155 K than at 100 K, as revealed by a negative feature in the difference Fourier map (Fig. 1b ▶). The Cys67-Cys94 disulfide bond is the most stable one and is equally radiation-insensitive at both 100 and 155 K (data not shown). Another sulfur-containing group is the only free cysteine in TcAChE, the buried Cys231 residue. Figure 2 ▶ shows that its radiation sensitivity is strongly temperature-dependent: highly radiation-sensitive at 155 K, based on the negative feature seen in difference maps based on data sets collected at the different temperatures, 100 and 155 K (Fig. 2 ▶), yet completely radiation-insensitive at 100 K as judged by a featureless Fourier difference map between data sets A and E, contoured at a level of 3σ (data not shown). All 15 methionine residues in TcAChE are unaffected by radiation at 100 K (data not shown). At 155 K, two methionine residues, Met43 and Met83, display significant loss of electron density for the sulfur atoms as data collection proceeds, whereas the others remain unaffected (data not shown).
Fig. 1.

Differential temperature-dependence of radiation sensitivities for the Cys254-Cys265 and the Cys402-Cys521 disulfide bonds. Electron density and difference Fourier maps for the Cys254-Cys265 (a) and Cys402-Cys521 (b) disulfide bonds of TcAChE in the first (A), third (C), and fifth (E) data set collected at 100 K and 155 K. Electron density maps are contoured at a level of 1.5σ and difference Fourier maps, that is, Fobs (155 K) − Fobs (100 K), are contoured at 5σ (green) and −5σ (red).
Fig. 2.
Difference Fourier maps showing that the electron density defining the sulfur atom of Cys231 disappears at 155 K, compared to 100 K, under the influence of X-irradiation. The maps were calculated using the first (A), third (C), and last (E) data sets collected at 100 K and 155 K, that is, Fobs (155 K) − Fobs (100 K). Negative (−5σ) contours are shown in red.
Our observation that damage to sulfur-containing groups can be temperature-dependent shows that the underlying process is of a secondary, not primary nature; that is, it is caused by an attack by secondary radicals, produced either in the solvent or in the protein region, that reach radiation-sensitive groups. Primary damage, that is, direct interaction between X-ray photons and atoms in the sample, should be independent of temperature.
Upon irradiation of proteins, a variety of radicals are formed, and their relative amounts and stability are a function of temperature (Taub et al. 1979). However, other factors are likely to be at the origin of the difference in temperature-dependent radiation sensitivity of the chemically identical disulfide bonds. There may indeed be a correlation between solvent accessibility and radiation sensitivity, since Cys254-Cys265 is both the disulfide bond most exposed to solvent and the most sensitive to radiation. Regarding methionine residues, our results suggest strong connections among their solvent accessibility, radiation sensitivity, and the solvent viscosity, since the two solvent exposed residues, Met43 and Met83, are damaged at 155 K but not at 100 K. Burmeister (2000) reported that one free cysteine in crystalline myrosinase is completely stable at 100 K, whereas another is radiation-sensitive. The first is buried, the latter partly accessible to solvent. Other studies using frozen solutions also show that solvent-exposed parts of a protein are the most radiation-sensitive (Filali-Mouhim et al. 1997; Audette et al. 2000). However, sulfur-containing groups differ not only in their solvent accessibility but also in their structural environment, which has been proposed to play an important role in defining the stability of a disulfide bond towards free radical attack (Ravelli and McSweeney 2000; Lmouméne et al. 2000).
What causes the observed differences in temperature-dependent radiation sensitivity of different disulfide bonds? To address this question it is helpful to summarize the sequence of processes leading to the rupture of such bonds. The first step consists of electron capture by the disulfide to form a disulfide radical anion, RSSR•−, which can undergo spontaneous and reversible S-S bond rupture. Upon protonation of the disulfide radical anion the equilibrium is shifted towards the broken state (Armstrong 1990), yielding a thiol, RSH, and a thiyl radical, RS• (von Sonntag 1990). The equilibrium between the open and the closed form is believed to be influenced by the conformational flexibility of the bond partners (von Sonntag 1990; Favaudon et al. 1990). Irreversible S-S bond rupture is therefore favored in a flexible local environment. Which of these steps might explain the observed temperature dependence of certain disulfide bonds? Electrons certainly reach disulfide bonds at both 100 K and 155 K, since electron tunneling in proteins has been reported to occur at temperatures as low as 5 K (Dick et al. 1998; for a recent review see Winkler 2000). The availability of other radicals such as OH• and H•, created by radiolysis of water in the solvent area, as well as that of protons, might be temperature-dependent because protons are known to become mobile in amorphous ice at ∼115 K on the time scale of our data collection series (i.e., on an hour-time scale ; Fisher and Devlin 1995). The flexibility of the partners forming the disulfide bond is certainly temperature-dependent. The Cys254-Cys265 disulfide breaks readily at 100 K, implying that even at 100 K radicals must be able to reach this group, and that the environment of the bond is sufficiently flexible. The observed temperature-dependent radiation sensitivity of the more buried disulfide Cys402-Cys521 might be explained by the temperature-dependence of either the transport of protons and radicals through the protein matrix or the flexibility of its environment. Both characteristics are expected to be influenced by protein dynamics, because proton transport through the protein matrix will also demand flexibility of the components of its trajectory (Chen et al. 2000). Thus, our observation that the Cys402-Cys521 disulfide bond breaks at 155 K indicates that the protein has acquired substantial local conformational flexibility relative to 100 K, even though we cannot determine which process causes the temperature dependence.
Our observations that all methionine residues are unaffected at 100 K and that only the solvent-exposed ones are affected at 155 K suggest that radiation damage to methionine residues involves radicals such as OH• that are larger than electrons and protons. These radicals, created in the solvent area of the crystal by radiolysis of water, are trapped at 100 K and mobile at 155 K (Symons 1999). However, even though they are mobile in the solvent at 155 K they apparently cannot reach buried groups within the protein at that temperature. Another possible explanation is that radiation damage to methionine residues involves cleavage of the CG–SD bond and that the resulting radical can diffuse away at 155 K but not at 100 K.
Conformational changes in the active site above the solvent glass transition
The active site of TcAChE is formed by a catalytic triad characteristic of serine hydrolases, comprised of Ser200, His440, and Glu327, which are hydrogen-bonded in the resting state of the enzyme. Figure 3 ▶ shows difference Fourier maps of the active site between the first (A) and the last (E) data sets collected at 100 K and 155 K. At 100 K (Fig. 3a ▶), a negative difference density peak at Glu199, indicating radiation-induced decarboxylation, is the most prominent feature. At 155 K (Fig. 3b ▶), the picture is more complex. In addition to an enlarged negative peak at Glu199, a pair of positive and negative difference peaks associated with the catalytic His440 indicates that this residue undergoes a conformational change at 155 K under the influence of X-irradiation. This conformational change, absent at 100 K, is not a gradual movement but corresponds to the increasing population of a second state as data collection proceeds (as seen in difference Fourier maps calculated between data sets with increasing intervals at 155 K). The two conformations were not modelled independently due to the limited resolution of the data. In the experiment shown, a relatively low X-ray dose was used and the movement of the model of His440 is small. However, when the X-ray dose was increased in a series of data sets collected at 155 K, or when the temperature was raised above 155 K, His440NE2 (the nitrogen facing Ser200OG) moved as much as 1.6 Å away from its position in the undamaged structure — a position which was close to the positive difference density peak in Fig. 3b ▶ (data not shown). As a consequence, the hydrogen bond between His440 and Ser200 was broken. This conformational change, induced by X-irradiation, thus produces a nonfunctional conformation, and Fig. 3b ▶ effectively displays radiation-inactivation of an enzyme at the atomic level.
Fig. 3.
Sequential difference Fourier maps showing structural changes in the active site of TcAChE in response to radiation damage. The maps in panels a and b were calculated between the first (A) and the last (E) data sets collected at 100 K (Fig. 1a ▶) and at 155 K (Fig. 1b ▶), that is, FobsE (100 K) − FobsA (100 K) and FobsE (155 K) − FobsA (155 K). Positive (5σ) and negative (−5σ) contours are shown in green and red, respectively. The red difference density for Glu199 indicates decarboxylation, and the pair of positive and negative difference densities close to His440 in panel b indicates a conformational change. Panel c shows a difference Fourier map calculated between the last (E) data sets at 100 K and 155 K, FobsE (155 K) − FobsE (100 K), contoured at 4σ. Panel d shows a difference Fourier map calculated between the last (E) data set at 100 K and the third (C) data set at 155 K, FobsC (155 K) − FobsE (100 K), contoured at 4σ. The protein model as determined in data sets A at 100 K and at 155 K is shown in panels a and b, respectively, and the one determined in data set E at 100 K in panels c and d.
The objection might be raised that the second conformation of His440 would be solely a consequence of increased damage to Glu199 at 155 K, which is evident from a negative peak at Glu199 in a difference map between data sets E at both temperatures (Fig. 3c ▶). This is ruled out by a difference map between data set E at 100 K and data set C at 155 K, which shows a conformational change for His440 but no difference feature at Glu199 (Fig. 3d ▶). However, decarboxylation of Glu199 is likely to be a necessary condition for His440 to move at 155 K. X-ray crystallographic (Millard et al. 1999) and NMR studies (Massiah et al. 2001) of AChE have suggested that His440 can form alternate hydrogen bonds to Glu199 and Glu327. This suggests that His440 is a mobile residue, which is in line with our observations.
It should be emphasized that the difference features shown in Fig. 3b ▶ associated with the catalytic histidine, together with those representing radiation damage to the three disulfide bonds, to Cys231 and to Glu199, are by far the strongest peaks in the difference Fourier map of the entire TcAChE molecule. This shows that the active site of TcAChE is particularly radiation-sensitive. It has been suggested that radiation damage to the active-site histidine leads to inactivation of ribonuclease (Adams et al. 1972). Indeed, OH• radicals can transform histidine in proteins to aspartate (Dean et al. 1989) or 2-oxo-histidine (Uchida and Kawakishi 1993). In the present study, however, the electron density of His440 excludes the formation of either compound, and the difference density features represent a movement of His440.
Intense X-ray irradiation produces changes in the energy landscape of proteins via alterations such as decarboxylation of Glu199. Our results show that at 155 K, but not at 100 K, the active-site residues have sufficient flexibility to relax in order to adapt to the altered energy landscape. The active-site residues in TcAChE display substantial flexibility at a temperature well below the temperature range in which proteins undergo a global dynamical transition (180–230 K). This might either be evidence for a `local' dynamical transition, for protein flexibility triggered by the solvent glass transition of trigonal TcAChE crystals, or for differential global dynamical behavior of TcAChE with respect to other proteins.
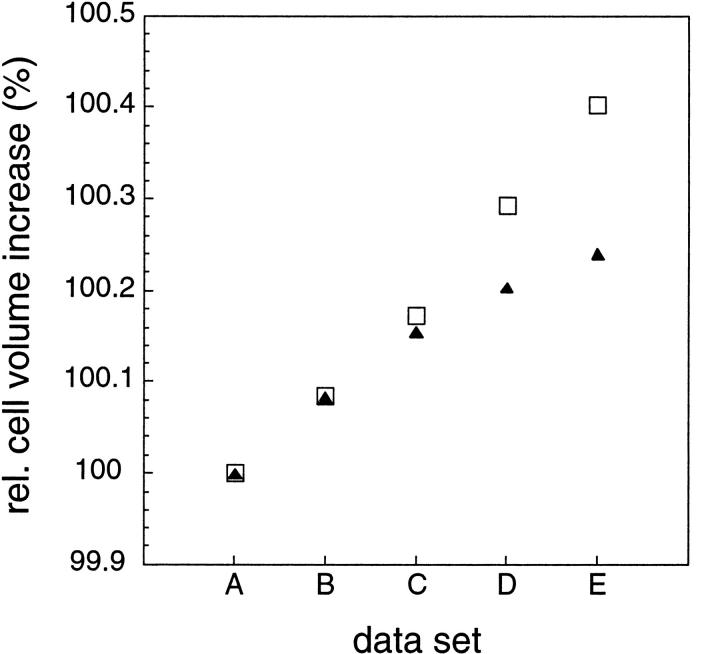
Unit cell volume increase
Irradiation of protein crystals with highly intense synchrotron radiation has been reported to result in an increase in unit cell dimensions (Yonath et al. 1998; Burmeister 2000; Ravelli and McSweeney 2000). The absolute unit cell volume in data set A of the present study is 0.8% higher at 155 K than at 100 K (Table 1), mainly due to thermal expansion of the crystal (Weik et al. 2001). Figure 4 ▶ shows the relative cell volume increases at 100 and 155 K. The increase in cell volume at constant temperature is due solely to X-ray irradiation, because no ice rings were detected in the diffraction patterns at either 100 or 155 K. The formation of ice rings in TcAChE crystals would have indicated crystallization of the solvent which would have increased the unit cell volume drastically (Weik et al. 2001). The fact that no ice formation was detected in the 155 K series is attributed to the fact that it occurs slowly at this temperature on the experimental time scale employed. The cell volume increase for data set E relative to data set A is larger at 100 K than at 155 K (Fig. 4 ▶). This shows that the radiation-induced increase in unit cell volume does not reflect the degree of specific damage. The mechanism(s) leading to the unit cell volume increase and the mechanism(s) leading to specific damage are, therefore, distinct. We suggest that the first is caused mainly by primary damage, whereas the latter is caused mainly by secondary damage. At 100 K, radicals created in the solvent area are mostly trapped, and their creation increases the unit cell volume linearly with dose. At 155 K, above the solvent glass transition, radicals are able to diffuse and recombine, which could explain the nonlinear unit cell volume increase (Fig. 4 ▶). This is in line with observations that OH• radicals produced at lower temperatures recombine upon raising the temperature to 130 K (Symons 1999).
Table 1.
Diffraction data statistics
| Data set | 100K (A) | 100K (B) | 100K (C) | 100K (D) | 100K (E) | 155K (A) | 155K (B) | 155K (C) | 155K (D) | 155K (E) |
| Resolution (Å) | 30–2.5 | 30–2.6 | 30–2.6 | 30–2.6 | 30–2.5 | 30–2.3 | 30–2.35 | 30–2.4 | 30–2.5 | 30–2.5 |
| No. unique reflections | 35414 | 31555 | 31563 | 31567 | 35416 | 45803 | 43005 | 40435 | 35832 | 35823 |
| Redundancy | 4.3 | 4.3 | 4.3 | 4.3 | 4.3 | 4.2 | 4.2 | 4.2 | 4.2 | 4.2 |
| <I>/<σ(I)>a | 14.5 (2.4) | 14.8 (2.6) | 13.7 (2.3) | 15.7 (2.9) | 18.2 (3.5) | 15.1 (2.3) | 14.9 (2.3) | 14.8 (2.4) | 15.2 (2.6) | 16.9 (3.4) |
| Completeness (%)a | 99.1 (99.4) | 99.1 (99.5) | 99.1 (99.4) | 99.1 (99.5) | 99.1 (99.3) | 99.6 (99.6) | 99.6 (99.8) | 99.6 (99.8) | 99.6 (99.7) | 99.5 (99.6) |
| Rmerge (%)b | 9.6 | 9.4 | 10.2 | 9.2 | 7.6 | 9.0 | 9.2 | 9.4 | 9.0 | 8.4 |
| Mosaicity (degrees) | 0.22 | 0.23 | 0.23 | 0.22 | 0.24 | 0.26 | 0.26 | 0.27 | 0.28 | 0.29 |
| Unit cell parameters | a = 112.18 | a = 112.21 | a = 112.25 | a = 112.30 | a = 112.33 | a = 112.51 | a = 112.53 | a = 112.55 | a = 112.56 | a = 112.57 |
| (Å) | c = 137.60 | c = 137.62 | c = 137.67 | c = 137.71 | c = 137.77 | c = 137.88 | c = 137.94 | c = 138.00 | c = 138.04 | c = 138.08 |
| Unit cell volume relative to A (%) | 100 | 100.08 | 100.17 | 100.29 | 100.40 | 100 | 100.08 | 100.16 | 100.20 | 100.24 |
a Numbers in parentheses indicate statistics for highest resolution shells.
b Rmerge = ∑hkl∑i|Ii(hkl)-<I(hkl)>|/∑hkl∑iIi(hkl).
Fig. 4.
Unit cell volume increase (relative to data sets A) at 100 K (open squares) and 155 K (closed triangles). No errors are given since they are difficult to assess. However, the characteristic unit cell volume increase displayed in this figure proved to be reproducible.
Conclusions
The aim of this study was to gain insight into the dynamical behavior of a protein near the glass transition of its surrounding solvent. At 100 K, the solvent in trigonal crystals of the enzyme TcAChE is amorphous and rigid, whereas it is liquid-like at 155 K after having undergone a glass transition. Temperature-dependent specific damage provided by intense X-irradiation was shown to be a marker for protein dynamics. At 155 K, the active site residue His440 undergoes a conformational change, and certain sulfur-containing groups show markedly increased radiation sensitivity compared to that at 100 K. This shows that at 155 K the protein has gained substantial local flexibility. We do not yet know whether this happened at a discrete temperature between 100 and 155 K or gradually, as experiments were done only at the two temperatures. TcAChE gains local flexibility, namely in the active site, at a temperature well below the global dynamical transitions of other proteins that mark the onset of large-amplitude motions such as side-chain movements and rotations and that have been reported to take place at 180–230 K (Parak et al. 1982; Doster et al. 1989; Ferrand et al. 1993). This might be explained by protein flexibility, local or global, triggered by the solvent glass transition in trigonal TcAChE crystals. Another possibility is that specific local flexibility onsets at lower temperatures than the global flexibility characterizing the entire protein, which would correspond in Frauenfelder's energy landscape to dynamical transitions in different tiers. It is also possible that TcAChE, by virtue of its high turnover number, displays global dynamical behavior different from that of other proteins investigated. The latter issue will be addressed by studying cholinesterases in neutron spectroscopy experiments, which have allowed mapping of the internal flexibility of macromolecules in relation to their function (Zaccai 2000). X-ray crystallographic experiments at various temperatures in solvents displaying different glass transition temperatures will provide further insight into the degree of coupling between protein and solvent dynamics.
Our results imply an important role for protein and solvent dynamics in chemical modifications of biological macromolecules such as specific radiation damage to proteins. At 155 K, certain sulfur-containing groups show greatly increased radiation sensitivity relative to that at 100 K. This can be explained either by increased mobility of radicals or by increased flexibility of the local environment of the radiation-sensitive bonds. In particular, the differential unit cell volume increase at 100 and 155 K points towards increased mobility of radicals above the solvent glass transition. At 155 K, the catalytic histidine undergoes a conformational change producing a nonfunctional active-site conformation. We observe that the most prominent radiation-induced alterations include the ones in the active site of TcAChE, which might hold true for enzymes in general. Temperature-dependent crystallographic radiation damage studies of other proteins crystallized under various conditions and as different crystal forms may provide further insight into the factors that control radiation sensitivity and stability of functional groups in proteins.
Materials and methods
Crystallization
Trigonal TcAChE crystals of space group P3121 were grown in 32–34% (v/v) polyethyleneglycol (PEG) 200 / 0.3 M 2-morpholinoethanesulfonic acid (MES), pH 5.8, at 4°C (Raves et al. 1997).
Data collection, processing, and structure refinement and analysis
Crystals were soaked for 24 h in 36% (v/v) PEG 200 / 0.3 M MES, pH 5.8, at 4°C and then transferred to a drop of mineral oil for about 1 min, mounted in a standard cryoloop and flash-cooled in liquid N2. Precooled crystals were mounted in the cryostream of a cooling device (600 series, Oxford Cryosystems, Oxford, UK) operating at 100 K on the microdiffractometer installed on beam line ID14–EH2 of the ESRF (Grenoble). Five successive data sets were collected on single crystals at 100 K and 155 K (reading of the cooling device). The beam, of wavelength 0.933 Å, was defined by a pinhole 50 μm in diameter, and was provided by the synchrotron operating in the 16-bunch filling mode. The crystal used for data collection at 155 K was first mounted at 100 K. The temperature was then raised to 155 K at 360 K/h. Care was taken to start both data collection series at similar storage ring intensities in order to minimize differences in total radiation dose per data collection series. Refill of the synchrotron storage ring took place during the collection of data set D in the 100 K series and between the collection of data sets D and E in the 155 K series. For each data set, 70 frames were collected, with an oscillation range of 1° and an exposure time of 5 sec per frame. Data collection took about 15 min per data set. Data sets were processed using DENZO and SCALEPACK (Otwinoski and Minor 1997), and the same free R set assigned using the CCP4 program suite (Collaborative Computational Project 1994). Molecular replacement, energy minimization, simulated annealing, and grouped (side-chain and main-chain atoms) thermal B-factor refinement were done using the program CNS (Brünger et al. 1998), taking the native TcAChE structure (PDB code ID 1EA5) as a starting model, after having omitted sugar and solvent molecules. All 6 cysteine residues taking part in intramolecular disulfide bonds in TcAChE were replaced by alanine residues in order to minimize model bias (For data collection and refinement statistics see Tables 1 and 2). All structures were refined in an identical, automated way. The programs molscript (Kraulis 1991), bobscript (Kraulis 1991; Esnouf 1999) and raster3d (Merritt and Bacon 1997) were used to produce figures from electron density maps calculated using cns. The solvent accessibility of atoms was calculated with the program surface (Collaborative Computational Project 1994), based on 1EA5 coordinates from which water and carbohydrate molecules had been omitted, and using a probe radius of 1.4 Å.
Table 2.
Refinement statistics
| Data set | 100K (A) | 100K (B) | 100K (C) | 100K (D) | 100K (E) | 155K (A) | 155K (B) | 155K (C) | 155K (D) | 155K (E) |
| R-factor (%) | 24.2 | 23.7 | 23.7 | 23.5 | 23.4 | 24.5 | 24.2 | 23.8 | 23.5 | 23.3 |
| Rfree (%) | 26.6 | 26.4 | 26.0 | 26.3 | 25.9 | 26.5 | 26.5 | 25.7 | 26.0 | 26.1 |
Dose calculation
A typical flux on beam line ID14–EH2 is 5 • 1012 photons s−1, through a collimator with a surface of 0.2 • 0.2 mm2, when the synchrotron is operating at a storage ring intensity of 200 mA (E. Mitchell, pers. comm.). Given the size of the collimator used, and an average storage ring intensity of 66 mA during our experiments, the beam intensity was estimated to be 4 • 1013 photons s−1 mm−2 at the sample position. We calculated the linear absorption coefficient to be 0.25 mm−1 at wavelength 0.95 Å, taking into account the contribution of the mother liquor. This led to a total absorption of 2.5% (assuming an average path length of 100 μm) and to an absorbed dose of about 4 • 106 Gy per data set based on the measured crystal size of 0.15 • 0.15 • 0.05 mm3 and a calculated crystal density of 1.133 g cm−3.
Temperature calibration and crystal heating
It is common in cryocrystallography to report the temperature as displayed on the cooling device. However, this temperature is measured in the gas stream within the nozzle, and might differ from that within the crystal during X-irradiation due both to a temperature gradient in the gas stream and to heating caused by X-ray absorption, especially if high-intensity synchrotron sources are employed. For the sake of coherence with earlier work, the cited temperatures, that is, 100 and 155 K, were the displayed ones. Nevertheless, we have attempted to estimate the real temperature at the crystal position during data collection. The temperature at about 10 mm from the nozzle is expected to be about 2 K higher than the one displayed (Mike Glazer, Oxford Cryosystems, pers. comm.). This figure was confirmed by monitoring characteristic changes in unit cell dimensions (Kobayashi et al. 1971) of ferroelectric crystals of potassium dihydrogen phosphate (KDP; Nelmes 1987) at its Curie temperature on a home source. The issue of crystal heating by the intense X-ray beams of third-generation synchrotron sources has recently been addressed theoretically (Kuzay et al. 2001; Nicholson et al. 2001). A 100 μm thick cryocooled (100 K) crystal in a beam of intensity 1013 photons s−1 mm−2 at 8 keV reaches a steady-state temperature 6 K above the ambient gas temperature after 4.5 s (Kuzay et al. 2001). This value can serve as an estimate for the heating of TcAChE crystals under our experimental conditions, since the higher beam intensity is compensated for by a lower total absorption, as compared to the test case calculated by Kuzay and coworkers. Nicholson and co-workers (2001) calculated that the temperature will rise under similar conditions by 4–6 K if nitrogen gas is used, depending on whether a cryoprotectant film is taken into account or not. If we assume the two corrections to be additive (i.e., neglecting crystal heating on a home source), the real temperature of the TcAChE crystals during X-irradiation under our experimental condition could be ∼8 K higher than the cited ones, that is, 108 and 163 K versus 100 and 155 K, still respectively well below and above the solvent glass transition. Experimental evidence that crystal heating did not exceed the calculated value more substantially was provided by the absence of ice formation in the data collection series at 155 K. Heating to temperatures well above 155 K would have resulted in solvent crystallization on the experimental time scale with concomitant formation of ice rings in the diffraction pattern.
Acknowledgments
We thank John Warman for a stimulating discussion and Giuseppe Zaccai and Chantal Houee-Levin for critical and constructive reading of the manuscript. Help from Ed Mitchell during data collection on beamline ID14–EH2 at the ESRF in Grenoble is gratefully acknowledged. We thank Lilly Toker for preparing the purified TcAChE and Toine Schreurs and Martin Lutz for conducting the temperature calibration experiments. M.W. was supported by an EMBO long-term fellowship. The study was supported by the U.S. Army Medical and Materiel Command under contract number DAMD17-97-2-7022, the EU 4th Framework Program in Biotechnology, and the Kimmelman Center for Biomolecular Structure and Assembly (Rehovot, Israel). I.S. is Bernstein-Mason Professor of Neurochemistry.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1101/ps.09801.
References
- Adams, G.E., Bisby, R.H., Cundall, R.B., Redpath, J.L., and Wilson, R.L. 1972. Selective free radical reactions with proteins and enzymes: The inactivation of ribonuclease. Radiat. Res. 49 290–299. [PubMed] [Google Scholar]
- Armstrong, D.A. 1990. Application of pulse radiolysis for the study of short-lived sulphur species. In Sulfur-centered reactive intermediates in chemistry and biology (eds. C. Chatgilialoglu and K.-D. Asmus), pp. 121–134. Plenum Press, New York.
- Audette, M., Chen, X., Houee-Levin, C., Potier, M., and Le Maire, M. 2000. Protein γ-radiolysis in frozen solution is a macromolecular surface phenomenon: Fragmentation of lysozyme, citrate synthase and α-lactalbumin in native or denatured states. Int. J. Radiat. Biol. 76 673–681. [DOI] [PubMed] [Google Scholar]
- Brünger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. 1998. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54 905–921. [DOI] [PubMed] [Google Scholar]
- Burmeister, W.P. 2000. Structural changes in a cryo-cooled protein crystal owing to radiation damage. Acta Crystallogr. D Biol. Crystallogr. 56 328–41. [DOI] [PubMed] [Google Scholar]
- Chen, K., Hirst, J., Camba, R., Bonagura, C.A., Stout, C.D., Burgess, B.K., and Armstrong, F.A. 2000. Atomically defined mechanism for proton transfer to a buried redox centre in a protein. Nature 405 814–817. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4. 1994. The CCP4 Suite: Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50 760–763. [DOI] [PubMed] [Google Scholar]
- Daniel, R.M., Smith, J.C., Ferrand, M., Hery, S., Dunn, R., and Finney, J. 1998. Enzyme activity below the dynamical transition at 220 K. Biophys. J. 75 2504–2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean, R.T., Wolff, S.P., and McElligott, M.A. 1989. Histidine and proline are important sites of free radical damage to proteins. Free Radic. Res. Commun. 7 97–103. [DOI] [PubMed] [Google Scholar]
- Dick, L.A., Malfant, I., Kuila, D., Nebolsky, S., Nocek, J.M., Hoffman, B.M., and Ratner, M.A. 1998. Cryogenic electron tunneling within mixed-metal hemoglobin hybrids: Protein glassing and electron-transfer energetics. J. Am. Chem. Soc. 120 11401–11407. [Google Scholar]
- Doster, W., Cusack, S., and Petry, W. 1989. Dynamical transition of myoglobin revealed by inelastic neutron scattering. Nature 337 754–756. [DOI] [PubMed] [Google Scholar]
- Esnouf, R.M. 1999. Further additions to MolScript version 1.4, including reading and contouring of electron-density maps. Acta Crystallogr. D Biol. Crystallogr. 55 938–940. [DOI] [PubMed] [Google Scholar]
- Favaudon, V., Tourbez, H., Houee-Levin, C., and Lhoste, J.M. 1990. CO2 - radical induced cleavage of disulfide bonds in proteins. A gamma-ray and pulse radiolysis mechanistic investigation. Biochemistry 29 10978–10989. [DOI] [PubMed] [Google Scholar]
- Ferrand, M., Dianoux, A.J., Petry, W., and Zaccai, G. 1993. Thermal motions and function of bacteriorhodopsin in purple membranes: Effects of temperature and hydration studied by neutron scattering. Proc. Natl. Acad. Sci. 90 9668–9672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filali-Mouhim, A., Audette, M., St-Louis, M., Thauvette, L., Denoroy, L., Penin, F., Chen, X., Rouleau, N., Le Caer, J.P., Rossier, J., et al. 1997. Lysozyme fragmentation induced by γ-radiolysis. Int. J. Radiat. Biol. 72 63–70. [DOI] [PubMed] [Google Scholar]
- Fisher, M. and Devlin, J.P. 1995. Defect activity in amorphous ice from isotopic exchange data: Insight into the glass transition. J. Phys. Chem. 99 11584–11590. [Google Scholar]
- Frauenfelder, H., Sligar, S.G., and Wolynes, P.G. 1991. The energy landscapes and motions of proteins. Science 254 1598–1603. [DOI] [PubMed] [Google Scholar]
- Garman, E. F. and Schneider, T.R. 1997. Macromolecular cryocrystallography. J. Appl. Cryst. 30 211–237. [Google Scholar]
- Johari, G.P., Hallbrucker, A., and Mayer, E. 1987. The glass-liquid transition of hyperquenched water. Nature 330 552–553. [DOI] [PubMed] [Google Scholar]
- Kobayashi, J., Uesu, Y., and Enomoto, Y. 1971. X-ray dilatometric study on ferroelectric phase transition of KH2PO4. Phys. Stat. Sol. 45 293–304. [Google Scholar]
- Kraulis, P. 1991. MOLSCRIPT: A program to produce both detailed and schematic plots of protein structure. J. Appl. Cryst. 24 946–950. [Google Scholar]
- Kuzay, T.M., Kazmierczak, M., and Hsieh, B.J. 2001. X-ray beam/biomaterial thermal interactions in third-generation synchrotron sources. Acta Crystallogr. D Biol. Crystallogr. 57 69–81. [DOI] [PubMed] [Google Scholar]
- Lmouméne, C.E., Conte, D., Jacquot, J.P., and Houee-Levin, C. 2000. Redox properties of protein disulfide bond in oxidized thioredoxin and lysozyme: A pulse radiolysis study. Biochemistry 39 9295–9301. [DOI] [PubMed] [Google Scholar]
- Massiah, M.A., Viragh, C., Reddy, P.M., Kovach, I.M., Johnson, J., Rosenberry, T.L., and Mildvan, A.S. 2001. Short, strong hydrogen bonds at the active site of human acetylcholinesterase: Proton NMR studies. Biochemistry 40 5682–5690. [DOI] [PubMed] [Google Scholar]
- McMillan, J.A. and Los, S.C. 1965. Vitreous ice: Irreversible transformations during warm-up. Nature 206 806–807. [Google Scholar]
- Merritt, E.A. and Bacon, D.J. 1997. Raster3D—photorealistic molecular graphics. Methods Enzymol. 277 505–524. [DOI] [PubMed] [Google Scholar]
- Millard, C.B., Koellner, G., Ordentlich, A., Shafferman, A., Silman, I., and Sussman, J.L. 1999. Reaction products of acetylcholinesterase and VX reveal a mobile histidine in the catalytic triad. J. Am. Chem. Soc. 121 9883–9884. [Google Scholar]
- Mishima, A. and Stanley, H.E. 1998. The relationship between liquid, supercooled and glassy water. Nature 396 329–335. [Google Scholar]
- Nelmes, R.J. 1987. Structural studies of KDP and the KDP-type transition by neutron and X-ray diffraction: 1970–1985. Ferroelectrics 71 87–123. [Google Scholar]
- Nicholson, J., Nave, C., Fayz, K., Fell, B., and Garman, E. 2001. Modelling heating effects in cryocooled protein crystals. Nuc. Instr. Meth. in press.
- Ostermann, A., Waschipky, R., Parak, F.G., and Nienhaus, G.U. 2000. Ligand binding and conformational motions in myoglobin. Nature 404 205–208. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. and Minor, W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276 307–326. [DOI] [PubMed] [Google Scholar]
- Parak, F., Knapp, E.W., and Kucheida, D. 1982. Protein dynamics. Mossbauer spectroscopy on deoxymyoglobin crystals. J. Mol. Biol. 161 177–194. [DOI] [PubMed] [Google Scholar]
- Rasmussen, B.F., Stock, A.M., Ringe, D., and Petsko, G.A. 1992. Crystalline ribonuclease A loses function below the dynamical transition at 220 K. Nature 357 423–424. [DOI] [PubMed] [Google Scholar]
- Ravelli, R.B. and Mcsweeney, S.M. 2000. The `fingerprint' that X-rays can leave on structures. Structure Fold. Des. 8 315–328. [DOI] [PubMed] [Google Scholar]
- Raves, M.L., Harel, M., Pang, Y.-P., Silman, I., Kozikowski, A.P., and Sussman, J.L. 1997. 3D structure of acetylcholinesterase complexed with the nootropic alkaloid (−)-huperzine. Nature Struct. Biol. 4 57–63. [DOI] [PubMed] [Google Scholar]
- Réat, V., Dunn, R., Ferrand, M., Finney, J.L., Daniel, R.M., and Smith, J.C. 2000. Solvent dependence of dynamic transitions in protein solutions. Proc. Natl. Acad. Sci. 97 9961–9966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, R.S., Huang, C., and Kay, B.D. 1997. Evidence for molecular translational diffusion during the crystallization of amorphous solid water. J. Phys. Chem. B 101 6123–6126. [Google Scholar]
- Sussman J.L., Harel M., Frolow F., Oefner C., Goldman A., Toker L., and Silman I. 1991. Atomic structure of acetylcholinesterase from Torpedo californica: A prototypic acetylcholine-binding protein. Science 253 872–879. [DOI] [PubMed] [Google Scholar]
- Symons, M.C.R. 1999. Mechanism of radiation damage to proteins and DNA – an EPR perspective. Progress in Reaction Kinetics and Mechanisms 24 139–164. [Google Scholar]
- Taub, I.A., Halliday, J.W., and Sevilla, M.D. 1979. Chemical reactions in proteins irradiated at subfreezing temperatures. Adv. Chem. Ser. 180 109–140. [Google Scholar]
- Uchida, K. and Kawakishi, S. 1993. 2-Oxo-histidine as a novel biological marker for oxidatively modified proteins. FEBS Lett. 332 208–210. [DOI] [PubMed] [Google Scholar]
- Vitkup, D., Ringe, D., Petsko, G.A., and Karplus, M. 2000. Solvent mobility and the protein `glass' transition. Nature Struct. Biol. 7 34–38. [DOI] [PubMed] [Google Scholar]
- von Sonntag, C. 1990. Free-radical reactions involving thiols and disulfides. In Sulfur-centered reactive intermediates in chemistry and biology (eds. C. Chatgilialoglu and K.-D. Asmus), pp. 359–366.Plenum Press, New York.
- Walser, R. and Van Gunsteren, W.F. 2001. Viscosity dependence of protein dynamics. Proteins 42 414–421. [PubMed] [Google Scholar]
- Weik, M., Kryger, G., Schreurs, A.M., Bouma, B., Silman, I., Sussman, J.L., Gros, P., and Kroon, J. 2001. Solvent behaviour in flash-cooled protein crystals at cryogenic temperatures. Acta Crystallogr. D Biol. Crystallogr. 57 566–573. [DOI] [PubMed] [Google Scholar]
- Weik, M., Ravelli, R.B., Kryger, G., Mcsweeney, S., Raves, M.L., Harel, M., Gros, P., Silman, I., Kroon, J., and Sussman, J.L. 2000. Specific chemical and structural damage to proteins produced by synchrotron radiation. Proc. Natl. Acad. Sci. 97 623–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler, J.R. 2000. Electron tunneling pathways in proteins. Curr. Opin. Chem. Biol. 4 192–198. [DOI] [PubMed] [Google Scholar]
- Yonath, A., Harms, J., Hansen, H.A., Bashan, A., Schlunzen, F., Levin, I., Koelln, I., Tocilj, A., Agmon, I., Peretz, M., et al. 1998. Crystallographic studies on the ribosome, a large macromolecular assembly exhibiting severe nonisomorphism, extreme beam sensitivity and no internal symmetry. Acta Crystollgr. A 54 945–955. [DOI] [PubMed] [Google Scholar]
- Zaccai, G. 2000. How soft is a protein? A protein dynamics force constant measured by neutron scattering. Science 288 1604–1607. [DOI] [PubMed] [Google Scholar]