Abstract
Objectives. We examined whether automated electronic laboratory reporting of notifiable-diseases results in information being delivered to public health departments more completely and quickly than is the case with spontaneous, paper-based reporting.
Methods. We used data from a local public health department, hospital infection control departments, and a community-wide health information exchange to identify all potential cases of notifiable conditions that occurred in Marion County, Ind, during the first quarter of 2001. We compared traditional spontaneous reporting to the health department with automated electronic laboratory reporting through the health information exchange.
Results. After reports obtained using the 2 methods had been matched, there were 4785 unique reports for 53 different conditions during the study period. Chlamydia was the most common condition, followed by hepatitis B, hepatitis C, and gonorrhea. Automated electronic laboratory reporting identified 4.4 times as many cases as traditional spontaneous, paper-based methods and identified those cases 7.9 days earlier than spontaneous reporting.
Conclusions. Automated electronic laboratory reporting improves the completeness and timeliness of disease surveillance, which will enhance public health awareness and reporting efficiency.
Increased public awareness of and continued media attention on potential natural disease outbreaks have focused attention in the United States on the need for more comprehensive and timely disease surveillance.1,2 This awareness and attention arose from concerns regarding pandemic influenza,3 intentionally initiated outbreaks such as the anthrax cases in October 2001,4 and large regional outbreaks of Escherichia coli O157 and salmonella in 2006 caused by food contamination. As a result, the Office of the National Coordinator for Healthcare Information Technology of the Department of Health and Human Services has designated biosurveillance as a “breakthrough area” for a nationwide health information network, calling for the nation to “implement real-time nationwide public health event monitoring and support rapid response management across public health and care delivery communities and other authorized government agencies.”5
Public health monitoring of disease outbreaks, including reports of notifiable conditions from laboratories and health care providers to public health authorities, is fundamental to the prevention and control of population-based disease.6,7 Unfortunately, current approaches to public health monitoring rely on manual, spontaneous reporting processes that lead to incomplete and delayed event notification.8,9
Clinical laboratory reporting (as opposed to reporting by health care providers) has become increasingly valuable in disease surveillance.10,11 In fact, most communicable disease reports received by health departments originate from clinical laboratories.12 Schramm et al.13 and Vogt14 evaluated laboratory reports of 11 communicable diseases made to the Vermont Department of Health. They found 2035 reports on 1636 infectious cases, 71% originating from laboratories, 10% from nurses, 10% from physician offices, and 9% from other sources. Laboratories reported 100% of the time. Laboratories supplied more than 80% of initial reports for enteric infections and 48% of initial reports for hepatitis A and B but supplied only a minority of initial reports for invasive Haemophilus influenza and meningococcal disease.
Harkess et al. compared laboratory diagnosis records with physician and laboratory reports of shigellosis; overall, 69 of 80 positive cultures (86%) were reported, 3 (4%) by physicians and 67 (84%) by laboratories (1 culture was reported by both).15 Stanchert found that active laboratory-based surveillance for Neisseria meningitides resulted in twice as many reports as spontaneous methods.16,17
Although laboratories cannot provide the same amount of case data as direct care providers (e.g., data on whether a patient has been treated for a specific infection), laboratory reporting can offer strong evidence that further investigation and evaluation are warranted. In addition, unlike most provider practices, laboratories have the necessary information infrastructure and processes to facilitate reporting.18 To simplify and accelerate adoption of electronic laboratory reporting (ELR), the Centers for Disease Control and Prevention (CDC) has adopted clinical information standards and implementation guides for clinical data exchange.19,20
There have been few evaluations of automated ELR in comparison with spontaneous (including laboratory) reporting, but many believe that ELR can be more effective.21 In an evaluation of ELR for a limited number of notifiable diseases from regional reference laboratories in Hawaii, Effler et al. found a 2.3-fold increase in the number of reports; they also found that the ELR reports contained more information than the reports produced via conventional methods.11 Examining culture reports from hospitals belonging to the same health system, Panackal et al.22 did not find an increase in number of reports but did find that public health departments received reports an average of 4 days earlier. Both of these studies were small, assessed only a limited number of conditions, and evaluated systems that were not based on the clinical information standards identified by CDC.23
To address the limitations just described, we compared the completeness and timeliness of automated, standards-based electronic laboratory reports and spontaneous, paper-based reporting for a broad spectrum of notifiable conditions across a large population.
METHODS
Setting
We examined data from Indianapolis, Ind (population = 791926), for the first quarter of 2001. Blacks account for 24.2% of the city’s population, Asians account for 1.4%, and Hispanic individuals of other racial/ethnic backgrounds account for 3.9%. We included data on notifiable conditions from 3 sources: (1) the Indiana Network for Patient Care (INPC) ELR notifiable condition database, (2) hospital infection-control department databases and paper-based records of conditions reported from hospitals to public health officials, and (3) the Marion County Health Department notifiable condition databases.
Automated Electronic Laboratory Reporting
Investigators at the Regenstrief Institute have been developing electronic medical record systems for almost 30 years.24 INPC represents the latest step in this process.25 INPC now links 24 hospitals as well as physician practices, laboratories, radiology centers, and public health departments in central Indiana, creating a shared electronic medical record that includes encounter information, transcribed reports, medication histories, electrocardiograms, and other data such as laboratory results. A common database structure is used to store results, and results are converted to common terminologies so that they can be compared across sources.26
We have previously described the automated ELR component of INPC.27 Briefly, the software compares the Logical Observation Identifiers Names and Codes (LOINC) coded test result label for each result to the entries in the CDC notifiable condition mapping tables (http://www.cdc.gov/phin/index.html). When observations match, the software compares the SNOMED (http://www.snomed.org) coded result (from the fifth field of the observation segment of a result message delivered in the Health Level Seven clinical message standard format) with the entries in the notifiable-condition mapping table, and if the algorithm finds an entry that identifies a notifiable condition, the result is stored in a notifiable-condition database. The database is replicated to state and county health departments each day.
At the time of our study, INPC received laboratory test result data from 9 of the 13 hospitals in the county (60% of laboratory test results). We extracted data for the first quarter of 2001 from the INPC notifiable-condition database directly into an Microsoft Access 2002 (Microsoft Corp, Redmond, Wash) database for analysis (we hereafter refer to this source as “INPC”).
Spontaneous Reporting
Indianapolis is served by the Marion County Health Department. The Marion County Health Department uses Indiana State Department of Health case definitions (http://www.in.gov/isdh/publications/comm_dis_rule.pdf), which specify 55 laboratory notifiable conditions similar to those specified in most other states.28 Providers such as hospitals and physicians are required to report cases of notifiable conditions to the Marion County Health Department. Laboratories are required to report results that could indicate the presence of a notifiable condition to the Indiana State Department of Health but may also report them directly to the local health department. The Indiana State Department of Health forwards these results to the local health department in the relevant county.
At the time of the study, both providers and laboratories reported notifiable conditions exclusively in paper format. Hospital infection control departments are typically responsible for reporting notifiable conditions, including those identified through the hospital laboratory, to public health departments. Infection control practitioners rely on a variety of case-finding methods, including reports from clinicians and reports of laboratory results that indicate the presence of a notifiable condition.
We contacted the responsible infection control practitioner at each of the hospitals delivering laboratory data to INPC at the time of the study and arranged to obtain copies of the spreadsheets (in Microsoft Excel 2003 version 11 [Microsoft Corp, Redmond, Wash]) or documents detailing the notifiable condition reports they made to Marion Country Health Department and the Indiana State Department of Health. The hospital data were almost exclusively recorded on paper, so we manually entered them into a Microsoft Access database created for this analysis (we hereafter refer to this source as “hospital”).
The Marion Country Health Department receives case reports from many sources, including national and regional laboratories, hospitals (and their laboratories), and physicians. Department personnel record report data in condition-specific databases (e.g., tuberculosis or sexually transmitted diseases), review the data, obtain additional data, and then determine whether the report meets the case definition. The Marion Country Health Department exports the data from its condition-specific databases as delimited text files, which we imported into the analysis database (we hereafter refer to this source as “public health department”). The cases from both the hospital and public health department data sources included spontaneous reports from providers as well as laboratories, whereas the cases from INPC included only those identified using automated ELR.
Data Analysis
In an effort to identify all unique notifiable condition reports for Marion County, we used condition name and patient medical record number, name, and date of birth information to link reports across the 3 sources (public health department, hospital, and INPC). In addition to agreement on notifiable condition name, we required at least 1 of the following agreement criteria to declare a match between reports: (1) reporting institution and patient’s medical record number at the institution; (2) patient’s last name, first name, and birth month; or (3) patient’s last name and full birth date. To improve name matching accuracy, we compared first and last names after applying the New York State Identification and Intelligence System phonetic transformation algorithm. We applied the capture–recapture method to the 3 data sources (INPC, hospital, public health department) to identify the universe of notifiable disease reports.29
To apply this method, we took the logical union of all reports of notifiable conditions from these 3 sources as the universe of potential reports, and we compared the completeness of ELR with spontaneous reports made to Marion Country Health Department. We also computed reporting-time lag (difference between report date from Marion Country Health Department or hospital databases and INPC receipt date) because INPC receives reports from laboratories in real time and these reports typically represent the earliest explicit evidence of the existence of the notifiable condition.
Finally, we estimated the frequency with which specific data elements required by local public health practitioners were included in the reports identified using either method. To make these estimates, we determined the presence of completed fields through manual reviews (for paper reports) and database queries (for electronic laboratory reports). To measure the rate at which relevant data elements were included in the reports, we determined the presence or absence of data for each of the elements included on the Marion Country Health Department’s paper-based reporting form.
RESULTS
During the 3-month study period, 4785 unique reports were made across 53 different notifiable conditions. Chlamydia was the most prevalent condition reported, followed by hepatitis B, hepatitis C, and gonorrhea. Table 1 ▶ details reporting completeness by condition for spontaneous reports and automated ELR.
TABLE 1—
Reporting Completeness for Traditional Methods and Electronic Laboratory Reporting, by Condition: Indianapolis, Ind, 2001
| Condition | Spontaneous Reporting (Public Health Department), No. (%) | Electronic Laboratory Reporting (INPC), No. (%) | No. of Unique Reports |
| Acute herpes | 0 (0.0) | 0 (0.0) | 1 |
| Amebiasis | 0 (0.0) | 1 (100.0) | 1 |
| Blastomycosis | 0 (0.0) | 1 (100.0) | 1 |
| Campylobacteriosis | 3 (60.0) | 3 (60.0) | 5 |
| Chickenpox | 0 (0.0) | 8 (88.9) | 9 |
| Chlamydial infection | 398 (52.5) | 719 (94.9) | 758 |
| Coccidioidomycosis | 0 (0.0) | 1 (100.0) | 1 |
| Cryptococcosis | 0 (0.0) | 9 (100.0) | 9 |
| Cryptosporidiosis | 2 (66.7) | 2 (66.7) | 3 |
| Cytomegalovirus | 0 (0.0) | 116 (100.0) | 116 |
| Ebola | 0 (0.0) | 4 (100.0) | 4 |
| Elevated carboxyhemoglobin level | 0 (0.0) | 65 (100.0) | 65 |
| Enterococcus, vancomycin resistant | 0 (0.0) | 44 (73.3) | 60 |
| Escherichia coli O157:H7 infection | 1 (50.0) | 2 (100.0) | 2 |
| Fifth disease | 0 (0.0) | 9 (100.0) | 9 |
| Giardiasis | 3 (42.9) | 6 (85.7) | 7 |
| Gonorrhea | 228 (56.7) | 383 (95.3) | 402 |
| Haemophilus influenzae | 0 (0.0) | 138 (100.0) | 138 |
| Hepatitis A | 6 (4.0) | 146 (97.3) | 150 |
| Hepatitis B | 20 (3.1) | 635 (99.4) | 639 |
| Hepatitis C | 164 (34.1) | 463 (96.3) | 481 |
| Hepatitis D | 0 (0.0) | 2 (100.0) | 2 |
| Herpes simplex | 0 (0.0) | 35 (100.0) | 35 |
| Herpes simplex type 1 | 0 (0.0) | 37 (100.0) | 37 |
| Herpes simplex type 2 | 0 (0.0) | 61 (100.0) | 61 |
| Histoplasmosis | 4 (12.9) | 30 (96.8) | 31 |
| HIV | 23 (3.7) | 616 (99.5) | 619 |
| Human T-lymphotropic virus | 0 (0.0) | 4 (100.0) | 4 |
| Influenza | 0 (0.0) | 71 (96.0) | 74 |
| Lead exposure | 5 (29.4) | 17 (100.0) | 17 |
| Lyme disease | 0 (0.0) | 2 (100.0) | 2 |
| Meningitis: bacterial | 0 (0.0) | 31 (88.6) | 35 |
| Meningitis: fungal | 0 (0.0) | 0 (0.0) | 1 |
| Meningitis: viral | 0 (0.0) | 0 (0.0) | 2 |
| Meningococcal disease | 0 (0.0) | 2 (100.0) | 2 |
| Mumps | 0 (0.0) | 13 (92.9) | 14 |
| Mycobacterium, nontuberculous | 0 (0.0) | 68 (100.0) | 68 |
| Other | 0 (0.0) | 0 (0.0) | 19 |
| Poliomyelitis | 0 (0.0) | 2 (100.0) | 2 |
| Rubella | 0 (0.0) | 22 (100.0) | 22 |
| Salmonellosis, nontyphoid | 3 (21.4) | 12 (85.7) | 14 |
| Shigellosis | 27 (84.4) | 28 (87.5) | 32 |
| Sickle cell disease | 0 (0.0) | 62 (100.0) | 62 |
| Streptococcus pneumoniae | 30 (53.6) | 45 (80.4) | 56 |
| Streptococcus: group A | 5 (1.3) | 380 (100.0) | 380 |
| Streptococcus: group B | 3 (2.2) | 136 (99.3) | 137 |
| Syphilis | 18 (13.3) | 133 (98.5) | 135 |
| Tetanus | 0 (0.0) | 8 (100.0) | 8 |
| Toxoplasmosis | 0 (0.0) | 4 (100.0) | 4 |
| Trichomoniasis | 0 (0.0) | 41 (100.0) | 41 |
| Tuberculosis | 1 (25.0) | 4 (100.0) | 4 |
| Typhoid fever | 0 (0.0) | 3 (100.0) | 3 |
| Yersiniosis, nonplague | 0 (0.0) | 1 (100.0) | 1 |
Note. INPC = Indiana Network for Patient Care.
ELR identified 4.4 times as many cases (n = 4625; 95.4% of unique reports) as traditional spontaneous reporting methods (n = 944; 20% of unique reports; Table 1 ▶). Figure 1 ▶ illustrates the overlap in reports from the 3 sources. Examination of the 818 cases identified by both methods showed that ELR identified cases 7.9 days earlier than did spontaneous reporting (Table 2 ▶).
FIGURE 1—
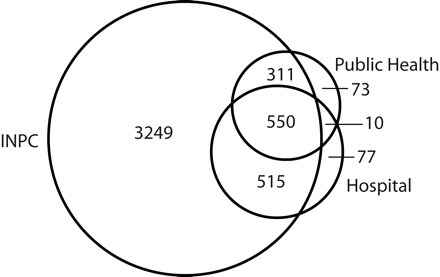
Overlap in 4785 unique cases identified by electronic laboratory reporting (Indiana Network for Patient Care [INPC]) and passive surveillance methods (public health department and hospital laboratories): Indianapolis, Ind, 2001.
Note. The 3 circles represent the cases identified by each of the 3 sources, and the numerals represent the number of cases for each area.
TABLE 2—
Reporting Timeliness for Traditional Methods and Electronic Laboratory Reporting, by Condition: Indianapolis, Ind, 2001
| Spontaneous Reporting (Public Health Department) | ||
| Condition | Average Lag Time, Days | No. of Cases |
| Campylobacteriosis | 0.0 | 1 |
| Chlamydial infection | 10.0 | 363 |
| Cryptosporidiosis | 0.0 | 1 |
| Escherichia coli O157:H7 infection | –1.0 | 1 |
| Giardiasis | 3.5 | 2 |
| Hepatitis A | 4.0 | 4 |
| Hepatitis B | 2.2 | 17 |
| Hepatitis C | 5.5 | 157 |
| Histoplasmosis | –8.5 | 4 |
| Salmonellosis, nontyphoid | –1.0 | 3 |
| Shigellosis | 0 | 24 |
| Streptococcus: group A | 1.2 | 5 |
| Streptococcus: group B | 20.0 | 2 |
| Syphilis | 4.4 | 17 |
Note. Negative values indicate that electronic laboratory reports were received later than were spontaneous reports on average. Only conditions for which timeliness could be calculated are included.
Table 3 ▶ details the frequency with which specific data elements were included in spontaneous and electronic laboratory reports. Patient name and medical record number were present at an equal rate for the 2 reporting methods, whereas physician name was more frequently present in spontaneous reports. Among the 18 fields examined, data for 5 were present more often in spontaneous reports, data for 10 were present more often in electronic laboratory reports, and data for 3 were present at an equal rate for the 2 methods.
TABLE 3—
Frequency With Which Selected Data Elements Were Included in Spontaneous and Electronic Laboratory Reports: Indianapolis, Ind, 2001
| Spontaneous Reports (Public Health Department), % | Electronic Laboratory Reports (INPC), % | |
| Patient | ||
| Medical record number | 100.0 | 100.0 |
| Last name | 100.0 | 100.0 |
| First name | 100.0 | 100.0 |
| Birth date | 83.0 | 93.1 |
| Gender | 78.0 | 99.5 |
| Street address | 82.0 | 66.3 |
| City | 54.0 | 66.3 |
| Zip code | 81.0 | 69.1 |
| Telephone number | 70.0 | 53.5 |
| Specimen | ||
| Collection date | 99.0 | 100.0 |
| Type | 27.0 | 41.2 |
| Physician | ||
| Last name | 83.0 | 55.7 |
| First name | 79.0 | 26.5 |
| Street address | 0.0 | 24.4 |
| City | 0.0 | 24.8 |
| State | 0.0 | 24.8 |
| Zip code | 0.0 | 24.4 |
| Telephone number | 0.0 | 4.0 |
Note. INPC = Indiana Network for Patient Care.
DISCUSSION
There were 2 key findings of our study. First, automated ELR results in more-complete and more-timely reporting of notifiable diseases than does traditional spontaneous reporting. Second, electronic reporting of notifiable conditions according to the standards recommended by CDC is feasible and scalable. These findings are particularly notable because they are based on nearly all notifiable conditions in a large population and reflect the performance of a system that relies on Public Health Information Network standards.
Completeness
We found an even larger increase in reports of notifiable cases than Effler et al. found in Hawaii.11 The reason may have been that we included a broader range of notifiable conditions, and reporting of sexually transmitted diseases and hepatitis is much more complete with automated ELR. In addition, we observed a significant degree of improved timeliness similar to that reported by Panackal et al.22 Of course, the importance of timeliness varies according to condition, with some conditions such as meningococcal meningitis demanding more-rapid reporting than, for example, Lyme disease.
We believe that the higher rate of completeness we observed with ELR resulted from a number of barriers being overcome, including lack of awareness on the part of providers, human error, lack of motivation, and ineffective processes. Laboratory personnel may not be as aware of notifiable conditions as hospital infection control practitioners, so they may not identify relevant positive test results. Even when laboratory personnel and infection control practitioners are aware that a positive result should be reported to the public health department, they may fail to report because the case has been overlooked because of human error or because they lack the motivation to report it. Poor system processes within laboratories may also contribute to underreporting. For example, a query that is intended to be initiated by hand each week may not be initiated when the technician is on vacation, or the query may lack the codes to identify all appropriate tests.
Timeliness
Traditional, spontaneous-reporting methods were more timely than were ELR for 3 notifiable conditions: Escherichi coli O157 infection, salmonellosis, and histoplasmosis. In several of these reports, the organisms isolated were confirmed and further characterized by public health laboratories before the hospital laboratories delivered the final microbiology reports through INPC. In the case of the remaining 13 conditions for which we could calculate timeliness, INPC was timelier than traditional reporting. In the case of certain conditions, more timely detection can reduce the mortality and morbidity resulting from an outbreak by allowing earlier public health intervention.30–32 Timely detection has also been strongly linked to the potential effectiveness of public health responses in cases of agents likely to be used to create an outbreak intentionally.33,34
Cost
Although we did not directly assess the cost of ELR, the cost is very low once standardized data are being captured through, for example, a health information exchange such as INPC. Software that costs approximately $40000 to develop initially can continuously monitor the data and report results at essentially no ongoing cost. In addition, because the software is written to operate on standardized data, it can be reused essentially as it is in other settings in which standardized data are captured. Of course, the improved completeness and timeliness of ELR reporting also lead to benefits in that public health interventions can be initiated at an earlier point, leading in turn to fewer lost workdays, fewer direct medical care costs, decreased probabilities that antimicrobial resistance will develop, and decreased mortality.
Lessons Learned
We learned a number of lessons that may help others who create ELR systems in the future. For example, creating an ELR system based on clinical information standards provided several benefits. One was that we were able to take direct advantage of CDC’s notifiable condition tables, which reduces the amount of maintenance required. By contrast, in the Effler et al. study,11 which used batch file transfer from laboratory systems, the systems failed to function almost one third of the time. In addition, we have been able to add ELR for new data sources simply by standardizing laboratory result messages. The notifiable condition processor then begins identifying notifiable conditions without further effort. Laboratories are readily able to provide the outbound HL7 messages that we require, although there are a number of issues with these messages, as outlined subsequently.
ELR incorrectly identified 4 reports of Ebola virus (the algorithm comparing organism names delivered from the laboratory as free text to SNOMED codes matched certain results to Ebola incorrectly). In addition, all 22 reports of rubella identified by ELR were simply seropositivity results from female patients screened for exposure as part of their prenatal care rather than because of suspicion that they had rubella.
Laboratory reporting regulations do not require that patients satisfy the full case definition. For example, a previously infected or immunized woman may have a positive rubella titer according to prenatal screening but does not meet the case definition. However, an infected individual will have a positive titer and does in fact meet the case definition. ELR alone cannot distinguish the positive rubella titers that represent cases and those that do not.
Because ELR can substantially increase the volume of reports, efforts must be made to ensure that most reports represent true cases. In some situations, data from clinical databases may be able to help distinguish the instances in which positive results are representative of cases. For example, a history of immunization for rubella would greatly increase the probability that a positive rubella titer does not represent an actual case. Likewise, a requirement for meeting the hepatitis case definition is a concomitant liver function test elevation rather than simply a positive viral antibody result.
Another issue that must be addressed is that of distinguishing new evidence of a notifiable condition from information about a known case based on a laboratory report. If a person with hepatitis B is retested (often because the clinician caring for the patient does not have the results of the previous test), this test needs to be identified as an additional test administered to a patient known to have the condition. The ELR system would include, in the data forwarded to the public health department, an indication that there is previous laboratory evidence of hepatitis B for this patient.
Although we were able to standardize results by mapping test names from multiple laboratories to LOINC codes, thereby simplifying analysis and processing, this process required approximately 6 person-months of effort on the part of an individual in each laboratory experienced in mapping. This effort would be distributed more widely in laboratories using LOINC codes as primary test result identifiers or at least as alternative identifiers; however, we have found that staff with specialized expertise are more efficient at performing this mapping work. The total amount of effort required is no different when laboratories are responsible for mapping; rather, the only change involves the source of the work. Taking advantage of mapping and using the data for other purposes help reduce the investment that must be supported by public health departments.
Microbiology cultures are the most challenging data to standardize for ELR because laboratories typically report results as freeform text (e.g., “positive,” “present,” or “Corynebacterium diphtheriae”) rather than SNOMED codes. Although we use a variety of parsing methods to map these results to SNOMED organism codes, this continues to be a source of inclusion errors (e.g., as with the Ebola cases described earlier) as well as exclusion errors (we recently identified an additional variant with respect to how methicillin-resistant Staphylococcus is reported, resulting in a marked increase in the number of cases identified). If laboratories used freely available standard SNOMED codes for microbiology findings, ELR would be less difficult and there would be fewer inappropriate cases identified.
In addition, many laboratories report the results of certain tests as negations—a list of organisms the laboratory did not find—as a way to explicitly communicate to the provider the organisms included in the laboratory tests. For example, laboratories typically report stool cultures from which they did not isolate any organisms as “no Shigella, salmonella, or E coli O157” rather than simply as “negative,” and thus the software must accommodate these negations as well.
Finally, in the case of a number of notifiable conditions, it would be particularly helpful if laboratories would use the appropriate HL7 fields to consistently flag results as normal or abnormal. When laboratories have available contextual information that allows them to flag results as abnormal, this allows the ELR system to improve the proportions of reports that represent actual cases. For example, if the stool culture reported as “no Shigella, salmonella, or E coli O157” were flagged as normal, then the logic for identifying notifiable cases could be greatly simplified.
We have not used all of the methods available to us to enhance information in electronic reports. For example, INPC receives patient registration data from hospitals that have complete patient demographic information on hand, which we could have used to augment the demographic data received from laboratories. We did add information from a database created through sources such as hospitals, medical associations, and the state licensing board in an attempt to improve the completeness of our provider data; however, we found that these data were often incorrect or out of date. Since then, we have invested considerable effort in creating a comprehensive, up-to-date, and accurate database that we can use to add provider demographic information to the results.
Limitations
Our study involved a number of limitations. Although we included hospital laboratories, which accounted for a large proportion of laboratory testing in the region, we did not include regional or national laboratories. We now include both of these types of laboratories and apply the same standards and processes used with hospital laboratories.
Some notifiable conditions are better suited for laboratory reporting than others. In particular, conditions that require a clinical context for diagnosis (such as meningitis) may be difficult to identify with a high degree of completeness and appropriateness using ELR alone. Patients with meningitis may have little or no laboratory evidence of disease, and not all patients with leukocytes in their cerebrospinal fluid have meningitis. In addition, some conditions require knowledge of multiple laboratory results, which can be difficult to implement.
INPC’s ELR system reports only final results. We made the decision not to include preliminary results because delivering a report that may subsequently change could complicate a public health practitioner’s workflow. In some cases, this decision results in significant reporting delays. Consider a hospital laboratory that transmits a preliminary report of Shigella sp isolated from a stool specimen. The isolate is subsequently sent to the Indiana State Department of Health laboratory; there the organism is typed, the result is sent back to the hospital laboratory, and the information is entered into the laboratory’s information system, which then transmits the final report.
As INPC’s ELR system has continued to operate and deliver information to the Indiana State Department of Health and local public health departments, these departments have continued to make increasing use of the data. For instance, they have developed processes in which they can use the data to initiate case investigations (e.g., of sexually transmitted diseases) and engage in population health surveillance (e.g., of lead levels). Developing an ELR reporting system requires more than standardizing and delivering the data to public health departments. It also requires adaptation of workflows, development of additional collaboration between providers and public health officials so that the data can be properly understood and interpreted, and creation of data analysis methods that will allow determination of the appropriate public health response.
Although current Indiana State Department of Health regulations allow laboratories to report electronically, many laboratories continue to report using traditional methods. (We recognize that markedly increasing reporting will place a greater burden on public health resources.) The Indiana State Department of Health is reviewing its guidance on this subject.
Conclusions
Automated ELR can improve the completeness and timeliness of laboratory reporting to public health departments and contribute to enhanced disease surveillance capability. Building this ELR capability is feasible and should facilitate rapid expansion. Finally, ELR allows tighter integration of public health information flows with clinical information flows, increasing the feasibility of creating a true nationwide health information network.
Acknowledgments
This work, conducted at the Regenstrief Institute, was supported by the National Library of Medicine (grants N01-LM-4-3410, N01-LM-6-3546, and G08 LM008232).
We gratefully acknowledge the assistance of the Marion County Health Department and the hospital infection control department staff members who helped us obtain the data and provided valuable reviews of the findings.
Human Participant Protection This study was approved by the institutional review board of Indiana University Purdue University at Indianapolis.
Peer Reviewed
Contributors J. M. Overhage originated the study, oversaw creation of the software, gathered the data, assisted in the analysis, and led the writing. S. Grannis assisted in the data collection, performed the analysis, and assisted in the writing. C. J. McDonald assisted in creation of the software and contributed to the analysis.
References
- 1.Henderson DA. The looming threat of bioterrorism. Science. 1999;283:1279–1282. [DOI] [PubMed] [Google Scholar]
- 2.Fine A, Layton M. Lessons from West Nile encephalitis outbreak in New York City, 1999: implications for bioterrorism preparedness. Clin Infect Dis. 2001;32:277–282. [DOI] [PubMed] [Google Scholar]
- 3.Falagas ME, Kiriaze IJ. Reaction to the threat of influenza pandemic: the mass media and the public. Crit Care. 2006;10:408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jernigan DB, Raghunathan PL, Bell BP, et al. Investigation of bioterrorism-related anthrax, United States, 2001: epidemiologic findings. Emerg Infect Dis. 2002;8:1019–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Office of the National Coordinator for Healthcare Information Technology. Harmonized use case for bio-surveillance. Available at http://www.hhs.gov/healthit/documents/BiosurveillanceUseCase.pdf. Accessed January 2, 2007.
- 6.Institute of Medicine. Emerging Infections: Microbial Threats to Health in the United States. Washington, DC: National Academy Press; 1992. [PubMed]
- 7.Chorba TL, Berkelman RL, Safford SK, Gibbs NP, Hull HF. Mandatory reporting of infectious diseases by clinicians. MMWR Morb Mortal Wkly Rep. 1990;39 (RR-9):1–17. [PubMed] [Google Scholar]
- 8.Doyle TJ, Glynn MK, Groseclose SL. Completeness of notifiable infectious disease reporting in the United States: an analytical literature review. Am J Epidemiol. 2002;155:866–874. [DOI] [PubMed] [Google Scholar]
- 9.Jajosky RA, Groseclose SL. Evaluation of reporting timeliness of public health surveillance systems for infectious diseases. BMC Public Health. 2004;4:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klaucke D, Buehler J, Thacker S, Parrish R, Trowbride F, Berkelman R. Guidelines for evaluating surveillance systems. MMWR Morb Mortal Wkly Rep. 1988;37:1–15. [Google Scholar]
- 11.Effler P, Ching-Lee M, Bogard A, Ieong M, Nekomoto T, Jernigan DB. Statewide system of electronic notifiable disease reporting from clinical laboratories: comparing automated reporting with conventional methods. JAMA. 1999;282:1845–1850. [DOI] [PubMed] [Google Scholar]
- 12.Godes JR, Hall WN, Dean AG, Morse CD. Laboratory-based disease surveillance: a survey of state laboratory directors. Minn Med. 1982;65:762–764. [PubMed] [Google Scholar]
- 13.Schramm M, Vogt R, Mamolen M. The surveillance of communicable disease in Vermont: who reports? Public Health Rep. 1991;106:95–97. [PMC free article] [PubMed] [Google Scholar]
- 14.Vogt R. Laboratory reporting and disease surveillance. J Public Health Manage Pract. 1996;2:28–30. [DOI] [PubMed] [Google Scholar]
- 15.Harkess JR, Gildon BA, Archer PW, Istre GR. Is passive surveillance always insensitive? An evaluation of shigellosis surveillance in Oklahoma. Am J Epidemiol. 1988;128:878–881. [DOI] [PubMed] [Google Scholar]
- 16.Standaert S, Lefkowitz I, Horan J, Huncheson R, Shaffner W. The reporting of communicable diseases: a controlled study of Neisseria meningitides and Haemophilus influenzae infections. Clin Infect Dis. 1995;20: 30–36. [DOI] [PubMed] [Google Scholar]
- 17.Backer HD, Bissell SR, Vugia DJ. Disease reporting from an automated laboratory-based reporting system to a state health department via local county health departments. Public Health Rep. 2001;116: 257–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McDonald CJ, Overhage JM, Dexter P, Takesue BY, Dwyer DM. A framework for capturing clinical data sets from computerized sources. Ann Intern Med. 1997; 127:675–682. [DOI] [PubMed] [Google Scholar]
- 19.Electronic Reporting of Laboratory Data for Public Health: Meeting Report and Recommendations. Atlanta, Ga: Centers for Disease Control and Prevention; 1997.
- 20.Electronic Reporting of Laboratory Information for Public Health: Microbiology Reporting Implementation Guide. Atlanta, Ga: Centers for Disease Control and Prevention; 2003.
- 21.Brennan PF. Recommendations for national health threat surveillance and response. J Am Med Inform Assoc. 2002;9:204–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Panackal AA, M’ikanatha NM, Tsui F-C, et al. Automatic electronic laboratory-based reporting of notifiable infectious diseases at a large health system. Emerg Infect Dis. 2002;8:685–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.M’ikanatha NM, Southwell B, Lautenbach E. Automated laboratory reporting of infectious diseases in a climate of bioterrorism. Available at: http://www.cdc.gov/ncidod/EID/vol9no9/02-0486.htm. Accessed November 5, 2007. [DOI] [PMC free article] [PubMed]
- 24.McDonald CJ, Overhage JM, Tierney WM. The Regenstrief Medical Record System: a quarter century experience. Int J Med Inform. 1999;54:225–253. [DOI] [PubMed] [Google Scholar]
- 25.Overhage JM, Tierney WM, McDonald CJ. Design and implementation of the Indianapolis Network for Patient Care and Research. Bull Med Libr Assoc. 1995; 83:48–56. [PMC free article] [PubMed] [Google Scholar]
- 26.McDonald CJ, Overhage JM, Barnes M, et al. The Indiana Network for Patient Care: a working local health information infrastructure. Health Aff. 2005; 224:1214–1220. [DOI] [PubMed] [Google Scholar]
- 27.Overhage JM, Suico J, McDonald CJ. Electronic laboratory reporting: barriers, solutions and findings. J Public Health Manage Pract. 2001;7:60–66. [DOI] [PubMed] [Google Scholar]
- 28.Roush S, Birkhead G, Koo D, Cobb A, Fleming D. Mandatory reporting of diseases and conditions by health care professionals and laboratories. JAMA. 1999;282:164–170. [DOI] [PubMed] [Google Scholar]
- 29.Wittes JT, Colton T, Sidel VW. Capture-recapture methods for assessing the completeness of case ascertainment when using multiple information sources. J Chronic Dis. 1974;27:25–36. [DOI] [PubMed] [Google Scholar]
- 30.Berkelman RL. Emerging infectious diseases in the United States, 1993. J Infect Dis. 1994;170: 272–277. [DOI] [PubMed] [Google Scholar]
- 31.Eisenberg MS, Bender TR. Botulism in Alaska, 1947 through 1974: early detection of cases and investigation of outbreaks as a means of reducing mortality. JAMA. 1976;235:35–38. [DOI] [PubMed] [Google Scholar]
- 32.Rice SK, Heinl RE, Thornton LL, Opal SM. Clinical characteristics, management strategies, and cost implications of a statewide outbreak of enterovirus meningitis. Clin Infect Dis. 1995;20:931–937. [DOI] [PubMed] [Google Scholar]
- 33.Kaufmann AF, Meltzer MI, Schmid GP. The economic impact of a bioterrorist attack: are prevention and postattack intervention programs justifiable? Emerg Infect Dis. 1997;3:83–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wein LM, Craft DL, Kaplan EH. Emergency response to an anthrax attack. Proc Natl Acad Sci U S A. 2003;100:4346–4351. [DOI] [PMC free article] [PubMed] [Google Scholar]


