Abstract
Microtubule spindle assembly in mitosis is stimulated by Ran·GTP, which is generated along condensed chromosomes by the guanine nucleotide exchange factor (GEF) RCC1. This relationship suggests that similar activities might modulate other microtubule structures. Interphase microtubules usually extend from the centrosome, although noncentrosomal microtubules function in some differentiated cells, including megakaryocytes. In these cells, platelet biogenesis requires massive mobilization of microtubules in the cell periphery, where they form proplatelets, the immediate precursors of platelets, in the apparent absence of centrioles. Here we identify a cytoplasmic Ran-binding protein, RanBP10, as a factor that binds β-tubulin and associates with megakaryocyte microtubules. Unexpectedly, RanBP10 harbors GEF activity toward Ran. A point mutation in the candidate GEF domain abolishes exchange activity, and our results implicate RanBP10 as a localized cytoplasmic Ran-GEF. RNA interference-mediated loss of RanBP10 in cultured megakaryocytes disrupts microtubule organization. These results lead us to propose that spatiotemporally restricted generation of cytoplasmic Ran·GTP may influence organization of the specialized microtubules required in thrombopoiesis and that RanBP10 might serve as a molecular link between Ran and noncentrosomal microtubules.
Microtubules (MTs)3 are essential components of the cytoskeleton in all cells, although specific fiber types and nucleation mechanisms differ according to cell lineage and over the cell cycle. Interphase cells typically contain a single MT-organizing center (MTOC) located within the centrosome. This structure contains two perpendicularly oriented centrioles and the γ-tubulin ring complex, which initiates assembly of radial MT arrays (1, 2). Centrioles duplicate during S phase and subsequently migrate to opposite cell poles, where they anchor the mitotic spindle and enable chromosome segregation (3-5). After cytokinesis, the centrioles serve again as origins for interphase MTs, which are relatively stable structures. In contrast, the considerably more dynamic MTs in the mitotic spindle are assembled through the activity of the small GTPase Ran (6-9).
Ran controls at least three essential functions at different stages in the cell cycle: nucleocytoplasmic traffic in interphase, spindle formation at mitosis, and nuclear envelope assembly during telophase (10). Each function requires GTP hydrolysis and exchange of GDP for GTP. Small GTPases have low intrinsic rates of nucleotide exchange, which are enhanced over 105-fold by guanine nucleotide exchange factors (GEFs), a diverse group of enzymes that show limited sequence similarity and considerable target protein specificity (11-14). RCC1, the only known Ran-GEF, is a chromatin-associated nuclear protein that activates Ran by loading GTP (15). Conversely, the cytoplasmic Ran-GTPase-activating protein (GAP) RanGAP1 acts to produce Ran·GDP (16), and the resulting interphase Ran·GTP concentration gradient drives macromolecule transport across the nuclear membrane (17). With nuclear envelope breakdown at mitosis, chromatin-associated RCC1 generates high local Ran·GTP levels, which promote MT assembly on condensed chromosomes through distinct effects on MT nucleation, dynamics, and motor activities (18, 19). Closely related mechanisms control macromolecule transport and mitotic spindle assembly, as Ran·GTP helps release both the cargo in nuclear import and spindle assembly factors in mitosis from the importin proteins α and β (20-22). The Ran·GTP gradients required to achieve these diverse functions have been visualized in Xenopus egg extracts (23). In somatic cells, these gradients rely on spatially segregated GEF and GAP activities, each of which is thought to be provided by one known enzyme. Here we report unexpected Ran-GEF activity in a novel cytoplasmic MT- and Ran-binding protein, RanBP10. This factor is expressed to high levels in megakaryocytes (MKs), the blood cell lineage in which we identified RanBP10 and studied its functions.
MKs are large polyploid cells that conclude their maturation by assembling nascent blood platelets within MT-based cytoplasmic extensions known as proplatelets (24, 25). These structures extend a considerable distance from the cell body and generate nascent platelets at their tips, where single MT filaments coil repeatedly to generate the platelet marginal band (24). The elaboration, growth, stability, and dynamic quality of proplatelets are strictly dependent on MTs and require de novo nucleation of MT filaments. However, centrioles are conspicuously absent from MKs during thrombopoiesis (25-27), and mechanisms of MT nucleation in terminally differentiated MKs are unclear. We show that a loss of RanBP10 disrupts MT organization, and its GEF activity provides a compelling basis to understand how the atypical MT structure of terminally differentiated MKs may be regulated. Our results also provide a novel link between Ran function and MTs other than those in the mitotic spindle.
EXPERIMENTAL PROCEDURES
Yeast Two-hybrid Assays—cDNA corresponding to the terminal 131 amino acids of mouse β1-tubulin (XP_283812.2) or 124 C-terminal amino acids of β5-tubulin (NP_035785) was cloned into pGBKT7 (Matchmaker System; Clontech) and introduced into the AH109 yeast strain; clones were selected by growth on Trp- medium, and protein expression was verified by immunoblotting. A cDNA library of 5 × 105 independent clones from cultured mouse MKs was created in the pGADT7 prey vector and introduced into β1-tubulin-expressing yeast, followed by selection on Trp-Leu-His- medium. 5 × 106 transformants yielded 110 clones that activated three independent reporter genes (histidine, β-galactosidase, and adenine at increasing levels of stringency). Three independent clones corresponded to a sequence represented in GenBank™ (BC024698), which was recently designated RanBP10 (NM_145824). Mouse RanBP10 constructs were derived from Image clone 3667000. All of the constructs created to map binding domains were generated by gap repair in yeast. Bait constructs to map interaction with Ran were generously provided by Iain Mattaj (EMBL, Heidelberg).
Co-immunoprecipitation, Immunoblotting, and Northern Analysis—293 cells were transfected with hemagglutinin (HA)-tagged RanBP10 plasmid, and cell lysates were precleared with protein G beads (Amersham Biosciences). Anti-HA Ab (12CA5; Roche Applied Science) was added for 2 h, and complexes captured using protein G beads were resolved by SDS-PAGE under nonreducing conditions. Immunoblot membranes were probed separately using β-tubulin (2-28-33; Sigma), actin, H-Ras, RhoA, RhoB (Santa Cruz Biotechnology), Ran (clone 20; BD Biosciences), or RanBP10 Ab. Platelet or mononuclear cell lysates were probed with Ran or GAPDH (Abcam) Ab. Recombinant His6-tagged Ran was purchased from Sigma-Aldrich; RanT24N and RanQ69L were prepared as described (28). To generate RanBP10 antisera, we immunized rabbits with the peptide SNGVASTKNKQNHSK and then purified the serum over affinity columns prepared with the peptide immunogen. Preimmune serum demonstrated no reactivity. For RNA expression analysis, a multiple-tissue Northern blot (Clontech) or ones we prepared with adult mouse bone marrow and other tissues were probed with a radiolabeled 1.6-kb fragment corresponding to the 3′-untranslated region of RanBP10.
Microtubule Association Assay—Fibroblasts were washed with PEM buffer (100 mm PIPES, 10 mm EGTA, 2 mm MgCl2, pH 7) prior to extraction with 0.5 mg/ml saponin in PEM buffer. The cells were washed twice in excess PEM buffer, and the proteins were solubilized in sample buffer before gel electrophoresis.
Cell Culture and MK Viral Infections—Fetal liver cells were cultured with thrombopoietin as described (29), and MKs were enriched over a discontinuous gradient of bovine serum albumin, which favors sedimentation of mature MKs based on cell density. Retroviruses were produced as described (30), using the pWZL expression vector to express EGFP-tagged β1-tubulin and RanBP10 as fusion proteins. Short hairpin (sh)RNAs directed against RanBP10 were introduced into MKs by infection with lentiviruses that also encoded EGFP. Two shRNAs directed against RanBP10, 1982 (target sequence AAGCCATGGGCAGAGTTCACT starting at position 1982 in Gen-Bank™ entry EU281316; supplemental Fig. S1) and 4971 (target AAACTGGCATCCTCCACAGTGT), were cloned in the pFCG vector. Only 1982 could deplete RanBP10; 4971 and empty virus served as negative controls. The cells were infected overnight with retroviruses encoding EGFP-fused β1-tubulin, RanBP10, or RanQ69L or with shRNA lentiviruses and cultured further until MKs were suitably mature. Green fluorescent cells were examined individually by fluorescence microscopy.
Immunofluorescence (IF)—The cells were cytocentrifuged onto coverslips, dried, fixed in 4% formaldehyde, permeabilized with 0.5% Triton X-100, and blocked with 1% goat serum. β-Tubulin, β1-tubulin, γ-tubulin (clone GTU-88; Sigma), Ran, RanGAP (Santa Cruz Biotechnology), or RanBP10 Ab were incubated for 30 min at room temperature, followed by fluorophore-conjugated secondary Ab for 30 min. Preimmune sera or isotype controls routinely yielded no signals. The cells were counterstained with Alexa Fluor 594-conjugated phalloidin (Molecular Probes, Eugene, OR) or DAPI to reveal the cytoplasm and nucleus, respectively. The coverslips were washed, mounted in Fluormount-G (Southern Biotech), and examined on the Olympus IX70 inverted fluorescence microscope. The images were acquired with a CM350 CCD camera (Applied Precision, Issaquah, WA) using a 60× (Olympus PLAN-APO 1.40 NA, 0.10 mm WD) or 100× (PLAN-APO 1.40NA, 0.10 mm WD) oil objective. 40-60 cross-sections were taken at 0.2-μm spacing, and the images were deconvolved using Delta-Vision software (Applied Precision, Issaquah, WA).
Sequence Comparisons—Alignments were performed using the Smart v4.0 data base and refined using ExPASy with a BLO-SUM 30 matrix, with gap open and gap extension penalties of 9 and 1.5, respectively. The E value threshold to prevent false positive results was set to 1, and the alignment between a Rho-GEF consensus sequence and RanBP10 returned an E-value of 0.025. Phylogenetic similarity analysis of GEF domains was performed using ClustalW tools.
Nucleotide Exchange Assay—Bacterially expressed glutathione S-transferase (GST)-fused RanBP10 and truncated forms lacking 95 or 159 N-terminal residues or point mutants were affinity-purified using glutathione beads (Amersham Biosciences). RanBP10 L301I mutant was generated using a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). Protein was eluted using excess glutathione and purified by fast protein liquid chromatography-mediated gel filtration (Superdex200 [10/300], Amersham Biosciences) in reaction buffer (30 mm potassium phosphate buffer containing 5 mm MgCl2 and 1 mm β-mercaptoethanol) (28); the isolated protein appeared as a single band on Coomassie Blue-stained gels. Aliquots resulting in a final concentration of 5 nm were added to recombinant Ran (Calbiochem; 1 μm), Rho1 (a gift of Michael Eck, Dana-Farber Cancer Institute, Boston, MA), or H-Ras (Calbiochem) in reaction buffer in the presence of a molar excess of mant-GDP or mant-GTP (Molecular Probes); the proteins were loaded with GDP or GTP (31) prior to purification by gel filtration chromatography. Nucleotide exchange was monitored at ambient temperature as fluorescence resonance energy transfer at 335 nm. GST and recombinant RCC1 (Calbiochem) were used as additional negative and positive controls, respectively. To calculate turnover rates and specific activities (1 unit is defined as the exchange of 1 nmol of Ran·GDP/min), we regarded dissociation rates to follow first order kinetics, as established previously (28) and substantiated in our experiments.
RESULTS
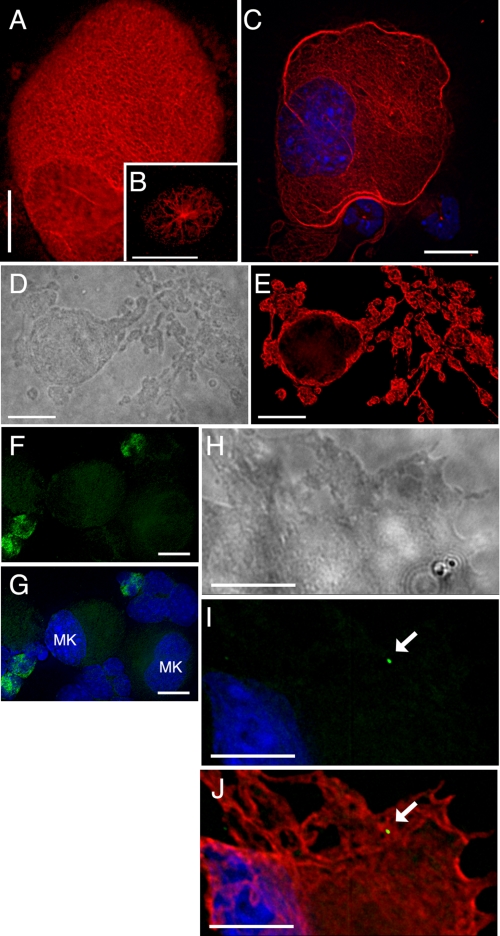
MT Modulation during MK Differentiation—Most interphase cells, including young MKs (Fig. 1B), carry a MTOC in the centrosome. As MKs mature, they increase significantly in size (routinely achieving >50 μm in diameter), become polyploid, and exhibit large, multi-lobed nuclei. By this time, astral MT arrays are replaced by characteristic complexes of interdigitating MT filaments organized in a reticular network (Fig. 1A). As the cells approach terminal differentiation and prepare to elaborate proplatelets, thick MT bundles accumulate in the cortex (Fig. 1C). Finally, new MT arrangements drive the multitude of proplatelets that are extended in the course of platelet assembly and release; this ordered sequence of events is illustrated with respect to MT morphologies in Fig. 1 (A-E). Centrioles have been difficult to detect in mature MKs (25, 27), which suggests atypical modes of MT assembly. To verify this premise, we tested for γ-tubulin in MKs derived from cultured fetal liver cells (32). Indirect IF revealed γ-tubulin concentrated in most small cells, representing immature MKs and other blood lineages, as judged by morphology (Fig. 1, F and G). In contrast, γ-tubulin levels dropped below detection (Fig. 1, F and G) and occasionally concentrated in a single cytoplasmic focus (Fig. 1, H-J) in well differentiated MKs, recognized by their size and multi-lobed nuclei. These traces of γ-tubulin cannot readily account for the plethora of MT filaments generated within proplatelets (Fig. 1, D and E).
FIGURE 1.
Organization of MTs and γ-tubulin in MKs. Cultured mouse MKs were cytocentrifuged and stained with β-tubulin or γ-tubulin Ab and TRITC-conjugated(β-tubulin) or fluorescein isothiocyanate-conjugated (γ-tubulin) secondary Ab. A, MT profile in a normal mature MK, revealing the most common (reticular) pattern of fibers. Younger cells (B) show MTs radiating from a MTOC, whereas terminally differentiated cells (C, nuclei stained with DAPI) reveal cortical accumulation of thick MT bundles prior to proplatelet formation. Single z layers are shown after image deconvolution. β-Tubulin immunostain (E) and the corresponding differential interference contrast image (D) of a single representative MK in the act of extending proplatelets reveal the extensive network of MT-based cytoplasmic processes in which nascent platelets assemble. γ-Tubulin immunofluorescence (F) and merger with the corresponding DAPI stain (G) verify the relative paucity of γ-tubulin in large (mature) MKs. This point is further illustrated with the cellular outline of a large, proplatelet-forming MK (H, differential interference contrast image) with corresponding DAPI and γ-tubulin immunostain (I) or the same stains together with β1-tubulin IF (J). Scale bars, 15 μm, except B (10 μm).
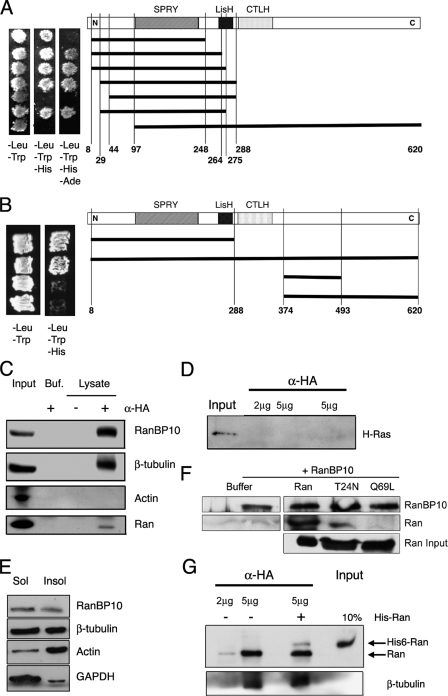
Isolation of RanBP10 as a β-Tubulin-binding Factor—Of the five mammalian β-tubulin isotypes, the most divergent, designated β1 in the mouse, is restricted to expression in MKs and platelets (29, 33, 34). β1-Tubulin is the predominant isoform in proplatelets and is required for efficient thrombopoiesis and discoid platelet shape (35). Much of its sequence divergence is concentrated in C-terminal helices, where MT-associated proteins are known to bind (36). Using a C-terminal fragment of β1-tubulin as the bait in a yeast two-hybrid screen of a mouse MK cDNA library (30), we isolated an interacting factor, RanBP10, which shares 67% amino acid sequence identity with the Ran-binding protein RanBPM/RanBP9 (37, 38). RanBP10 interacted equally well with the widely expressed β5-tubulin isoform in yeast cells (data not shown) and is hence nonselective for association with β-tubulins. Constructs missing 43 or 96 N-terminal amino acids failed to bind β-tubulin (Fig. 2A).
FIGURE 2.
RanBP10 is a β-tubulin- and Ran-binding protein. A, yeast two-hybrid mapping of the RanBP10 interaction with β-tubulin. RanBP10 plasmids, corresponding to the schematic shown to the right, were introduced into yeast strains expressing the C terminus of murine β1-tubulin, followed by selection on nutrient-depleted media. Proteins were considered to interact when yeast grew in the absence of histidine; growth in the absence of adenine implies stronger interactions. B, the N-terminal half of RanBP10 mediates interaction with Ran. For yeast two-hybrid analysis, RanBP10 and Ran plasmids were introduced into yeast cells, which were selected as described above. C, independent assessment of protein interactions. HA epitope-tagged RanBP10 was expressed in 293 cells. The lysates were precipitated with 5 μg of HA Ab and immunoblotted separately with Ab against β-tubulin and Ran or against RanBP10 and actin as positive and negative controls, respectively. 10% of the immunoprecipitated fraction was loaded in the Input lanes; Buf., buffer control, lacking cell lysate. D, lysates from fibroblasts expressing HA-RanBP10 were precipitated with 2 or 5 μg of HA Ab and immunoblotted for H-Ras, which was detected in the lysate but did not co-precipitate with RanBP10. E, RanBP10 is present almost equally in subcellular fractions enriched for monomeric and polymerized tubulins. Proteins were extracted (Sol) from saponin-permeabilized fibroblasts expressing HA-RanBP10, and residual cytoskeletal filaments (Insol) were solubilized in sample buffer. Both fractions were resolved by SDS-PAGE and immunoblotted with RanBP10, β-tubulin, actin, and GAPDH Ab. F, purified His6-tagged Ran, RanT24N, and RanQ69L were incubated in the presence of GST-tagged RanBP10 and complexes captured with glutathione beads prior to immunoblot analysis. RanBP10 binds preferentially to Ran and the T24N mutant in this biochemical assay. G, recombinant His6-tagged Ran was added to HA-RanBP10-expressing 293 cell lysates before precipitation with 2 or 5 μg of HA Ab and immunoblotting for Ran or β-tubulin. Partial exchange between exogenous Ran and the native protein did not alter tubulin binding.
Because RanBPM was originally identified as a Ran-binding factor (37), we asked whether its homolog RanBP10 would also interact with Ran. Yeast two-hybrid experiments verified the interaction between RanBP10 and Ran and revealed that it requires the N-terminal half of RanBP10 (Fig. 2B). To confirm the interaction, we expressed HA epitope-tagged RanBP10 in 293 cells; endogenous cellular β-tubulin co-precipitated with RanBP10 using HA antibody. RanBP10 also co-precipitated Ran but not actin, an abundant cellular protein (Fig. 2C). We detected no binding of RanBP10 to H-Ras (Fig. 2D), RhoA, or RhoB (data not shown), which argues against promiscuous interaction with small GTPases. To determine whether RanBP10 binds preferentially to monomeric or filamentous β-tubulin, we used fibroblasts expressing HA-tagged RanBP10 to separate polymerized MTs from monomeric β-tubulin. As expected, β-tubulin was present in roughly equal proportion in soluble and insoluble fractions from saponin-permeabilized cells. RanBP10 also appeared equally in the two fractions, in contrast to GAPDH, a marker for soluble cytosolic proteins, or actin, which delineated the insoluble cellular fraction (Fig. 2E).
Because Ran-RanBP10 interactions in these studies could potentially be bridged by factors present in cell lysates, we purified hexahistidine (His6)-tagged Ran, RanT24N, or RanQ69L from bacteria, added the proteins to recombinant GST-RanBP10 in Mg2+-free buffer, captured complexes on glutathione beads, and immunoblotted the precipitates. RanBP10 bound best to Ran-GDP and the T24N mutant, which reflects the nucleotide-depleted state (31), and poorly to RanQ69L (Fig. 2F). In the yeast two-hybrid assay, which may be less sensitive to such differences, RanBP10 seemed to interact equally well with the Ran mutants T24N and Q69L (data not shown).
To ask whether binding of Ran and β-tubulin may be mutually exclusive, we added His6-Ran to lysates from fibroblasts expressing HA-tagged RanBP10; the two Ran forms could be discriminated by their electrophoretic migration (Fig. 2G). Excess His6-Ran did not materially influence RanBP10 binding to either endogenous Ran or β-tubulin, which suggests that RanBP10 may interact with both proteins simultaneously. Pointing further to a connection with MTs, RanBP10 contains a lissencephaly homology domain, which in other proteins influences MT dynamics (39). Moreover, forced expression of a truncated form of the related protein RanBPM induces ectopic MT assembly in cultured fibroblasts (37). Taken together, these findings implicate RanBP10 as a protein that interacts with both β-tubulin and Ran. Besides the lissencephaly homology domain, RanBP10 contains a SPRY motif, which may mediate protein interactions (38, 40, 41).
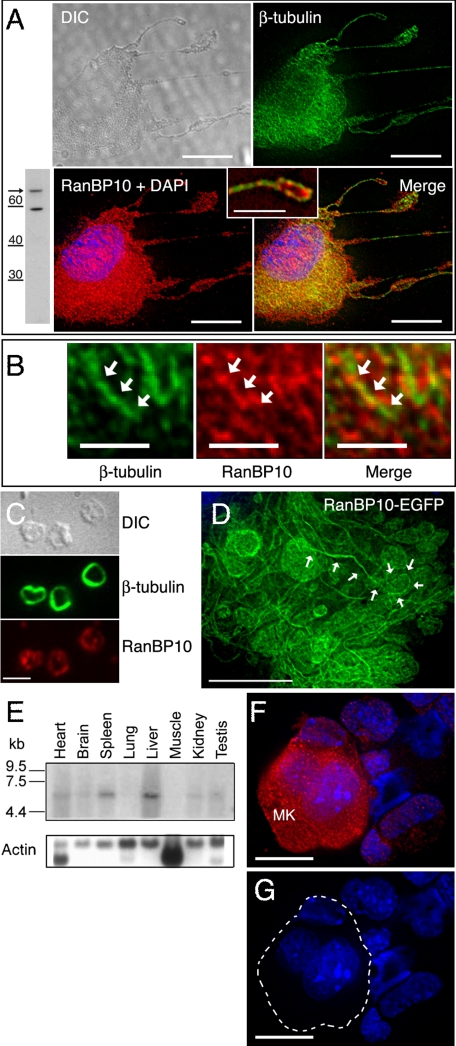
Expression of RanBP10 and Ran in MKs and Platelets—We generated RanBP10 antiserum using a peptide sequence unrelated to RanBPM, and immunoblotting of MK cell lysates detected a Mr ∼ 68,000 protein (Fig. 3A). Later in this report (see Fig. 6, A-C), we show that treatment of cells with RanBP10-specific shRNA significantly reduced antiserum reactivity, attesting to its specificity. Indirect IF revealed a higher concentration of endogenous RanBP10 in primary MKs along cytoplasmic filaments that stain with β-tubulin antibody. Association with MTs was especially evident in cells with features of advanced differentiation and along proplatelets (Fig. 3A, inset), whereas RanBP10 staining in young MKs was weak and diffuse (Fig. 3F and data not shown). At high magnification (Fig. 3B), we noted that RanBP10 staining followed the outline, largely but not exclusively, of cellular MT filaments. Like its localization in proplatelets, RanBP10 also appeared at the periphery in circulating mouse platelets, in similar distribution to β-tubulin in MT marginal bands (Fig. 3C). Finally, in cultured primary MKs, EGFP-tagged RanBP10 was targeted to proplatelets, which are highly enriched for MTs (Fig. 3D), and in permeabilized fibroblasts RanBP10 associates significantly with an insoluble cell fraction containing polymerized MTs (Fig. 2E). These observations collectively provide cellular correlations for the molecular interaction of RanBP10 with β-tubulin and reveal its affinity for both free tubulin and cellular MT filaments.
FIGURE 3.
RanBP10 is a tissue-restricted protein that associates with cytoplasmic MTs in mature MKs and platelets. A, immunoblot showing limited reactivity of RanBP10 Ab, including with a protein of the predicted size (arrow; numbers represent Mr), and IF detection of co-localization of endogenous RanBP10 (red) with MTs, stained with β-tubulin Ab (green), in a representative proplatelet-forming MK. The outline of the cell is revealed in the differential interference contrast image, and the inset shows a magnified portion of the merged panel, highlighting a single proplatelet. B, high resolution (scale bar, 2 μm) deconvolved images of the cytoplasm in a cell similar to that shown in A, emphasizing that RanBP10 distribution is structured and associated with MT filaments. C, in blood platelets, RanBP10 (bottom panel) is detected at the periphery, near the marginal MT band (middle panel), which is revealed by IF for β-tubulin. D, EGFP-tagged RanBP10, forcibly expressed in cultured MKs, appears abundantly in proplatelet extensions, one of which is outlined by white arrows. E, Northern analysis shows highest RanBP10 mRNA expression in adult mouse spleen and liver. F, endogenous RanBP10 levels, detected by IF, are especially high in mature MKs compared with other blood lineages or immature MKs. A single z layer is shown after image deconvolution, and in the corresponding DAPI stain (G), the large, well differentiated MK is outlined in white. Scale bars, A, D, and F, 10 μm; C, 3 μm. A, inset, 4 μm.
FIGURE 6.
Consequences of reduced RanBP10 expression in primary MKs. A-C, cells infected with lentivirus to express RanBP10-specific or control shRNA were identified by EGFP expression (A), and the reduction in RanBP10 protein (B, red signal) was monitored by IF. C, merged image. D-G, β-tubulin IF shows aberrant MT organization in the face of RanBP10 deficiency. Control cells display the characteristic reticular MT array (D), which is significantly disrupted upon RanBP10 depletion (E, nucleus stained with DAPI). High resolution (scale bars, 5 μm) images contrast the filamentous morphology (arrows in F) of MTs in control-infected MKs with filament paucity and numerous punctate deposits (arrows in G) seen with absence of RanBP10. H-J, although RanBP10 IF shows loss of the target protein after shRNA treatment (H, cell border outlined in white), Ran IF (red) and DAPI (blue) stains co-localize (I and J), indicating that Ran is not redistributed. K, x/z planes and cross-sectional intensity profiles after three-dimensional reconstruction verify the overlap in DAPI and Ran signals within the nucleus. Scale bars, A-E and H-J, 15 μm.
Expression of the 5.5-kb RanBP10 mRNA is limited to a few adult mouse organs, including spleen (Fig. 3E) and bone marrow (data not shown), sites of hematopoiesis. In fetal liver cultures, mature MKs express substantially more RanBP10 than do other blood cell lineages, as judged by indirect IF; Fig. 3 (F and G) shows representative z sections after three-dimensional reconstruction and deconvolution. Thus, RanBP10 expression is tissue-restricted and regulated.
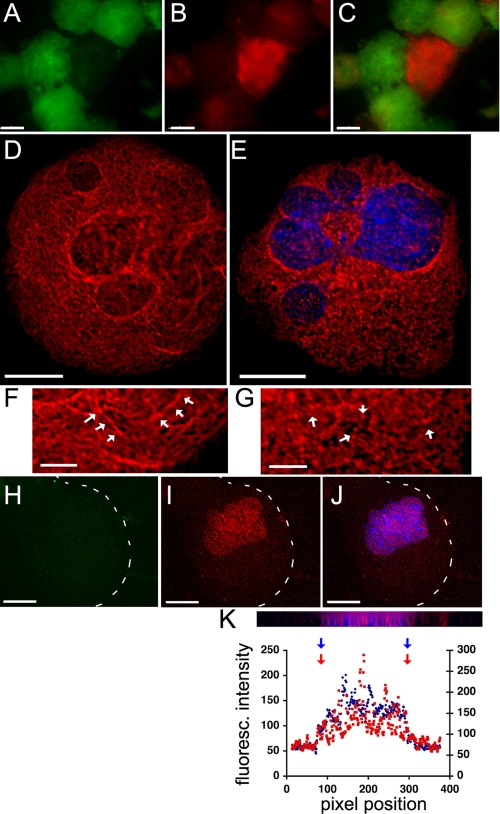
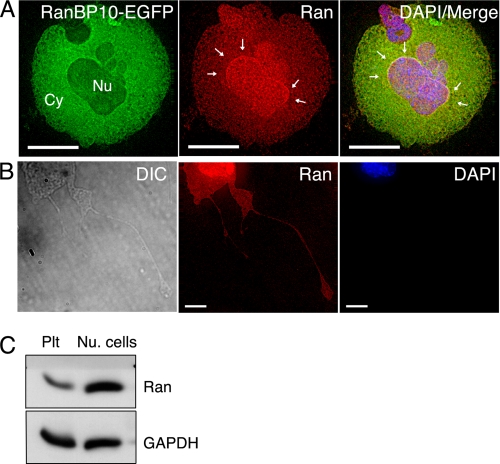
Whereas RanBP10 associates with cytoplasmic MTs, most cellular Ran is present in the nucleus in interphase cells. By forcing expression of GFP-tagged RanBP10 in cultured mouse MKs (Fig. 4A) or 293 cells (data not shown), we confirmed the superficially different distributions of the two proteins. Moreover, the fraction of cellular Ran co-immunoprecipitated with RanBP10 is much lower than that of β-tubulin (Fig. 2C), which is consistent with the predominantly nuclear location of Ran. However, for the molecular interaction between Ran and RanBP10 to have a physiologic role, the two proteins would need to reside within the same compartment under some circumstances. Indeed, although the bulk of cellular Ran appeared in the nucleus as expected, we found significant levels in the cytoplasm, especially in cytologically mature MKs (Fig. 4A); furthermore, like RanBP10 and β1-tubulin, Ran was observed in proplatelets (Fig. 4B). We also detected significant amounts of Ran by immunoblotting (Fig. 4C) and IF (data not shown) in platelet preparations that were devoid of contaminating nucleated cells, although the levels appear lower than in nucleated cells (Fig. 4C). Taken together, these observations support the possibility that RanBP10, Ran and MTs might interact in the cytoplasm. Because blood platelets are anucleate and thus lack the need for known Ran functions, its expression in platelets might represent a vestige of thrombopoiesis or an unidentified role.
FIGURE 4.
Ran is present in the MK cytoplasm and in anucleate platelets. A, EGFP-tagged RanBP10 localizes mainly in the MK cytoplasm (Cy), whereas indirect IF shows Ran predominantly but not exclusively in the nucleus (Nu). Excess RanBP10 causes perinuclear Ran accumulation (arrows), as discussed in the legend for Fig. 6 and in the text. B, in proplatelet-forming MKs (outlined by a corresponding differential interference contrast image to the left), IF staining detects Ran throughout the cytoplasm, including proplatelets (middle panel); DAPI stain is shown to the right. C, in circulating blood platelets (Plt), Ran is again detected by immunoblotting, though possibly at lower levels than those found in mononuclear blood cells (Nu. cells). GAPDH provides a loading control. Scale bars, A, 15 μm; B, 5 μm.
Unexpected GEF Activity in a Ran-binding Protein—RanBP10 and RanBPM are more similar to each other than they are to other Ran-binding factors. An early report placed Ran-BPM in the centrosome (37). Further work from the same laboratory refuted that conclusion (42), but the claim that RanBPM promotes MT assembly (37) stands, and we find independently that RanBP10 binds MTs. Because molecular functions of RanBP10 are not obvious from its currently annotated motifs, we sought other functional domains. Amino acid sequence comparisons using the alignment program Smart uncovered similarities between a conserved region in the RanBP10 sequence and a consensus Rho-GEF domain.
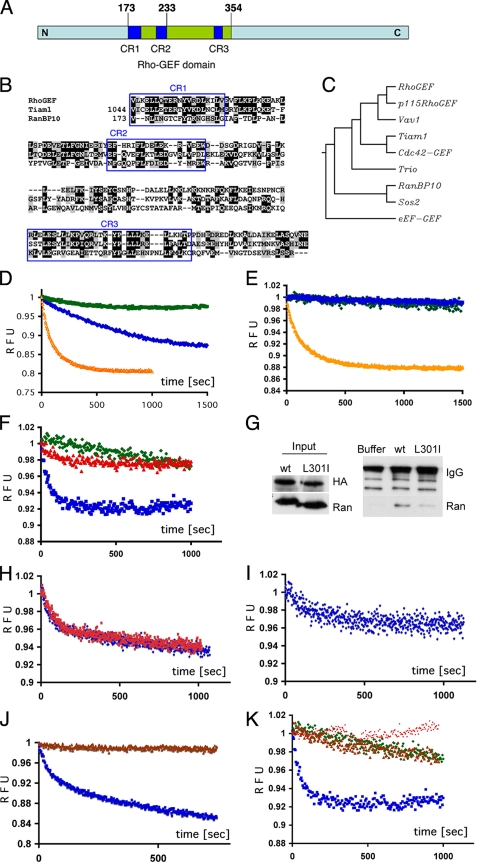
Overall similarity between RanBP10 and the consensus Rho-GEF domain in the region of overlap is about 30%. Three conserved areas, which constitute previously recognized subdomains (43), share about 50% similarity (Fig. 5, A and B), which approximates that observed between other GEF domains. The sequence and three-dimensional structure within the active domains of distinct GEFs can vary substantially (11, 44), although the diversity belies a common molecular mechanism for nucleotide exchange (13, 14, 44). We also compared the putative GEF domain sequence in RanBP10 with those of the Rho-GEF consensus sequence and seven well characterized GEF domains. As expected, proteins with GEF activity toward the Rac/Rho/Cdc42 family (Vav1, Tiam1, and Trio) clustered together, whereas sequences derived from Sos2, a Ras-GEF, and eEF-GEF are less similar to the consensus (Fig. 5C). Within this framework, the putative Rho-GEF domain in RanBP10 aligns closest with Sos2, implying that it may influence a substrate other than the Rac/Rho family. Because RanBP10 interacts with Ran, we considered the possibility that RanBP10 may function as a Ran-GEF.
FIGURE 5.
RanBP10 is a GEF for Ran. A, schematic illustration of the region of homology between a central portion of RanBP10 and a Rho-GEF consensus sequence. CR1-CR3 represent conserved subregions (see text). B, amino acid alignment of a Rho-GEF consensus domain (smart00325.10) with the known and putative GEF domains of Tiam1 (T-cell lymphoma invasion and metastasis-1) and RanBP10, respectively; the three highly conserved subregions (CR) are boxed. C, phylogenetic similarity analysis (ClustalW) of seven GEF domains and the RhoGEF consensus sequence suggests that RanBP10 is more closely related to the Ras-GEF Sos2 than to Vav1 or Tiam1. D and E, like recombinant RCC1 (orange curve), bacterially expressed RanBP10 (blue curve) displays intrinsic GEF activity toward a recombinant Ran substrate. Intrinsic fluorescence of GDP-loaded Ran was measured at 335 nm in the presence of excess mant-GDP. GEF activity results in fluorescence resonance energy transfer, monitored over 1000-1500 s and expressed here in relative fluorescence units (RFU). The low intrinsic nucleotide exchange rate is revealed with buffer or GST (green) controls and a RanBP10 mutant lacking 95 N-terminal residues (E, blue curve). The results represent over six independent experiments. F, a L301I mutation within the putative GEF domain of RanBP10 (red curve) renders inactivity compared with the wild-type protein (blue curve). G, overexpressed HA-tagged wild-type (wt) and L301I mutant RanBP10 were precipitated with HA Ab and immunoblotted for Ran. In three independent experiments, interaction of the L301I mutant with endogenous Ran was weaker than that of the wild-type protein. H, in the presence of RanBP10, Ran-associated GDP exchanges equally well with mant-GDP (blue curve) or mant-GTP (red curve). I, RanBP10 exchanges Ran-bound GTP for mant-GDP (blue curve). J, addition of RanT24N (brown curve) blocks RanBP10 exchange activity for Ran-GDP (blue curve). K, RanBP10 shows substrate selectivity toward Ran (blue curve) and does not manifest GEF activity for either Rho1 (brown curve) or H-Ras (red curve). Buffer controls are represented in green.
Repeated attempts to express full-length RanBP10 as a GST-fused protein in bacteria were frustrated by proteolysis. We therefore expressed the N-terminal 407 amino acids and measured enzymatic activity of the recombinant protein, purified by gel filtration chromatography, in a well established nucleotide exchange assay (28). When excited at 295 nm, tryptophan residues near the Ran nucleotide-binding pocket emit light at 335 nm. Recombinant GDP-loaded Ran was mixed with fluorescent mant-GDP, which absorbs light at 335 nm and, when bound to Ran, quenches emission by fluorescence resonance energy transfer. GEF activity is accurately monitored by a decrease in fluorescence intensity at 335 nm, as shown in Fig. 5D using recombinant RCC1. The calculated turnover rate with RCC1 was 76 pmol/min, which is close to a reported value of 37 pmol/min (28). GST-tagged RanBP10 also harbors Ran-GEF activity (Fig. 5D). The reaction output is comparable with that of RCC1, although the turnover rate of 8 pmol/min is slower; GST alone never demonstrated such effect. The specific activity of RanBP10 was 5-10-fold lower than that of recombinant RCC1; other RanBP10 fragments or the full-length protein might express higher activity, and additional factors probably stabilize RanBP10 or enhance its function in cells.
To confirm the validity of these results, we mutated RanBP10. We deleted 95 N-terminal residues, which make up part of the Ran interaction domain (Fig. 2C), and based on crucial residues identified in other Rho-GEFs (45), generated a L301I point mutant. Both changes eliminated GEF activity (Fig. 5, E and F), which indicates that it is genuine, mediated by the RhoGEF homology domain and requires interaction with Ran. Because the L301I mutation abolished Ran-GEF activity, we asked whether the mutant protein can interact with Ran. In lysates prepared from fibroblasts expressing HA-tagged wildtype or mutant RanBP10, less Ran protein co-precipitated with the L301I mutant than with the wild-type protein (Fig. 5G); equal input amounts of Ran, transfected RanBP10, and IgG were evident. These results indicate that the putative GEF domain within RanBP10 may mediate part of the interaction with Ran and that L301I abrogates enzyme activity in part by diminished interaction.
GEFs catalyze the release of bound nucleotides, stabilize the nucleotide-depleted transition state, and thus facilitate nucleotide re-entry. To test these activities, we repeated the exchange assay with GDP-loaded Ran and RanBP10 in the presence of excess mant-GTP. GEF activity remained evident (Fig. 5H), indicating a lack of a nucleotide preference for exchange. We also loaded Ran with GTP and repeated the experiments using mant-GDP. Although activity was reduced in comparison with Ran-GDP, exchange occurred independent of the nucleotide form (Fig. 5I). This lower activity is consistent with reduced binding of RanBP10 to the Q69L mutant (Fig. 2F), which mimics the GTP-bound state. Finally, a 5-fold excess of constitutively inactive RanT24N blocked all RanBP10 exchange activity toward Ran (Fig. 5J), and nucleotide-loaded recombinant Rho1 and H-Ras could not serve as RanBP10 substrates (Fig. 5K). Our results hence reveal RanBP10 as a Ran-specific GEF with two additional properties: residence outside the nucleus and interaction with cellular MTs.
RanBP10 Deficiency Changes Gross MT Morphology—Centrosome-associated interphase arrays and the mitotic spindle represent two distinct classes of MT filaments, with prominent differences in thickness, length, and MT dynamics; Ran·GTP plays a particular role in mitotic spindle assembly. Additionally, interphase MTs in some specialized mammalian cells do not track to centrosomes, but the alternative structures are not fully characterized (46). The thickness and unusual flexibility of the proplatelet cytoskeleton suggests a specialized or hybrid type of MTs, and their propagation in the absence of MTOCs is reminiscent of mitotic spindle assembly. Based on its location in mature MKs and binding to β-tubulin, we postulated that RanBP10 might regulate MT structure or function.
To test this hypothesis, we depleted endogenous RanBP10 in MKs by introducing specific shRNA in a lentiviral vector, followed by RanBP10 and tubulin IF analysis of single cells. Expression of EGFP by the lentiviral shRNA constructs allowed identification of infected MKs, some of which showed almost no residual RanBP10 staining (Fig. 6, A-C), and we analyzed large cells in which the RanBP10 signal was reduced to background levels. A second RanBP10-specific shRNA failed to deplete the endogenous protein and, together with empty virus, served as a negative control. Loss of RanBP10 did not interfere with early MK maturation. Among well differentiated MKs, however, 40% of RanBP10-depleted cells (n = 37) displayed prominent MT anomalies, which were readily distinguished from any of the only three morphologies (Fig. 1, A-C) we have encountered with tubulin IF in hundreds of GFP-expressing or uninfected MKs. In place of a reticular array of long MT filaments (Figs. 1A and 6, D and F), MKs depleted of RanBP10 showed prominent disruption of the MT cytoskeleton, with a paucity of intact filaments and numerous punctate deposits that suggest an abundance of short MT fragments (Fig. 6, E and G). About 30% of cells harbored scattered tubulin filaments interspersed with short MT fragments and punctate foci, suggesting substantial disruption of the reticular MT network. Another 10% of RanBP10-depleted cells displayed a nearly complete loss of filamentous tubulin staining. We never observed either of these two phenotypes in uninfected or control shRNA-infected MKs, and in each case the presence of an intact nucleus confirmed cell viability (Fig. 6E).
Control EGFP virus-infected MKs showed Ran distributed uniformly in the nucleus and co-localized with DAPI chromatin stain, as confirmed in perpendicular reconstructions of IF images (data not shown). Similar to the overlap between DAPI and Ran signals in control cells, MKs lacking RanBP10 (Fig. 6H) showed typical intranuclear Ran localization (Fig. 6, I-K). Thus, loss of RanBP10 disrupts MT structure without affecting the cellular distribution of Ran.
A free cytosolic Ran-GEF could, in principle, antagonize Ran-GAP and drive futile cycles of nucleotide exchange. Thus, native RanBP10 and its inherent Ran-GEF activity must either be sequestered away from Ran-GAP or controlled to protect the transnuclear Ran gradient. In mature MKs, Ran-GAP concentrates at the nuclear envelope and is virtually absent from the cytoplasm (supplemental Fig. S2A). Additionally, whereas RanBP10 levels increase in mixed blood cell populations enriched for mature MKs (Fig. 3, F and G), Ran-GAP1 mRNA levels show a corresponding decline (supplemental Fig. S2B). Thus, Ran-GAP and RanBP10 proteins are present in distinct compartments, and the relative mRNA ratio appears to favor RanBP10 in mature MKs. These results suggest that cytosolic or MT-associated Ran-GEF activity is unlikely to be short-circuited by excess Ran-GAP.
DISCUSSION
At the conclusion of their life as nucleated cells, MKs generate a profusion of MT filaments in the cell periphery and elaborate proplatelets. These MTs contain the platelet- and MK-specific isotype β1 tubulin (29) and polymerize in the apparent absence of centrioles or significant γ-tubulin stores. Several mechanisms could potentially explain the formation of these structures, including release from centrosomes, de novo cytoplasmic assembly, breakage or severing, and nucleation at other sites (46). Even in the absence of direct evidence, some of these possibilities can be excluded, because mobilization of preassembled fibers cannot account for the abundance of dynamic filaments seen during proplatelet morphogenesis. In contrast, MTs do assemble in some cells in the absence of centrosomes by mechanisms that are understood incompletely (46). Our findings suggest one possible mechanism and also hint at an unexpected role for Ran in the cytoplasm. They reveal an unanticipated nucleotide exchange function for the cytosolic Ran-binding protein RanBP10 and implicate it in organization of nonmitotic MTs, although the latter function does not exclude a role for traces of γ-tubulin. The functions we propose for RanBP10 on MTs likely reflect protein-protein interactions, although other mechanisms are also possible. The enzymatic function we describe for RanBP10 differs from that reported for Mog1, a guanine nucleotide release factor that regulates nuclear protein import by binding to Ran·GTP (47). Mog1 is a nuclear protein that, unlike exchange factors, stabilizes nucleotide-depleted Ran and prevents nucleotide reloading (48).
RanBP10 is a β-tubulin- and Ran-binding protein that carries intrinsic Ran-GEF activity, is highly expressed in the cytoplasm of mature MKs, and influences MT structure in these cells. These features combine to suggest a model wherein a tubulin-associated cytoplasmic protein might generate local pools of Ran·GTP that, in turn, stimulate other factors to regulate atypical MTs. This model draws on known properties of Ran and has general implications for Ran function. RCC1-generated Ran·GTP, concentrated on condensed chromosomes, releases spindle assembly factors such as Tpx2 and NuMA from importin proteins and thereby promotes local assembly of chromatin-dependent MTs (19-21). We propose that cytoplasmic RanBP10 might operate in a manner analogous to RCC1 to regulate the extensive MT network that coordinates platelet assembly and release. The enzymatic GEF properties of RanBP10 are similar to RCC1, which shows no preference for GDP- or GTP-bound Ran states (31). Consistent with its structural similarity to the RhoGEF family, RanBP10 activity would likely help load nucleotide-depleted Ran with the most abundant guanine nucleotide, which in the cytoplasm is GTP (Refs. 31 and 49 and references therein). Although we performed most experiments in primary MKs, broader RanBP10 expression suggests a parallel role in other cell types.
Although the MT modulation we attribute to RanBP10 is influenced by knowledge of RCC1, the only other known Ran-GEF, it is also distinct in concept. Whereas mitotic MTs are only targets of locally generated Ran·GTP, our data implicate RanBP10 as both a Ran exchange factor and one that binds cytoplasmic MTs, the presumptive targets of Ran function in our model. In contrast, RCC1 binds chromatin and not MTs, which it influences through secondary factors like Tpx2 and NuMA. MT binding and GEF activities map to distinct RanBP10 domains. We envision that the N terminus serves to localize RanBP10 on peripheral MTs, positioning the central enzyme domain for Ran nucleotide exchange. The resulting local enrichment of Ran·GTP might then recruit other regional factors to influence stability or dynamic assembly of the thick, bipolar MT bundles in the MK periphery (50). MT filaments in the mitotic spindle differ substantially from those in mature MKs, where they are multi-focal, highly dynamic, mixed in polarity, and probably do not rely on a single focal Ran·GTP source; nor is Ran·GTP likely to be the sole determinant of proplatelet MT assembly.
A functional link between RanBP10 and cytoplasmic MTs is supported both by the data in this report and by the precedent for the role of Ran in regulating MTs. Ran-GEFs suppress yeast α-tubulin mutations that induce excessive cytoplasmic MTs and consequent cell cycle arrest (51). Increased Ran·GTP levels in Xenopus egg extracts induce robust MT aster and spindle formation (6-9). The essential role of Ran in mitotic spindle assembly reflects independent effects on MT nucleation, polymerization, stability, and balance of motor activities (18, 19). Cytoplasmic pools of Ran might similarly regulate other MTs, although such influence must avoid interference with interphase concentration gradients.
In the cytoplasm, RanGAP1 rapidly converts GTP-charged Ran to Ran·GDP, which travels to the nucleus, and interphase segregation of GEF and GAP activities by the nuclear envelope maintains the Ran concentration gradient required for nucleocytoplasmic transport. A free cytosolic Ran-GEF could, in principle, antagonize RanGAP1 and drive futile cycles of nucleotide exchange. Thus, native RanBP10 and its inherent Ran-GEF activity must either be sequestered away from Ran-GAP or controlled to protect the trans-nuclear Ran gradient. Because RanBP10 expression is tissue-restricted and, in MKs, confined to cells of advanced differentiation, there may be few instances where cells encounter this problem, and in any case, its nucleotide exchange activity is probably tightly regulated. The presence of auto-inhibitory domains in many Rho-GEFs (14), interactions with extraneous modulators (52), and a requirement for RCC1 phosphorylation in mitosis (53) point to some mechanisms by which cells restrict GEF activities. In our GEF assays, the specific activity of RanBP10 was 5-10-fold lower than that of recombinant RCC1 and may have been restrained by the construct design or absence of a co-factor. Immunostaining of mature MKs confirmed that Ran-GAP concentrates at the nuclear envelope and is virtually absent from the cytoplasm (supplemental Fig. S2A), where RanBP10 resides. Finally, RanBP10 levels rise with MK maturation (Fig. 3F), whereas RanGAP1 mRNA levels may fall or stay constant (supplemental Fig. S2B). These observations combine to suggest that Ran-GEF activity by cytoplasmic or MT-associated RanBP10 can avoid being short-circuited by excess RanGAP1.
We speculate that RanBP10 enrichment along cytoplasmic MTs acts to restrict the physical range of Ran-GEF activity. Such compartmentalization of a cytoplasmic Ran-GEF may confine Ran·GTP to the immediate vicinity of MTs and regulate the cytoskeleton with minimal impact on the global Ran·GDP/GTP concentration gradient. Ran-GAP activity concentrates at the cytosolic face of the nuclear envelope, in complex with RanBP2 and nuclear pore proteins (54, 55) and in a distribution distinct from that of RanBP10. Thus, RanBP10 association with cellular MTs suggests both a mode of sequestration and a physical basis to regulate the MT cytoskeleton.
Supplementary Material
Acknowledgments
We thank Iain Mattaj for Ran plasmids; Florence Poy, Azin Nezami, Daniel Lietha, and Michael Eck (Dana-Farber Cancer Institute, Boston) and Martin Ruehl, Martin Heck, and Silke Fleischhauer (Charité, Berlin) for help with protein expression, purification, and fluorimetry; Scott Parkinson and John Alberta for help with lentivirus production and deconvolution microscopy, respectively; and Jeremy Green, John Hartwig, and David Pellman for helpful discussions and critical review of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant R01HL63143. This work was also supported by a Scholar Award from the Leukemia and Lymphoma Society (to R. A. S.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
Footnotes
The abbreviations used are: MT, microtubule; MTOC, MT-organizing center; GEF, guanine nucleotide exchange factor; GAP, GTPase-activating protein; MK, megakaryocyte; HA, hemagglutinin; Ab, antibody; PIPES, 1,4-piperazinediethanesulfonic acid; EGFP, enhanced green fluorescent protein; shRNA, short hairpin RNA; IF, immunofluorescence; DAPI, 4′,6′-diamino-2-phenylindole; GST, glutathione S-transferase; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; TRITC, tetramethylrhodamine isothiocyanate.
References
- 1.Schiebel, E. (2000) Curr. Opin. Cell. Biol. 12 113-118 [DOI] [PubMed] [Google Scholar]
- 2.Moritz, M., and Agard, D. A. (2001) Curr. Opin. Struct. Biol. 11 174-181 [DOI] [PubMed] [Google Scholar]
- 3.Urbani, L., and Stearns, T. (1999) Curr. Biol. 9 R315-R317 [DOI] [PubMed] [Google Scholar]
- 4.Meraldi, P., and Nigg, E. A. (2002) FEBS Lett. 521 9-13 [DOI] [PubMed] [Google Scholar]
- 5.Barr, F. A., Sillje, H. H., and Nigg, E. A. (2004) Nat. Rev. Mol. Cell. Biol. 5 429-440 [DOI] [PubMed] [Google Scholar]
- 6.Carazo-Salas, R. E., Guarguaglini, G., Gruss, O. J., Segref, A., Karsenti, E., and Mattaj, I. W. (1999) Nature 400 178-181 [DOI] [PubMed] [Google Scholar]
- 7.Kalab, P., Pu, R. T., and Dasso, M. (1999) Curr. Biol. 9 481-484 [DOI] [PubMed] [Google Scholar]
- 8.Ohba, T., Nakamura, M., Nishitani, H., and Nishimoto, T. (1999) Science 284 1356-1358 [DOI] [PubMed] [Google Scholar]
- 9.Wilde, A., and Zheng, Y. (1999) Science 284 1359-1362 [DOI] [PubMed] [Google Scholar]
- 10.Dasso, M. (2002) Curr. Biol. 12 R502-508 [DOI] [PubMed] [Google Scholar]
- 11.Boguski, M. S., and McCormick, F. (1993) Nature 366 643-654 [DOI] [PubMed] [Google Scholar]
- 12.Meller, N., Merlot, S., and Guda, C. (2005) J. Cell Sci. 118 4937-4946 [DOI] [PubMed] [Google Scholar]
- 13.Renault, L., Kuhlmann, J., Henkel, A., and Wittinghofer, A. (2001) Cell 105 245-255 [DOI] [PubMed] [Google Scholar]
- 14.Rossman, K. L., Der, C. J., and Sondek, J. (2005) Nat. Rev. Mol. Cell. Biol. 6 167-180 [DOI] [PubMed] [Google Scholar]
- 15.Bischoff, F. R., and Ponstingl, H. (1991) Nature 354 80-82 [DOI] [PubMed] [Google Scholar]
- 16.Bischoff, F. R., Klebe, C., Kretschmer, J., Wittinghofer, A., and Ponstingl, H. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 2587-2591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Izaurralde, E., Kutay, U., von Kobbe, C., Mattaj, I. W., and Gorlich, D. (1997) EMBO J. 16 6535-6547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carazo-Salas, R. E., Gruss, O. J., Mattaj, I. W., and Karsenti, E. (2001) Nat. Cell. Biol. 3 228-234 [DOI] [PubMed] [Google Scholar]
- 19.Wilde, A., Lizarraga, S. B., Zhang, L., Wiese, C., Gliksman, N. R., Walczak, C. E., and Zheng, Y. (2001) Nat. Cell. Biol. 3 221-227 [DOI] [PubMed] [Google Scholar]
- 20.Gruss, O. J., Carazo-Salas, R. E., Schatz, C. A., Guarguaglini, G., Kast, J., Wilm, M., Le Bot, N., Vernos, I., Karsenti, E., and Mattaj, I. W. (2001) Cell 104 83-93 [DOI] [PubMed] [Google Scholar]
- 21.Nachury, M. V., Maresca, T. J., Salmon, W. C., Waterman-Storer, C. M., Heald, R., and Weis, K. (2001) Cell 104 95-106 [DOI] [PubMed] [Google Scholar]
- 22.Wiese, C., Wilde, A., Moore, M. S., Adam, S. A., Merdes, A., and Zheng, Y. (2001) Science 291 653-656 [DOI] [PubMed] [Google Scholar]
- 23.Kalab, P., Weis, K., and Heald, R. (2002) Science 295 2452-2456 [DOI] [PubMed] [Google Scholar]
- 24.Italiano, J. E., Jr., Lecine, P., Shivdasani, R. A., and Hartwig, J. H. (1999) J. Cell Biol. 147 1299-1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Radley, J. M., and Scurfield, G. (1980) Blood 56 996-999 [PubMed] [Google Scholar]
- 26.Moskvin-Tarkhanov, M. I., and Onishchenko, G. E. (1978) Tsitologiia 20 1436-1438 [PubMed] [Google Scholar]
- 27.Nagata, Y., Muro, Y., and Todokoro, K. (1997) J. Cell Biol. 139 449-457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klebe, C., Nishimoto, T., and Wittinghofer, F. (1993) Biochemistry 32 11923-11928 [DOI] [PubMed] [Google Scholar]
- 29.Lecine, P., Italiano, J. E., Jr., Kim, S. W., Villeval, J. L., and Shivdasani, R. A. (2000) Blood 96 1366-1373 [PubMed] [Google Scholar]
- 30.Schulze, H., Korpal, M., Bergmeier, W., Italiano, J. E., Jr., Wahl, S. M., and Shivdasani, R. A. (2004) Blood 104 3949-3957 [DOI] [PubMed] [Google Scholar]
- 31.Klebe, C., Bischoff, F. R., Ponstingl, H., and Wittinghofer, A. (1995) Biochemistry 34 639-647 [DOI] [PubMed] [Google Scholar]
- 32.Lecine, P., Blank, V., and Shivdasani, R. (1998) J. Biol. Chem. 273 7572-7578 [DOI] [PubMed] [Google Scholar]
- 33.Lewis, S. A., Gu, W., and Cowan, N. J. (1987) Cell 49 539-548 [DOI] [PubMed] [Google Scholar]
- 34.Wang, D., Villasante, A., Lewis, S. A., and Cowan, N. J. (1986) J. Cell Biol. 103 1903-1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwer, H. D., Lecine, P., Tiwari, S., Italiano, J. E., Jr., Hartwig, J. H., and Shivdasani, R. A. (2001) Curr. Biol. 11 579-586 [DOI] [PubMed] [Google Scholar]
- 36.Nogales, E. (2001) Annu. Rev. Biophys. Biomol. Struct. 30 397-420 [DOI] [PubMed] [Google Scholar]
- 37.Nakamura, M., Masuda, H., Horii, J., Kuma, K., Yokoyama, N., Ohba, T., Nishitani, H., Miyata, T., Tanaka, M., and Nishimoto, T. (1998) J. Cell Biol. 143 1041-1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang, D., Li, Z., Schoen, S. R., Messing, E. M., and Wu, G. (2004) Biochem. Biophys. Res. Commun. 313 320-326 [DOI] [PubMed] [Google Scholar]
- 39.Emes, R. D., and Ponting, C. P. (2001) Hum. Mol. Genet. 10 2813-2820 [DOI] [PubMed] [Google Scholar]
- 40.Ponting, C., Schultz, J., and Bork, P. (1997) Trends Biochem. Sci. 22 193-194 [DOI] [PubMed] [Google Scholar]
- 41.Wang, D., Li, Z., Messing, E. M., and Wu, G. (2002) J. Biol. Chem. 277 36216-36222 [DOI] [PubMed] [Google Scholar]
- 42.Nishitani, H., Hirose, E., Uchimura, Y., Nakamura, M., Umeda, M., Nishii, K., Mori, N., and Nishimoto, T. (2001) Gene (Amst.) 272 25-33 [DOI] [PubMed] [Google Scholar]
- 43.Whitehead, I. P., Campbell, S., Rossman, K. L., and Der, C. J. (1997) Biochim. Biophys. Acta 1332 F1-F23 [DOI] [PubMed] [Google Scholar]
- 44.Sprang, S. R., and Coleman, D. E. (1998) Cell 95 155-158 [DOI] [PubMed] [Google Scholar]
- 45.Cheng, L., Rossman, K. L., Mahon, G. M., Worthylake, D. K., Korus, M., Sondek, J., and Whitehead, I. P. (2002) Mol. Cell. Biol. 22 6895-6905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Keating, T. J., and Borisy, G. G. (1999) Biol. Cell 91 321-329 [PubMed] [Google Scholar]
- 47.Nicolas, F. J., Moore, W. J., Zhang, C., and Clarke, P. R. (2001) J. Cell Sci. 114 3013-3023 [DOI] [PubMed] [Google Scholar]
- 48.Steggerda, S. M., and Paschal, B. M. (2000) J. Biol. Chem. 275 23175-23180 [DOI] [PubMed] [Google Scholar]
- 49.Zheng, Y. (2001) Trends Biochem. Sci. 26 724-732 [DOI] [PubMed] [Google Scholar]
- 50.Patel, S. R., Richardson, J. L., Schulze, H., Kahle, E., Galjart, N., Drabek, K., Shivdasani, R. A., Hartwig, J. H., and Italiano, J. E., Jr. (2005) Blood 106 4076-4085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kirkpatrick, D., and Solomon, F. (1994) Genetics 137 381-392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wells, C. D., Gutowski, S., Bollag, G., and Sternweis, P. C. (2001) J. Biol. Chem. 276 28897-28905 [DOI] [PubMed] [Google Scholar]
- 53.Li, H. Y., and Zheng, Y. (2004) Genes Dev. 18 512-527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mahajan, R., Delphin, C., Guan, T., Gerace, L., and Melchior, F. (1997) Cell 88 97-107 [DOI] [PubMed] [Google Scholar]
- 55.Matunis, M. J., Coutavas, E., and Blobel, G. (1996) J. Cell Biol. 135 1457-1470 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.