Abstract
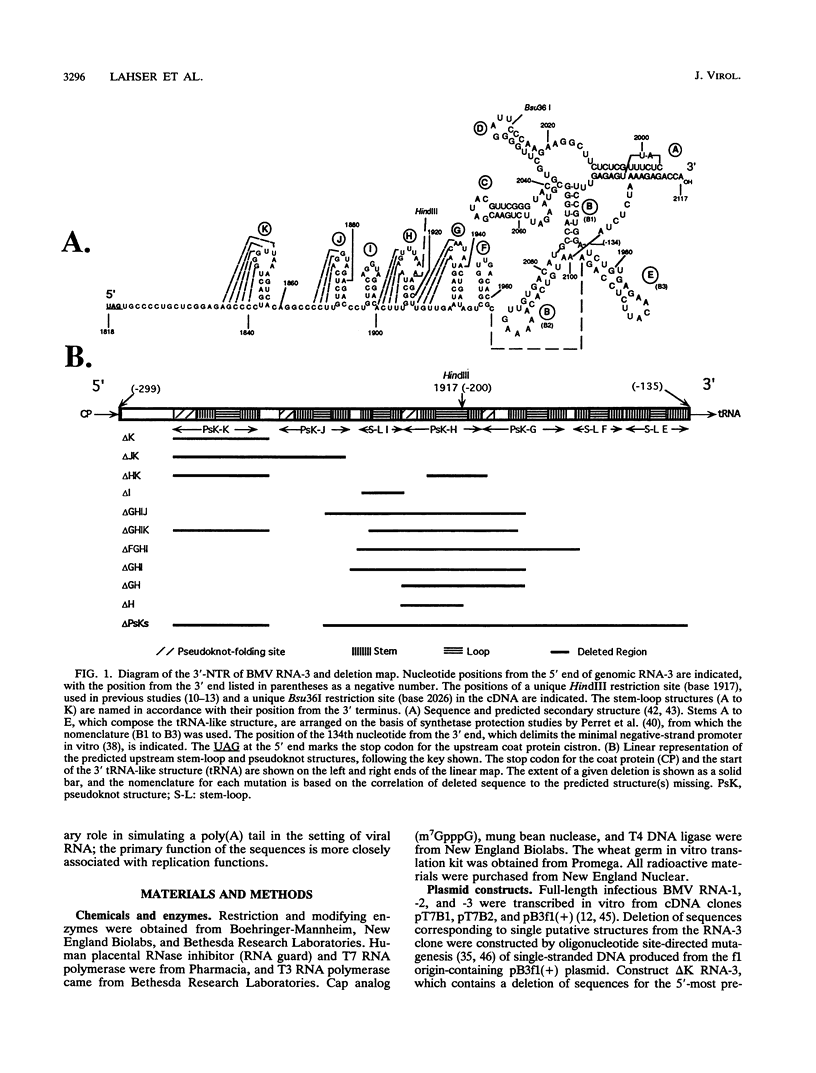
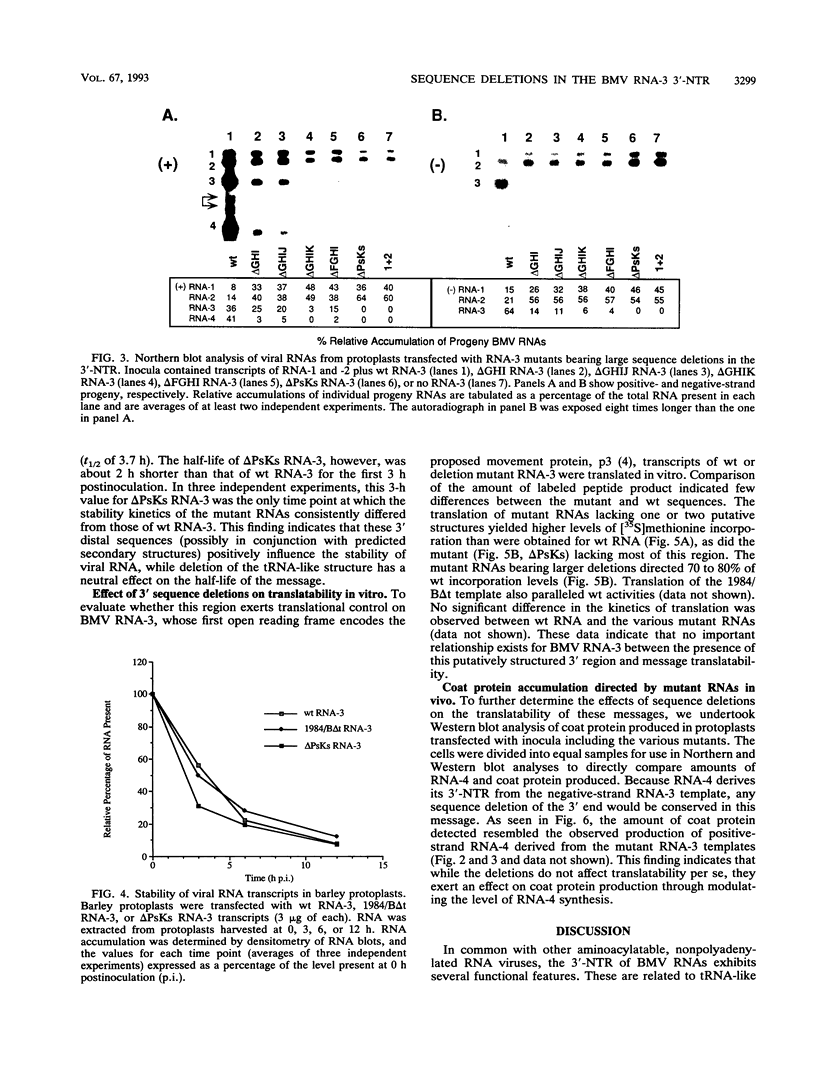
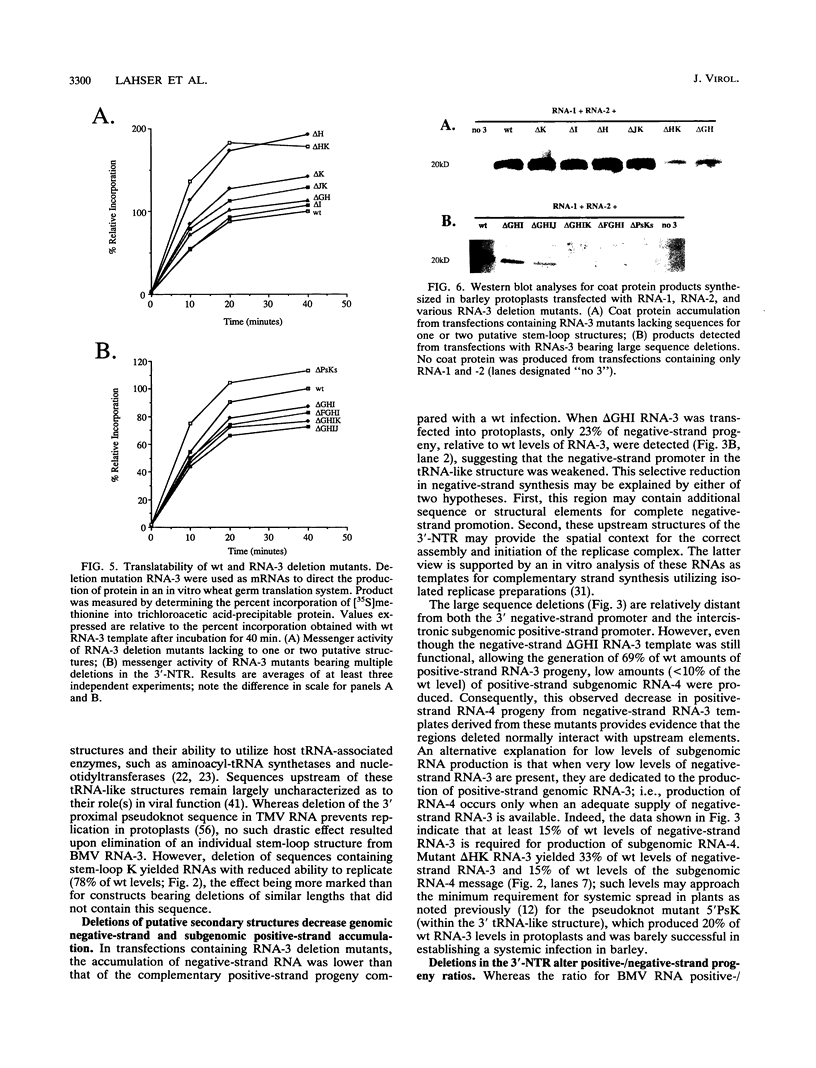
Sequences upstream of the 3'-terminal tRNA-like structure of brome mosaic virus RNAs have been predicted to fold into several stem-loop and pseudoknot structures. To elucidate the functional role of this upstream region, a series of deletions was made in cDNA clones of RNA-3, a genomic component not required for replication. These deletion mutants were transcribed in vitro and cotransfected with RNA-1 and RNA-2 into barley protoplasts. Deletion of single stem-loop structures gave progeny retaining near-wild-type accumulation levels. Constructions representing deletion of two or three stem-loops substantially lowered the accumulation of progeny RNA-3 relative to wild-type levels. RNA-3 mutants bearing deletions of longer sequences or of the entire region (delta PsKs RNA-3) replicated poorly, yielding no detectable RNA-3 or RNA-4 progeny. Levels of RNA-1 and RNA-2, in the presence of a mutant RNA-3, were found to increase relative to the accumulation observed in a complete wild-type transfection. The stability of delta PsKs RNA-3 in protoplasts was somewhat lower than that of wild-type RNA during the first 3 h postinoculation. Little difference in translatability in vitro of wild-type and RNA-3 constructs bearing deletions within the stem-loop region was observed, and Western immunoblot analysis of viral coat protein produced in transfected protoplasts showed that protein accumulation paralleled the amount of RNA-4 message produced from the various sequences evaluated. These results indicate that the RNA-3 pseudoknot region plays a minor role in translational control but contributes substantially to the overall replication of the brome mosaic virus genome.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abrahams J. P., van den Berg M., van Batenburg E., Pleij C. Prediction of RNA secondary structure, including pseudoknotting, by computer simulation. Nucleic Acids Res. 1990 May 25;18(10):3035–3044. doi: 10.1093/nar/18.10.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams C. C., Stern D. B. Control of mRNA stability in chloroplasts by 3' inverted repeats: effects of stem and loop mutations on degradation of psbA mRNA in vitro. Nucleic Acids Res. 1990 Oct 25;18(20):6003–6010. doi: 10.1093/nar/18.20.6003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahlquist P., Dasgupta R., Kaesberg P. Near identity of 3- RNA secondary structure in bromoviruses and cucumber mosaic virus. Cell. 1981 Jan;23(1):183–189. doi: 10.1016/0092-8674(81)90283-x. [DOI] [PubMed] [Google Scholar]
- Allison R., Thompson C., Ahlquist P. Regeneration of a functional RNA virus genome by recombination between deletion mutants and requirement for cowpea chlorotic mottle virus 3a and coat genes for systemic infection. Proc Natl Acad Sci U S A. 1990 Mar;87(5):1820–1824. doi: 10.1073/pnas.87.5.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein P., Ross J. Poly(A), poly(A) binding protein and the regulation of mRNA stability. Trends Biochem Sci. 1989 Sep;14(9):373–377. doi: 10.1016/0968-0004(89)90011-x. [DOI] [PubMed] [Google Scholar]
- Brown P. H., Tiley L. S., Cullen B. R. Effect of RNA secondary structure on polyadenylation site selection. Genes Dev. 1991 Jul;5(7):1277–1284. doi: 10.1101/gad.5.7.1277. [DOI] [PubMed] [Google Scholar]
- Doel M. T., Carey N. H. The translational capacity of deadenylated ovalbumin messenger RNA. Cell. 1976 May;8(1):51–58. doi: 10.1016/0092-8674(76)90184-7. [DOI] [PubMed] [Google Scholar]
- Dreher T. W., Hall T. C. Mutational analysis of the sequence and structural requirements in brome mosaic virus RNA for minus strand promoter activity. J Mol Biol. 1988 May 5;201(1):31–40. doi: 10.1016/0022-2836(88)90436-6. [DOI] [PubMed] [Google Scholar]
- Dreher T. W., Hall T. C. Mutational analysis of the tRNA mimicry of brome mosaic virus RNA. Sequence and structural requirements for aminoacylation and 3'-adenylation. J Mol Biol. 1988 May 5;201(1):41–55. doi: 10.1016/0022-2836(88)90437-8. [DOI] [PubMed] [Google Scholar]
- Dreher T. W., Rao A. L., Hall T. C. Replication in vivo of mutant brome mosaic virus RNAs defective in aminoacylation. J Mol Biol. 1989 Apr 5;206(3):425–438. doi: 10.1016/0022-2836(89)90491-9. [DOI] [PubMed] [Google Scholar]
- Duggal R., Rao A. L., Hall T. C. Unique nucleotide differences in the conserved 3' termini of brome mosaic virus RNAs are maintained through their optimization of genome replication. Virology. 1992 Mar;187(1):261–270. doi: 10.1016/0042-6822(92)90314-f. [DOI] [PubMed] [Google Scholar]
- Gallie D. R., Feder J. N., Schimke R. T., Walbot V. Functional analysis of the tobacco mosaic virus tRNA-like structure in cytoplasmic gene regulation. Nucleic Acids Res. 1991 Sep 25;19(18):5031–5036. doi: 10.1093/nar/19.18.5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie D. R., Feder J. N., Schimke R. T., Walbot V. Post-transcriptional regulation in higher eukaryotes: the role of the reporter gene in controlling expression. Mol Gen Genet. 1991 Aug;228(1-2):258–264. doi: 10.1007/BF00282474. [DOI] [PubMed] [Google Scholar]
- Gallie D. R. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 1991 Nov;5(11):2108–2116. doi: 10.1101/gad.5.11.2108. [DOI] [PubMed] [Google Scholar]
- Gallie D. R., Walbot V. RNA pseudoknot domain of tobacco mosaic virus can functionally substitute for a poly(A) tail in plant and animal cells. Genes Dev. 1990 Jul;4(7):1149–1157. doi: 10.1101/gad.4.7.1149. [DOI] [PubMed] [Google Scholar]
- Grens A., Scheffler I. E. The 5'- and 3'-untranslated regions of ornithine decarboxylase mRNA affect the translational efficiency. J Biol Chem. 1990 Jul 15;265(20):11810–11816. [PubMed] [Google Scholar]
- Gultyaev A. P. The computer simulation of RNA folding involving pseudoknot formation. Nucleic Acids Res. 1991 May 11;19(9):2489–2494. doi: 10.1093/nar/19.9.2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall T. C., Marsh L. E., Dreher T. W. Analysis of brome mosaic virus replication and aminoacylation functions by site-specific mutagenesis. J Cell Sci Suppl. 1987;7:287–301. doi: 10.1242/jcs.1987.supplement_7.20. [DOI] [PubMed] [Google Scholar]
- Hall T. C., Shih D. S., Kaesberg P. Enzyme-mediated binding of tyrosine to brome-mosaic-virus ribonucleic acid. Biochem J. 1972 Oct;129(4):969–976. doi: 10.1042/bj1290969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall T. C. Transfer RNA-like structures in viral genomes. Int Rev Cytol. 1979;60:1–26. doi: 10.1016/s0074-7696(08)61257-7. [DOI] [PubMed] [Google Scholar]
- Hatfield D., Oroszlan S. The where, what and how of ribosomal frameshifting in retroviral protein synthesis. Trends Biochem Sci. 1990 May;15(5):186–190. doi: 10.1016/0968-0004(90)90159-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingelbrecht I. L., Herman L. M., Dekeyser R. A., Van Montagu M. C., Depicker A. G. Different 3' end regions strongly influence the level of gene expression in plant cells. Plant Cell. 1989 Jul;1(7):671–680. doi: 10.1105/tpc.1.7.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki K., Temin H. M. The efficiency of RNA 3'-end formation is determined by the distance between the cap site and the poly(A) site in spleen necrosis virus. Genes Dev. 1990 Dec;4(12B):2299–2307. doi: 10.1101/gad.4.12b.2299. [DOI] [PubMed] [Google Scholar]
- Joshi R. L., Joshi S., Chapeville F., Haenni A. L. tRNA-like structures of plant viral RNAs: conformational requirements for adenylation and aminoacylation. EMBO J. 1983;2(7):1123–1127. doi: 10.1002/j.1460-2075.1983.tb01556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruys V., Wathelet M., Poupart P., Contreras R., Fiers W., Content J., Huez G. The 3' untranslated region of the human interferon-beta mRNA has an inhibitory effect on translation. Proc Natl Acad Sci U S A. 1987 Sep;84(17):6030–6034. doi: 10.1073/pnas.84.17.6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskey R. A., Mills A. D. Enhanced autoradiographic detection of 32P and 125I using intensifying screens and hypersensitized film. FEBS Lett. 1977 Oct 15;82(2):314–316. doi: 10.1016/0014-5793(77)80609-1. [DOI] [PubMed] [Google Scholar]
- Marsh L. E., Dreher T. W., Hall T. C. Mutational analysis of the core and modulator sequences of the BMV RNA3 subgenomic promoter. Nucleic Acids Res. 1988 Feb 11;16(3):981–995. doi: 10.1093/nar/16.3.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh L. E., Huntley C. C., Pogue G. P., Connell J. P., Hall T. C. Regulation of (+):(-)-strand asymmetry in replication of brome mosaic virus RNA. Virology. 1991 May;182(1):76–83. doi: 10.1016/0042-6822(91)90650-z. [DOI] [PubMed] [Google Scholar]
- McLeester R. C., Hall T. C. Simplification of amino acid incorporation and other assays using filter paper techniques. Anal Biochem. 1977 May 1;79(1-2):627–630. doi: 10.1016/0003-2697(77)90447-x. [DOI] [PubMed] [Google Scholar]
- Miller W. A., Bujarski J. J., Dreher T. W., Hall T. C. Minus-strand initiation by brome mosaic virus replicase within the 3' tRNA-like structure of native and modified RNA templates. J Mol Biol. 1986 Feb 20;187(4):537–546. doi: 10.1016/0022-2836(86)90332-3. [DOI] [PubMed] [Google Scholar]
- Pandey N. B., Marzluff W. F. The stem-loop structure at the 3' end of histone mRNA is necessary and sufficient for regulation of histone mRNA stability. Mol Cell Biol. 1987 Dec;7(12):4557–4559. doi: 10.1128/mcb.7.12.4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perret V., Florentz C., Dreher T., Giege R. Structural analogies between the 3' tRNA-like structure of brome mosaic virus RNA and yeast tRNATyr revealed by protection studies with yeast tyrosyl-tRNA synthetase. Eur J Biochem. 1989 Nov 6;185(2):331–339. doi: 10.1111/j.1432-1033.1989.tb15120.x. [DOI] [PubMed] [Google Scholar]
- Pleij C. W. Pseudoknots: a new motif in the RNA game. Trends Biochem Sci. 1990 Apr;15(4):143–147. doi: 10.1016/0968-0004(90)90214-v. [DOI] [PubMed] [Google Scholar]
- Pogue G. P., Hall T. C. The requirement for a 5' stem-loop structure in brome mosaic virus replication supports a new model for viral positive-strand RNA initiation. J Virol. 1992 Feb;66(2):674–684. doi: 10.1128/jvi.66.2.674-684.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogue G. P., Marsh L. E., Connell J. P., Hall T. C. Requirement for ICR-like sequences in the replication of brome mosaic virus genomic RNA. Virology. 1992 Jun;188(2):742–753. doi: 10.1016/0042-6822(92)90529-x. [DOI] [PubMed] [Google Scholar]
- Pogue G. P., Marsh L. E., Hall T. C. Point mutations in the ICR2 motif of brome mosaic virus RNAs debilitate (+)-strand replication. Virology. 1990 Sep;178(1):152–160. doi: 10.1016/0042-6822(90)90388-8. [DOI] [PubMed] [Google Scholar]
- Rao A. L., Dreher T. W., Marsh L. E., Hall T. C. Telomeric function of the tRNA-like structure of brome mosaic virus RNA. Proc Natl Acad Sci U S A. 1989 Jul;86(14):5335–5339. doi: 10.1073/pnas.86.14.5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietveld K., Linschooten K., Pleij C. W., Bosch L. The three-dimensional folding of the tRNA-like structure of tobacco mosaic virus RNA. A new building principle applied twice. EMBO J. 1984 Nov;3(11):2613–2619. doi: 10.1002/j.1460-2075.1984.tb02182.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietveld K., Pleij C. W., Bosch L. Three-dimensional models of the tRNA-like 3' termini of some plant viral RNAs. EMBO J. 1983;2(7):1079–1085. doi: 10.1002/j.1460-2075.1983.tb01549.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietveld K., Van Poelgeest R., Pleij C. W., Van Boom J. H., Bosch L. The tRNA-like structure at the 3' terminus of turnip yellow mosaic virus RNA. Differences and similarities with canonical tRNA. Nucleic Acids Res. 1982 Mar 25;10(6):1929–1946. doi: 10.1093/nar/10.6.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmel P. RNA pseudoknots that interact with components of the translation apparatus. Cell. 1989 Jul 14;58(1):9–12. doi: 10.1016/0092-8674(89)90395-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern D. B., Radwanski E. R., Kindle K. L. A 3' stem/loop structure of the Chlamydomonas chloroplast atpB gene regulates mRNA accumulation in vivo. Plant Cell. 1991 Mar;3(3):285–297. doi: 10.1105/tpc.3.3.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern S., Weiser B., Noller H. F. Model for the three-dimensional folding of 16 S ribosomal RNA. J Mol Biol. 1988 Nov 20;204(2):447–481. doi: 10.1016/0022-2836(88)90588-8. [DOI] [PubMed] [Google Scholar]
- Takamatsu N., Watanabe Y., Meshi T., Okada Y. Mutational analysis of the pseudoknot region in the 3' noncoding region of tobacco mosaic virus RNA. J Virol. 1990 Aug;64(8):3686–3693. doi: 10.1128/jvi.64.8.3686-3693.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C. K., Draper D. E. Unusual mRNA pseudoknot structure is recognized by a protein translational repressor. Cell. 1989 May 19;57(4):531–536. doi: 10.1016/0092-8674(89)90123-2. [DOI] [PubMed] [Google Scholar]
- ten Dam E. B., Pleij C. W., Bosch L. RNA pseudoknots: translational frameshifting and readthrough on viral RNAs. Virus Genes. 1990 Jul;4(2):121–136. doi: 10.1007/BF00678404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Belkum A., Abrahams J. P., Pleij C. W., Bosch L. Five pseudoknots are present at the 204 nucleotides long 3' noncoding region of tobacco mosaic virus RNA. Nucleic Acids Res. 1985 Nov 11;13(21):7673–7686. doi: 10.1093/nar/13.21.7673. [DOI] [PMC free article] [PubMed] [Google Scholar]





