Abstract
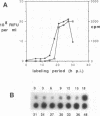
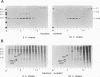
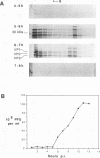
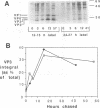
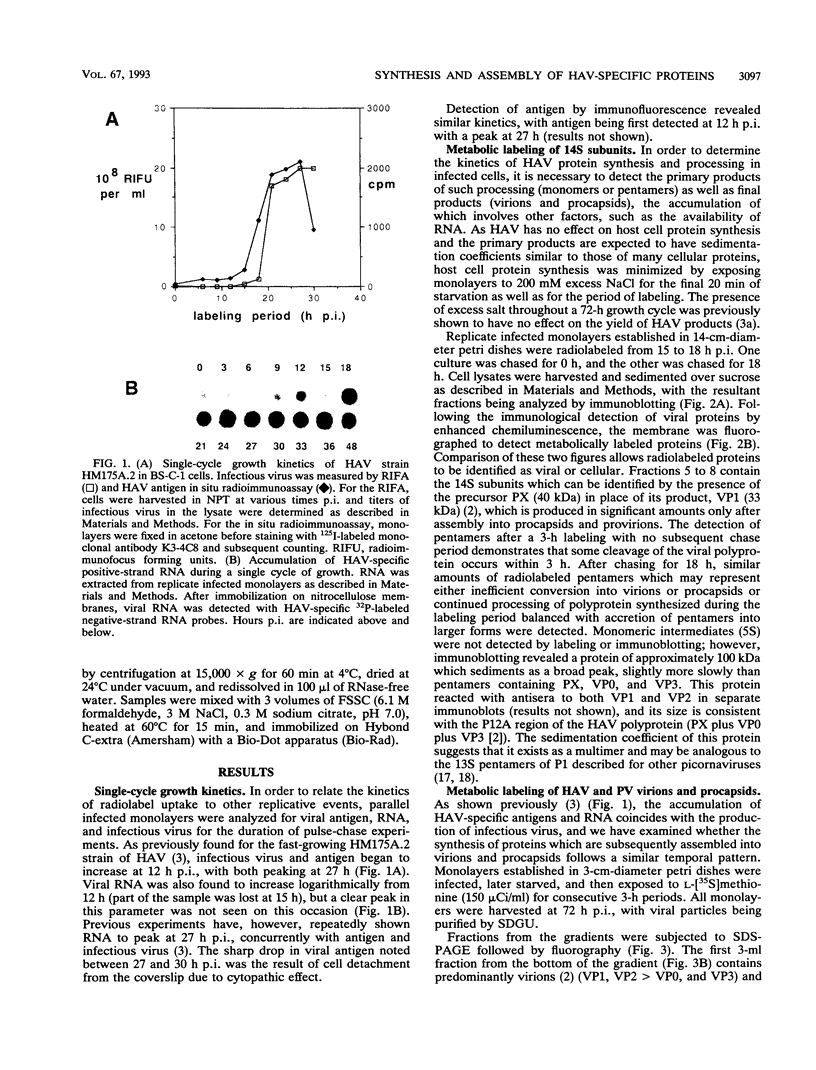
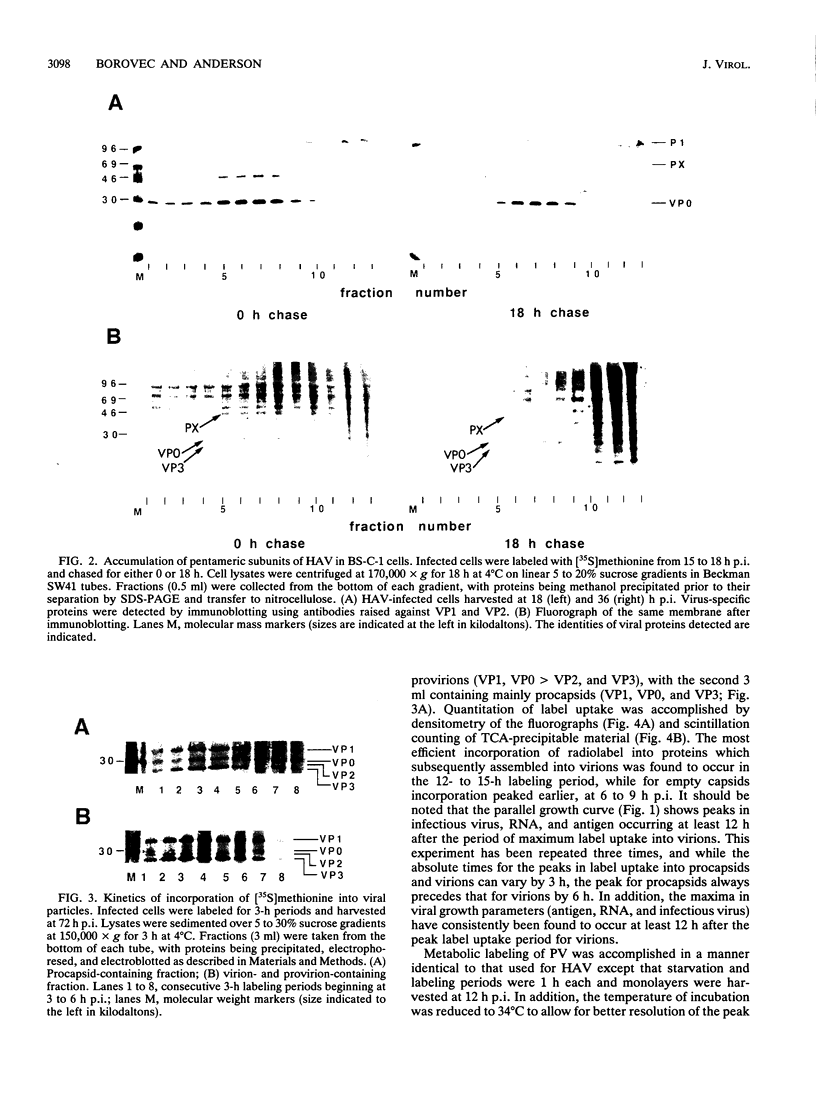
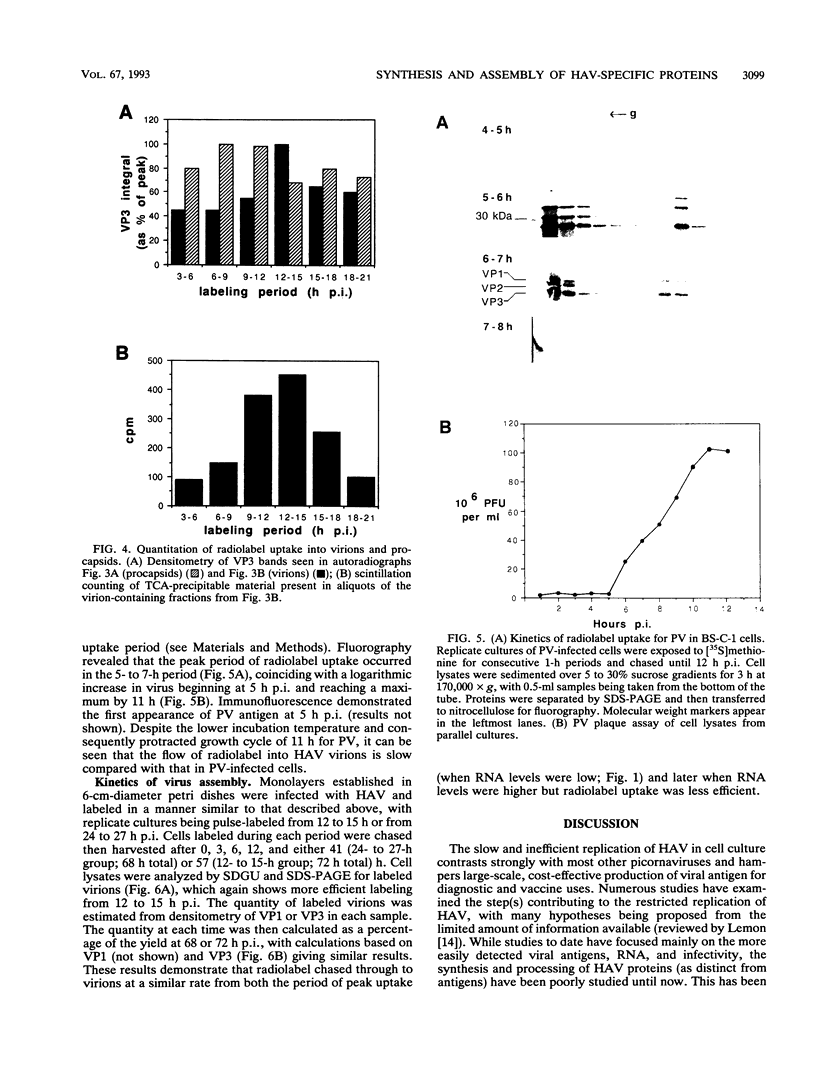
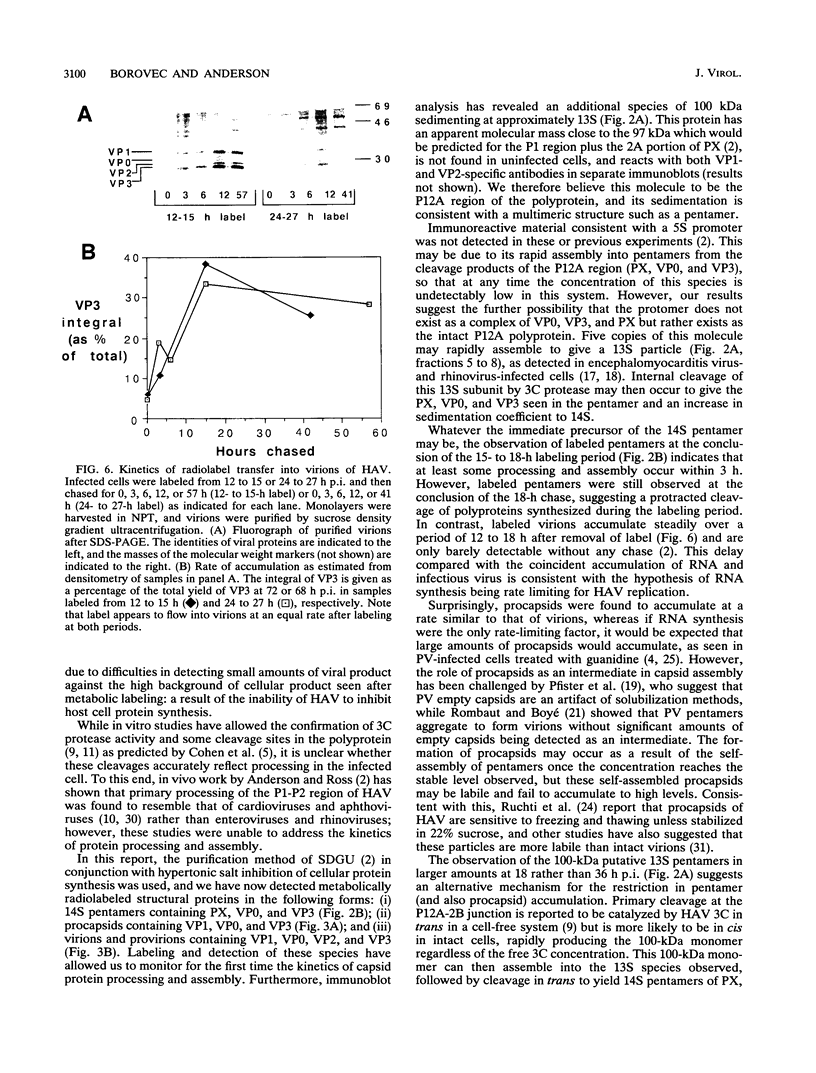
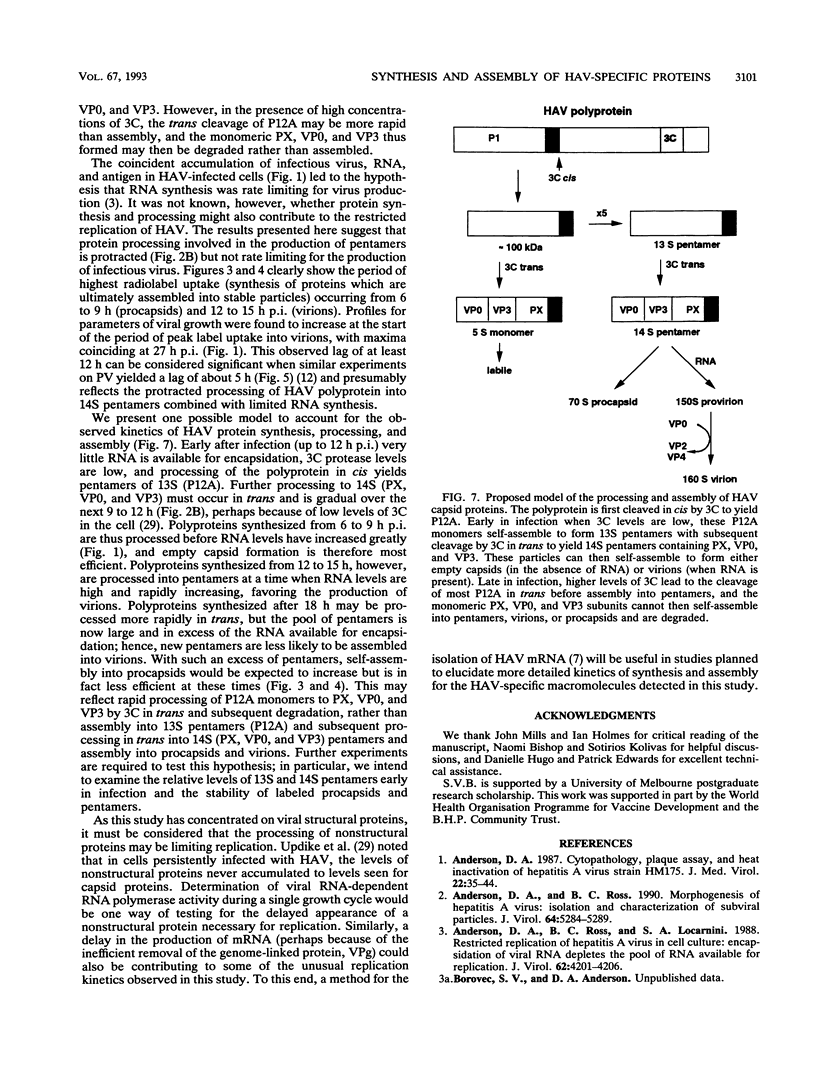
To determine the mechanism for the delayed and inefficient replication of the picornavirus hepatitis A virus in cell culture, we studied the kinetics of synthesis and assembly of virus-specific proteins by metabolic labeling of infected BS-C-1 cells with L-[35S]methionine and L-[35S]cysteine. Sedimentation, electrophoresis, and autoradiography revealed the presence of virions, provirions, procapsids, and 14S (pentameric) subunits. Virions and provirions contained VP1, VP0, VP2, and VP3; procapsids contained VP1, VP0, and VP3; and pentamers contained PX, VP0, and VP3, as previously shown by immunoblotting (D.A. Anderson and B.C. Ross, J. Virol. 64:5284-5289, 1990). Under single-cycle growth conditions label was found in 14S subunits immediately after labeling from 15 to 18 h postinfection (p.i.); however, a proportion of labeled polyprotein was not cleaved and assembled into pentamers for a further 18 h. When analyzed at 72 h p.i., incorporation of label which flowed into virions was detected from 3 h p.i., with maximal uptake levels being observed from 12 to 15 h p.i. Viral antigen, infectious virus, and viral RNA were determined in parallel, with coincident peaks in these variables being observed 12 h after the period of maximum label uptake. It was also found that the lag between the synthesis of the viral polyprotein and assembly of viral particles was the same after labeling from either 12 to 15 or 27 to 30 h p.i. despite increased levels of viral RNA during this period, suggesting that factors additional to the level of RNA are involved in the restriction of viral replication. Sedimentation and immunoblot analysis revealed an additional protein of approximately 100 kDa containing both VP1- and VP2-reactive sequences and sedimenting slightly more slowly than 14S pentamers, which may represent intact P12A assembled into pentamers as has been reported for the P1 of some other picornaviruses (S. McGregor and R. R. Rueckert, J. Virol. 21:548-553, 1977). The results of this study suggest that cleavage of the hepatitis A virus polyprotein to produce pentamers is protracted (though not rate limiting) early in infection, while the assembly of pentamers into higher structures is a rapid process once sufficient viral RNA is produced for encapsidation.
Full text
PDF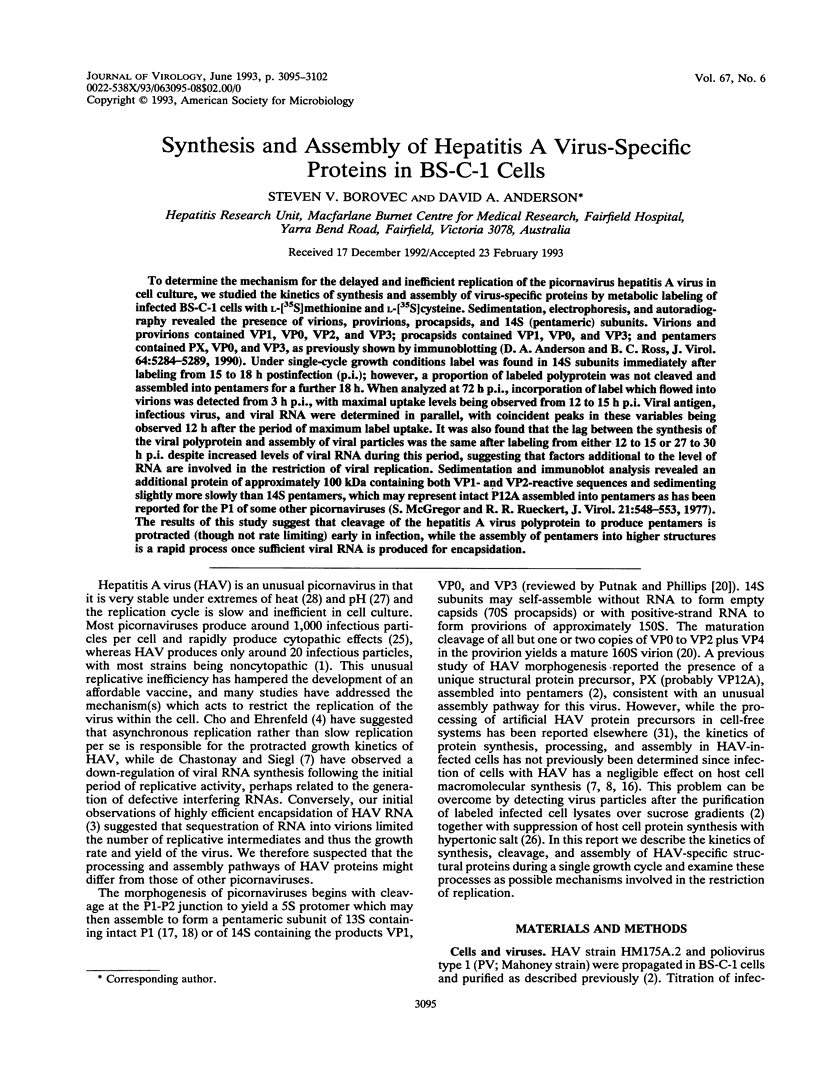


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson D. A. Cytopathology, plaque assay, and heat inactivation of hepatitis A virus strain HM175. J Med Virol. 1987 May;22(1):35–44. doi: 10.1002/jmv.1890220106. [DOI] [PubMed] [Google Scholar]
- Anderson D. A., Ross B. C., Locarnini S. A. Restricted replication of hepatitis A virus in cell culture: encapsidation of viral RNA depletes the pool of RNA available for replication. J Virol. 1988 Nov;62(11):4201–4206. doi: 10.1128/jvi.62.11.4201-4206.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson D. A., Ross B. C. Morphogenesis of hepatitis A virus: isolation and characterization of subviral particles. J Virol. 1990 Nov;64(11):5284–5289. doi: 10.1128/jvi.64.11.5284-5289.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho M. W., Ehrenfeld E. Rapid completion of the replication cycle of hepatitis A virus subsequent to reversal of guanidine inhibition. Virology. 1991 Feb;180(2):770–780. doi: 10.1016/0042-6822(91)90090-x. [DOI] [PubMed] [Google Scholar]
- Cohen J. I., Ticehurst J. R., Feinstone S. M., Rosenblum B., Purcell R. H. Hepatitis A virus cDNA and its RNA transcripts are infectious in cell culture. J Virol. 1987 Oct;61(10):3035–3039. doi: 10.1128/jvi.61.10.3035-3039.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulepis A. G., Tannock G. A., Locarnini S. A., Gust I. D. Evidence that the genome of hepatitis A virus consists of single-stranded RNA. J Virol. 1981 Jan;37(1):473–477. doi: 10.1128/jvi.37.1.473-477.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Chastonay J., Siegl G. Replicative events in hepatitis A virus-infected MRC-5 cells. Virology. 1987 Apr;157(2):268–275. doi: 10.1016/0042-6822(87)90269-8. [DOI] [PubMed] [Google Scholar]
- Gauss-Müller V., Deinhardt F. Effect of hepatitis A virus infection on cell metabolism in vitro. Proc Soc Exp Biol Med. 1984 Jan;175(1):10–15. doi: 10.3181/00379727-175-41757. [DOI] [PubMed] [Google Scholar]
- Harmon S. A., Updike W., Jia X. Y., Summers D. F., Ehrenfeld E. Polyprotein processing in cis and in trans by hepatitis A virus 3C protease cloned and expressed in Escherichia coli. J Virol. 1992 Sep;66(9):5242–5247. doi: 10.1128/jvi.66.9.5242-5247.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson R. J. A detailed kinetic analysis of the in vitro synthesis and processing of encephalomyocarditis virus products. Virology. 1986 Feb;149(1):114–127. doi: 10.1016/0042-6822(86)90092-9. [DOI] [PubMed] [Google Scholar]
- Jia X. Y., Ehrenfeld E., Summers D. F. Proteolytic activity of hepatitis A virus 3C protein. J Virol. 1991 May;65(5):2595–2600. doi: 10.1128/jvi.65.5.2595-2600.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korant B. D. Protein cleavage in virus-infected cells. Acta Biol Med Ger. 1977;36(11-12):1565–1573. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lemon S. M., Binn L. N., Marchwicki R. H. Radioimmunofocus assay for quantitation of hepatitis A virus in cell cultures. J Clin Microbiol. 1983 May;17(5):834–839. doi: 10.1128/jcm.17.5.834-839.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locarnini S. A., Coulepis A. G., Westaway E. G., Gust I. D. Restricted replication of human hepatitis A virus in cell culture: intracellular biochemical studies. J Virol. 1981 Jan;37(1):216–225. doi: 10.1128/jvi.37.1.216-225.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGregor S. Evidence for the existence of protomers in the assembly of encephalomyocarditis virus. J Virol. 1975 May;15(5):1107–1120. doi: 10.1128/jvi.15.5.1107-1120.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGregor S., Rueckert R. R. Picornaviral capsid assembly: similarity of rhinovirus and enterovirus precursor subunits. J Virol. 1977 Feb;21(2):548–553. doi: 10.1128/jvi.21.2.548-553.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister T., Pasamontes L., Troxler M., Egger D., Bienz K. Immunocytochemical localization of capsid-related particles in subcellular fractions of poliovirus-infected cells. Virology. 1992 Jun;188(2):676–684. doi: 10.1016/0042-6822(92)90522-q. [DOI] [PubMed] [Google Scholar]
- Putnak J. R., Phillips B. A. Picornaviral structure and assembly. Microbiol Rev. 1981 Jun;45(2):287–315. doi: 10.1128/mr.45.2.287-315.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rombaut B., Vrijsen R., Boeyé A. New evidence for the precursor role of 14 S subunits in poliovirus morphogenesis. Virology. 1990 Jul;177(1):411–414. doi: 10.1016/0042-6822(90)90502-i. [DOI] [PubMed] [Google Scholar]
- Ross B. C., Anderson D. A. Characterization of hepatitis A virus capsid proteins with antisera raised to recombinant antigens. J Virol Methods. 1991 May;32(2-3):213–220. doi: 10.1016/0166-0934(91)90052-2. [DOI] [PubMed] [Google Scholar]
- Ruchti F., Siegl G., Weitz M. Identification and characterization of incomplete hepatitis A virus particles. J Gen Virol. 1991 Sep;72(Pt 9):2159–2166. doi: 10.1099/0022-1317-72-9-2159. [DOI] [PubMed] [Google Scholar]
- Saborio J. L., Pong S. S., Koch G. Selective and reversible inhibition of initiation of protein synthesis in mammalian cells. J Mol Biol. 1974 May 15;85(2):195–211. doi: 10.1016/0022-2836(74)90360-x. [DOI] [PubMed] [Google Scholar]
- Scholz E., Heinricy U., Flehmig B. Acid stability of hepatitis A virus. J Gen Virol. 1989 Sep;70(Pt 9):2481–2485. doi: 10.1099/0022-1317-70-9-2481. [DOI] [PubMed] [Google Scholar]
- Siegl G., Weitz M., Kronauer G. Stability of hepatitis A virus. Intervirology. 1984;22(4):218–226. doi: 10.1159/000149554. [DOI] [PubMed] [Google Scholar]
- Updike W. S., Tesar M., Ehrenfeld E. Detection of hepatitis A virus proteins in infected BS-C-1 cells. Virology. 1991 Nov;185(1):411–418. doi: 10.1016/0042-6822(91)90789-e. [DOI] [PubMed] [Google Scholar]
- Vakharia V. N., Devaney M. A., Moore D. M., Dunn J. J., Grubman M. J. Proteolytic processing of foot-and-mouth disease virus polyproteins expressed in a cell-free system from clone-derived transcripts. J Virol. 1987 Oct;61(10):3199–3207. doi: 10.1128/jvi.61.10.3199-3207.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winokur P. L., McLinden J. H., Stapleton J. T. The hepatitis A virus polyprotein expressed by a recombinant vaccinia virus undergoes proteolytic processing and assembly into viruslike particles. J Virol. 1991 Sep;65(9):5029–5036. doi: 10.1128/jvi.65.9.5029-5036.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]