Abstract
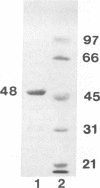
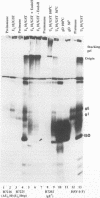
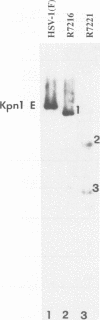
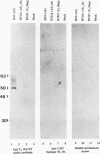
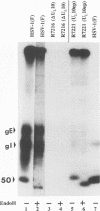
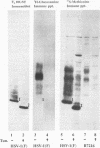
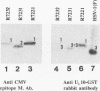
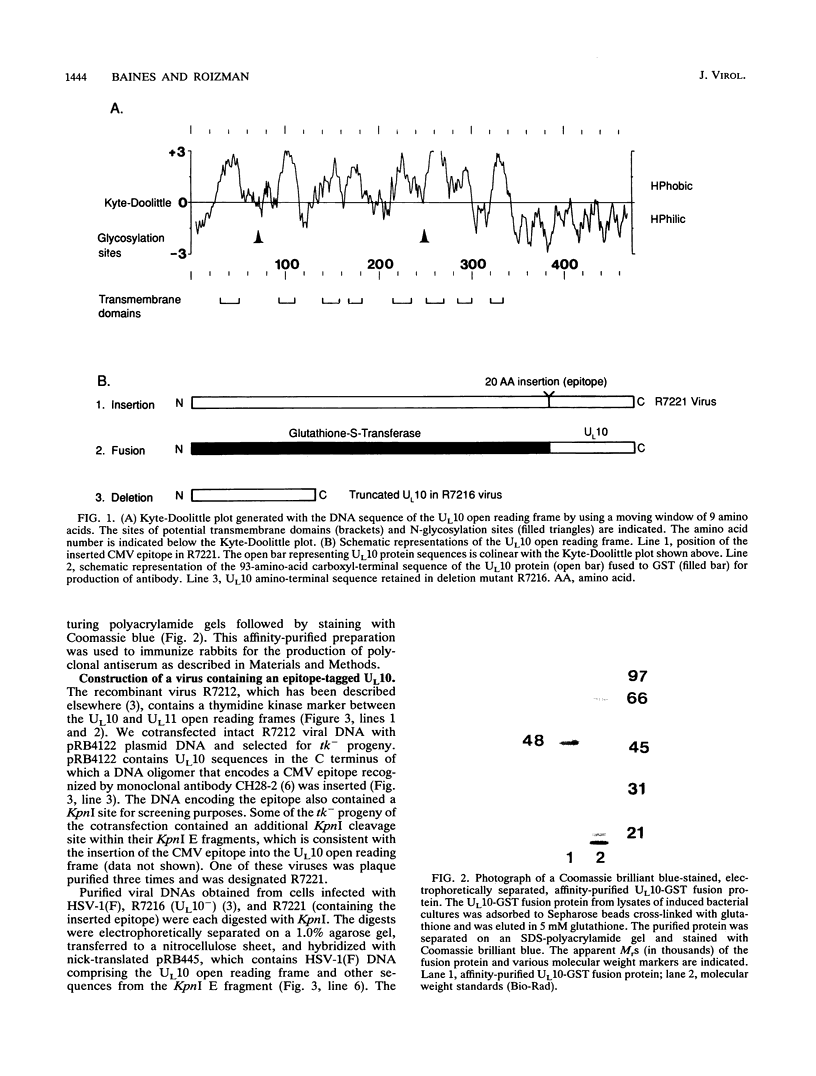
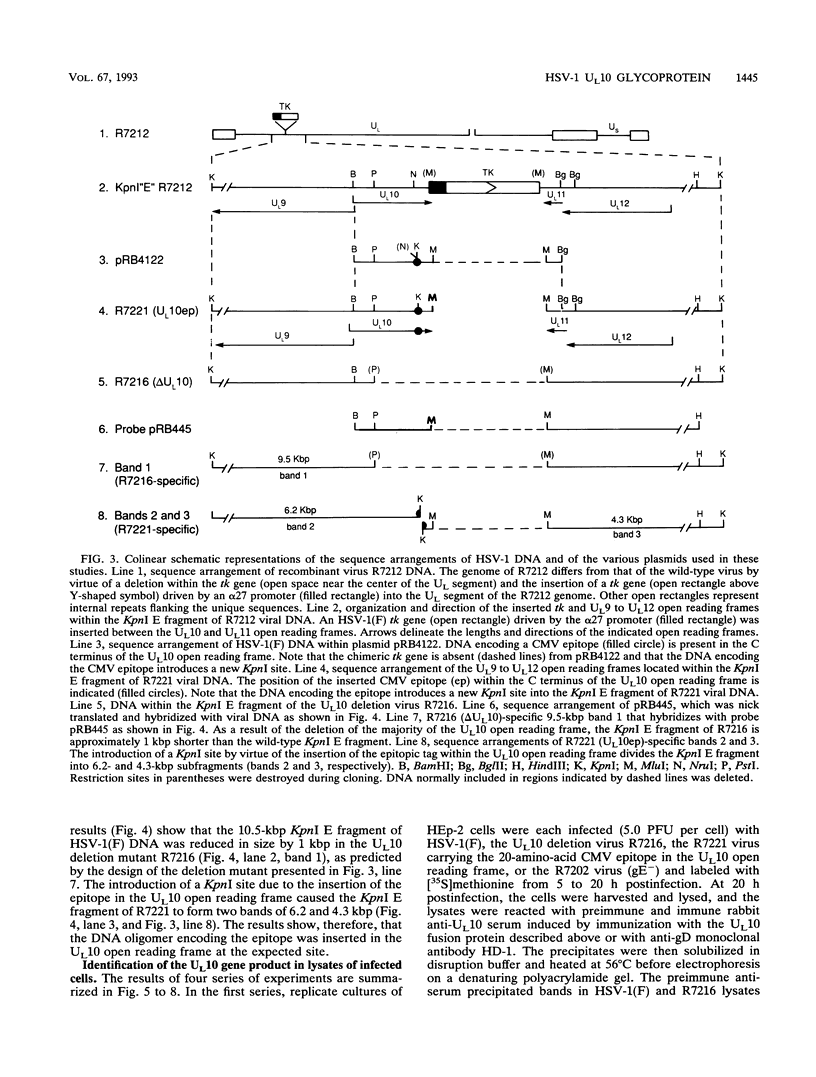
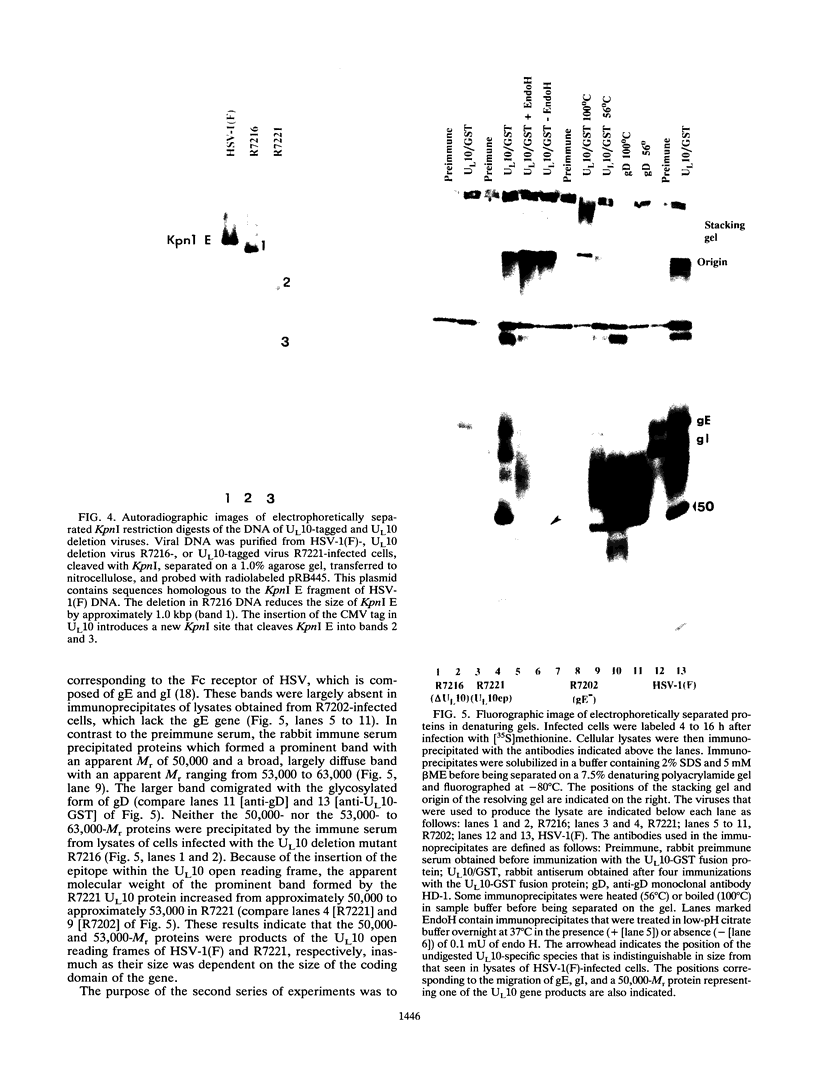
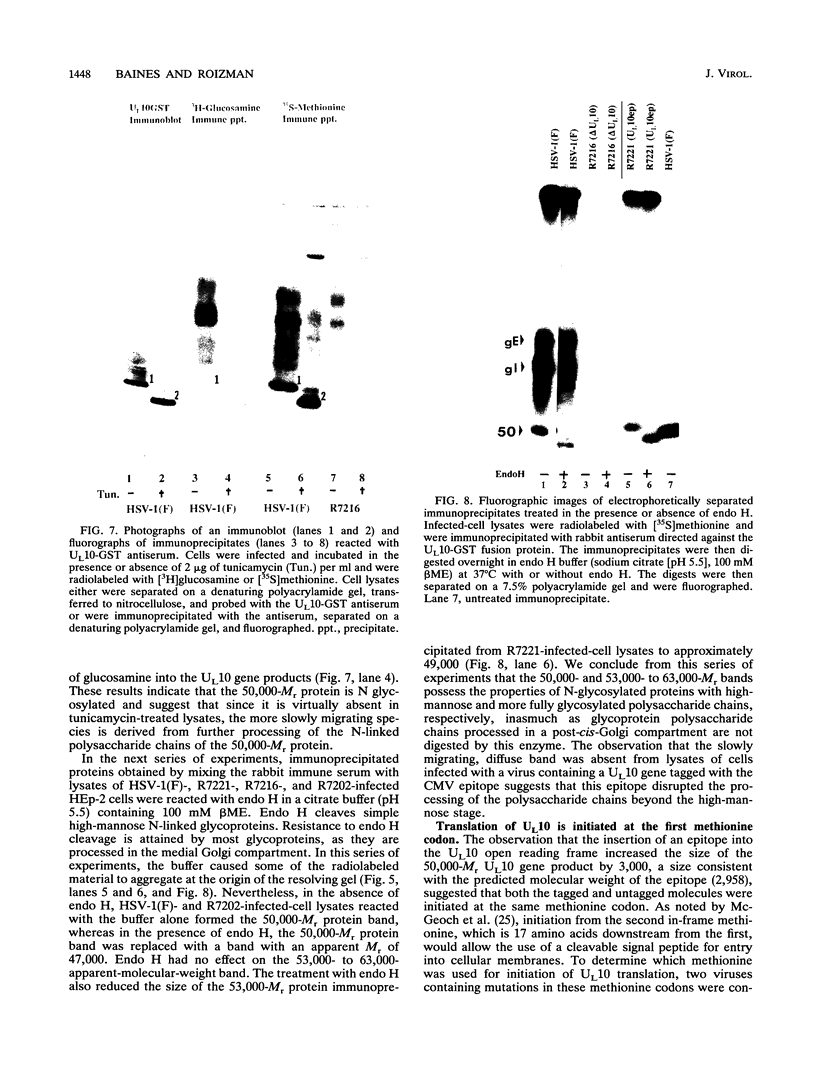
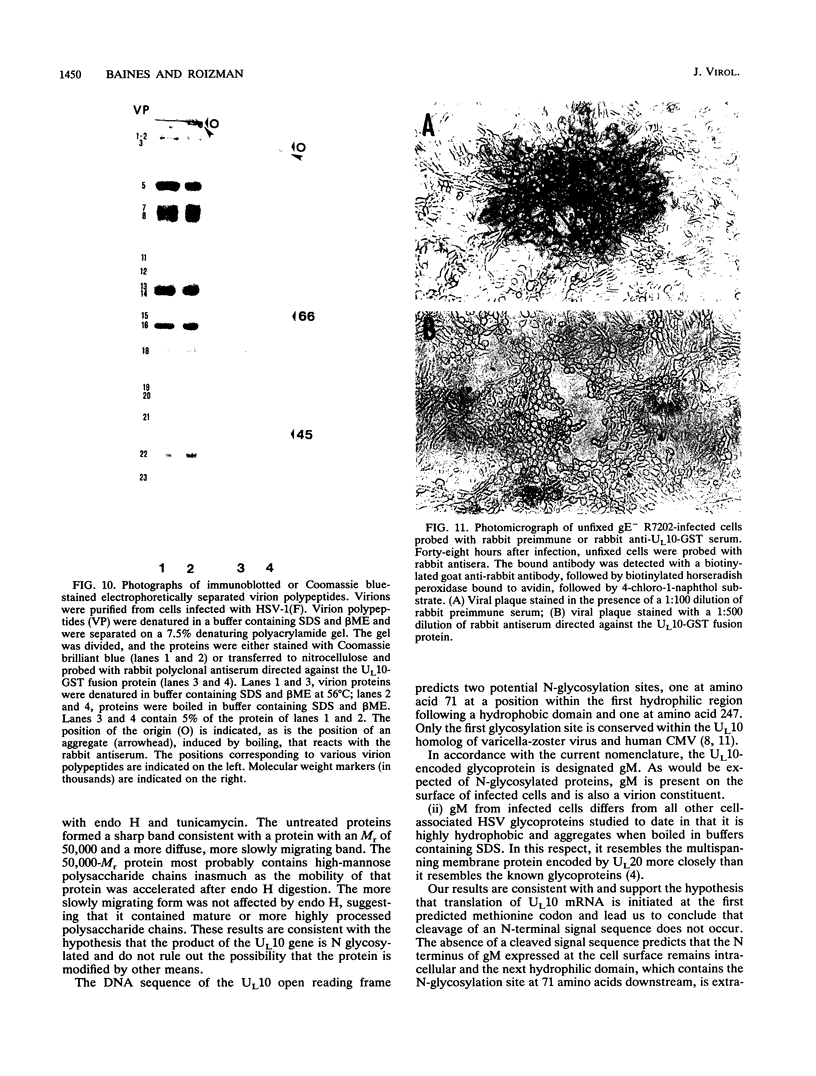
The herpes simplex virus 1 UL10 gene encodes a hydrophobic membrane protein dispensable for viral replication in cell culture (J.D. Baines and B. Roizman, J. Virol. 65:938-944, 1991). We report the following. (i) A fusion protein consisting of glutathione S-transferase fused to the C-terminal 93 amino acids of the UL10 protein was used to produce a rabbit polyclonal antiserum. The antiserum reacted with infected-cell proteins which formed in denaturing polyacrylamide gels a sharp band (apparent M(r) of 50,000) and a very broad band (M(r) of 53,000 to 63,000). These bands were not formed by lysates of UL10- virus or by lysates of infected cells boiled in the presence of sodium dodecyl sulfate before electrophoresis. (ii) The proteins forming both bands were labeled by [3H]glucosamine, indicating that they were glycosylated. (iii) The UL10 protein in cells treated with tunicamycin formed a single band (apparent M(r) of 47,000) reactive with the anti-UL10 antibody, indicating that the 47,000-M(r) protein was a precursor of N-glycosylated, more slowly migrating forms of UL10. Treatment of the immunoprecipitate with endoglycosidase H increased the electrophoretic mobility of the 50,000-M(r) species to that of the 47,000-M(r) species, indicating that the 50,000-M(r) species contained high-mannose polysaccharide chains, whereas the proteins forming the 53,000- to 63,000-M(r) bands contained mature chains inasmuch as they were resistant to digestion by the enzyme. (iv) The UL10 protein of R7221 carrying a 20-amino-acid epitope formed only one band with an M(r) of 53,000. This band was sensitive to endoglycosidase H, suggesting that the epitope inserted in the R7221 UL10 protein may have interfered with glycosylation. (v) The UL10 protein does not contain a cleavable signal sequence inasmuch as the first UL10 methionine codon was reflected in the 50,000-M(r) protein. (vi) The UL10 protein is present in virions and plasma membranes of unfixed cells that were reacted with the polyclonal rabbit antibody. In accordance with the current nomenclature, the UL10 protein is designated glycoprotein M.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ackermann M., Chou J., Sarmiento M., Lerner R. A., Roizman B. Identification by antibody to a synthetic peptide of a protein specified by a diploid gene located in the terminal repeats of the L component of herpes simplex virus genome. J Virol. 1986 Jun;58(3):843–850. doi: 10.1128/jvi.58.3.843-850.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackermann M., Longnecker R., Roizman B., Pereira L. Identification, properties, and gene location of a novel glycoprotein specified by herpes simplex virus 1. Virology. 1986 Apr 15;150(1):207–220. doi: 10.1016/0042-6822(86)90280-1. [DOI] [PubMed] [Google Scholar]
- Baines J. D., Roizman B. The open reading frames UL3, UL4, UL10, and UL16 are dispensable for the replication of herpes simplex virus 1 in cell culture. J Virol. 1991 Feb;65(2):938–944. doi: 10.1128/jvi.65.2.938-944.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baines J. D., Ward P. L., Campadelli-Fiume G., Roizman B. The UL20 gene of herpes simplex virus 1 encodes a function necessary for viral egress. J Virol. 1991 Dec;65(12):6414–6424. doi: 10.1128/jvi.65.12.6414-6424.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker D. E., Roizman B. The unique sequence of the herpes simplex virus 1 L component contains an additional translated open reading frame designated UL49.5. J Virol. 1992 Jan;66(1):562–566. doi: 10.1128/jvi.66.1.562-566.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basgoz N., Qadri I., Navarro D., Sears A., Lennette E., Youngblom J., Pereira L. The amino terminus of human cytomegalovirus glycoprotein B contains epitopes that vary among strains. J Gen Virol. 1992 Apr;73(Pt 4):983–988. doi: 10.1099/0022-1317-73-4-983. [DOI] [PubMed] [Google Scholar]
- Buckmaster E. A., Gompels U., Minson A. Characterisation and physical mapping of an HSV-1 glycoprotein of approximately 115 X 10(3) molecular weight. Virology. 1984 Dec;139(2):408–413. doi: 10.1016/0042-6822(84)90387-8. [DOI] [PubMed] [Google Scholar]
- Chee M. S., Bankier A. T., Beck S., Bohni R., Brown C. M., Cerny R., Horsnell T., Hutchison C. A., 3rd, Kouzarides T., Martignetti J. A. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr Top Microbiol Immunol. 1990;154:125–169. doi: 10.1007/978-3-642-74980-3_6. [DOI] [PubMed] [Google Scholar]
- Costa R. H., Devi B. G., Anderson K. P., Gaylord B. H., Wagner E. K. Characterization of a major late herpes simplex virus type 1 mRNA. J Virol. 1981 May;38(2):483–496. doi: 10.1128/jvi.38.2.483-496.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa R. H., Draper K. G., Kelly T. J., Wagner E. K. An unusual spliced herpes simplex virus type 1 transcript with sequence homology to Epstein-Barr virus DNA. J Virol. 1985 May;54(2):317–328. doi: 10.1128/jvi.54.2.317-328.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison A. J., Scott J. E. The complete DNA sequence of varicella-zoster virus. J Gen Virol. 1986 Sep;67(Pt 9):1759–1816. doi: 10.1099/0022-1317-67-9-1759. [DOI] [PubMed] [Google Scholar]
- Ejercito P. M., Kieff E. D., Roizman B. Characterization of herpes simplex virus strains differing in their effects on social behaviour of infected cells. J Gen Virol. 1968 May;2(3):357–364. doi: 10.1099/0022-1317-2-3-357. [DOI] [PubMed] [Google Scholar]
- Hayward G. S., Jacob R. J., Wadsworth S. C., Roizman B. Anatomy of herpes simplex virus DNA: evidence for four populations of molecules that differ in the relative orientations of their long and short components. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4243–4247. doi: 10.1073/pnas.72.11.4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heine J. W., Spear P. G., Roizman B. Proteins specified by herpes simplex virus. VI. Viral proteins in the plasma membrane. J Virol. 1972 Mar;9(3):431–439. doi: 10.1128/jvi.9.3.431-439.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson L., Browne H., Wargent V., Davis-Poynter N., Primorac S., Goldsmith K., Minson A. C., Johnson D. C. A novel herpes simplex virus glycoprotein, gL, forms a complex with glycoprotein H (gH) and affects normal folding and surface expression of gH. J Virol. 1992 Apr;66(4):2240–2250. doi: 10.1128/jvi.66.4.2240-2250.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson L., Goldsmith K., Snoddy D., Ghosh H., Graham F. L., Johnson D. C. Identification and characterization of a novel herpes simplex virus glycoprotein, gK, involved in cell fusion. J Virol. 1992 Sep;66(9):5603–5609. doi: 10.1128/jvi.66.9.5603-5609.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan L. Y., Jan Y. N. Tracing the roots of ion channels. Cell. 1992 May 29;69(5):715–718. doi: 10.1016/0092-8674(92)90280-p. [DOI] [PubMed] [Google Scholar]
- Johnson D. C., Frame M. C., Ligas M. W., Cross A. M., Stow N. D. Herpes simplex virus immunoglobulin G Fc receptor activity depends on a complex of two viral glycoproteins, gE and gI. J Virol. 1988 Apr;62(4):1347–1354. doi: 10.1128/jvi.62.4.1347-1354.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kousoulas K. G., Pellett P. E., Pereira L., Roizman B. Mutations affecting conformation or sequence of neutralizing epitopes identified by reactivity of viable plaques segregate from syn and ts domains of HSV-1(F) gB gene. Virology. 1984 Jun;135(2):379–394. doi: 10.1016/0042-6822(84)90194-6. [DOI] [PubMed] [Google Scholar]
- Lehner R., Meyer H., Mach M. Identification and characterization of a human cytomegalovirus gene coding for a membrane protein that is conserved among human herpesviruses. J Virol. 1989 Sep;63(9):3792–3800. doi: 10.1128/jvi.63.9.3792-3800.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F. Y., Roizman B. The promoter, transcriptional unit, and coding sequence of herpes simplex virus 1 family 35 proteins are contained within and in frame with the UL26 open reading frame. J Virol. 1991 Jan;65(1):206–212. doi: 10.1128/jvi.65.1.206-212.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longnecker R., Chatterjee S., Whitley R. J., Roizman B. Identification of a herpes simplex virus 1 glycoprotein gene within a gene cluster dispensable for growth in cell culture. Proc Natl Acad Sci U S A. 1987 Jun;84(12):4303–4307. doi: 10.1073/pnas.84.12.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longnecker R., Roizman B. Generation of an inverting herpes simplex virus 1 mutant lacking the L-S junction a sequences, an origin of DNA synthesis, and several genes including those specifying glycoprotein E and the alpha 47 gene. J Virol. 1986 May;58(2):583–591. doi: 10.1128/jvi.58.2.583-591.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean C. A., Efstathiou S., Elliott M. L., Jamieson F. E., McGeoch D. J. Investigation of herpes simplex virus type 1 genes encoding multiply inserted membrane proteins. J Gen Virol. 1991 Apr;72(Pt 4):897–906. doi: 10.1099/0022-1317-72-4-897. [DOI] [PubMed] [Google Scholar]
- McGeoch D. J., Dalrymple M. A., Davison A. J., Dolan A., Frame M. C., McNab D., Perry L. J., Scott J. E., Taylor P. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J Gen Virol. 1988 Jul;69(Pt 7):1531–1574. doi: 10.1099/0022-1317-69-7-1531. [DOI] [PubMed] [Google Scholar]
- McGeoch D. J., Dolan A., Donald S., Brauer D. H. Complete DNA sequence of the short repeat region in the genome of herpes simplex virus type 1. Nucleic Acids Res. 1986 Feb 25;14(4):1727–1745. doi: 10.1093/nar/14.4.1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeoch D. J., Dolan A., Donald S., Rixon F. J. Sequence determination and genetic content of the short unique region in the genome of herpes simplex virus type 1. J Mol Biol. 1985 Jan 5;181(1):1–13. doi: 10.1016/0022-2836(85)90320-1. [DOI] [PubMed] [Google Scholar]
- Para M. F., Zezulak K. M., Conley A. J., Weinberger M., Snitzer K., Spear P. G. Use of monoclonal antibodies against two 75,000-molecular-weight glycoproteins specified by herpes simplex virus type 2 in glycoprotein identification and gene mapping. J Virol. 1983 Mar;45(3):1223–1227. doi: 10.1128/jvi.45.3.1223-1227.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post L. E., Roizman B. A generalized technique for deletion of specific genes in large genomes: alpha gene 22 of herpes simplex virus 1 is not essential for growth. Cell. 1981 Jul;25(1):227–232. doi: 10.1016/0092-8674(81)90247-6. [DOI] [PubMed] [Google Scholar]
- Purves F. C., Spector D., Roizman B. UL34, the target of the herpes simplex virus U(S)3 protein kinase, is a membrane protein which in its unphosphorylated state associates with novel phosphoproteins. J Virol. 1992 Jul;66(7):4295–4303. doi: 10.1128/jvi.66.7.4295-4303.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza J. C., Hardwick K. G., Dean N., Pelham H. R. ERD2, a yeast gene required for the receptor-mediated retrieval of luminal ER proteins from the secretory pathway. Cell. 1990 Jun 29;61(7):1349–1357. doi: 10.1016/0092-8674(90)90698-e. [DOI] [PubMed] [Google Scholar]
- Sheldrick P., Berthelot N. Inverted repetitions in the chromosome of herpes simplex virus. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 2):667–678. doi: 10.1101/sqb.1974.039.01.080. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Spear P. G. Membrane proteins specified by herpes simplex viruses. I. Identification of four glycoprotein precursors and their products in type 1-infected cells. J Virol. 1976 Mar;17(3):991–1008. doi: 10.1128/jvi.17.3.991-1008.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spear P. G., Roizman B. Proteins specified by herpes simplex virus. V. Purification and structural proteins of the herpesvirion. J Virol. 1972 Jan;9(1):143–159. doi: 10.1128/jvi.9.1.143-159.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tognon M., Cassai E., Rotola A., Roizman B. The heterogenous regions in herpes simplex virus 1 DNA. Microbiologica. 1983 Jul;6(3):191–198. [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Wadsworth S., Jacob R. J., Roizman B. Anatomy of herpes simplex virus DNA. II. Size, composition, and arrangement of inverted terminal repetitions. J Virol. 1975 Jun;15(6):1487–1497. doi: 10.1128/jvi.15.6.1487-1497.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walboomers J. M., Schegget J. T. A new method for the isolation of herpes simplex virus type 2 DNA. Virology. 1976 Oct 1;74(1):256–258. doi: 10.1016/0042-6822(76)90151-3. [DOI] [PubMed] [Google Scholar]