SUMMARY
Proteins in the largest subset of AraC/XylS family transcription activators, including RhaS and RhaR, have C-terminal domains (CTDs) that mediate DNA-binding and transcription activation, and N-terminal domains (NTDs) that mediate dimerization and effector binding. The mechanism of the allosteric effector response in this family has been identified only for AraC. Here, we investigated the mechanism by which RhaS and RhaR respond to their effector, L-rhamnose. Unlike AraC, N-terminal truncations suggested that RhaS and RhaR don’t use an N-terminal arm to inhibit activity in the absence of effector. We used random mutagenesis to isolate RhaS and RhaR variants with enhanced activation in the absence of L-rhamnose. NTD substitutions largely clustered around the predicted L-rhamnose-binding pockets, suggesting that they mimic the structural outcome of effector binding to the wild-type proteins. RhaS-CTD substitutions clustered in the first HTH motif, and suggested that L-rhamnose induces improved DNA binding. In contrast, RhaR-CTD substitutions clustered at a single residue in the second HTH motif, at a position consistent with improved RNAP contacts. We propose separate allosteric mechanisms for the two proteins: Without L-rhamnose, RhaS doesn’t effectively bind DNA while RhaR doesn’t effectively contact RNAP. Upon L-rhamnose binding, both proteins undergo structural changes that enable transcription activation.
Keywords: transcription activation, AraC/XylS, allosteric, effector, rhamnose
INTRODUCTION
The RhaS and RhaR proteins are AraC/XylS family transcription activators of the Escherichia coli L-rhamnose catabolic regulon and are 30% identical to each other (Egan and Schleif, 1993; Egan and Schleif, 1994; Tate et al., 1992; Tobin and Schleif, 1987). RhaS activates transcription of the rhaBAD and rhaT operons, which encode the L-rhamnose catabolic enzymes and the L-rhamnose transport protein, respectively (Power, 1967; Tate et al., 1992). RhaR activates transcription of the rhaSR operon, which encodes RhaS and RhaR (Tobin and Schleif, 1987). The protein levels as well as the protein activities of RhaS and RhaR each increase in the presence of L-rhamnose, with the protein activities increasing on the order of 300-fold and 7-fold, respectively (Egan and Schleif, 1993; Tobin and Schleif, 1990b; Via et al., 1996) (unpublished results). The DNA binding sites for RhaS and RhaR dimers consist of two 17 bp half-sites separated by 16 or 17 bp, respectively, and overlapping the −35 promoter hexamer by four bp (Egan and Schleif, 1994; Tobin and Schleif, 1990a). In addition to the transcription activators RhaS and RhaR, the cAMP receptor protein (CRP) is also required for the full activation of all three of the rha operons (Egan and Schleif, 1993; Holcroft and Egan, 2000; Via et al., 1996).
The AraC/XylS family of transcription regulatory proteins are defined by a 100-amino-acid region of sequence similarity that comprises the DNA binding domain of the family members (Egan, 2002; Gallegos et al., 1993; Gallegos et al., 1997; Ramos, 1990; Tobin and Schleif, 1987). The majority of the proteins in the family, including RhaS and RhaR, consist of the conserved DNA binding domain, as well as at least one additional domain (Egan, 2002; Gallegos et al., 1997). The crystal structures of the AraC/XylS family DNA binding domains of MarA and Rob have been solved (MarA consists of only the DNA binding domain while Rob also has a second domain) (Kwon et al., 2000; Rhee et al., 1998). The MarA-DNA complex serves as our model for the RhaS- and RhaR-CTD structures given that its two helix-turn-helix (HTH) motifs each contact the DNA (Kwon et al., 2000; Rhee et al., 1998), similar to our findings for RhaS (Bhende and Egan, 1999). Our identification of several amino acid-base pair contacts in RhaS, allowed us to identify the orientation of the RhaS monomers at rhaBAD (Bhende and Egan, 1999). The conserved DNA binding domains of many AraC/XylS proteins also activate transcription by directly contacting the RNA polymerase (RNAP) α-subunit C-terminal domain and/or residues near the C-terminal end of σ70 (Dangi et al., 2004; Grainger et al., 2004a; Grainger et al., 2004b; Jair et al., 1995; Jair et al., 1996a; Jair et al., 1996b; Landini et al., 1997; Lonetto et al., 1998; Martin et al., 2002; Ruiz et al., 2001; Shah and Wolf, 2004). We’ve identified two residues in RhaS and one in RhaR and the residues in σ70 that they each contact to activate transcription (Bhende and Egan, 2000; Wickstrum and Egan, 2004).
The other domain of AraC/XylS proteins (usually the N-terminal domain, NTD) is not conserved throughout the family; however, proteins that also share sequence similarity with AraC in this domain make up the largest subset of the family. Among proteins in this subset, including RhaS and RhaR as well as XylS, MelR, and UreR, this domain is generally required for dimerization and/or effector binding. RhaS- and RhaR-NTDs have approximately 15% amino acid identity and 38% similarity with the AraC-NTD. The tertiary structure of the dimerization and effector-binding domain of AraC has been solved (Soisson et al., 1997a, b; Weldon et al., 2007), and serves as a model for the RhaS- and RhaR-NTD structures. Our results indicate that the RhaS and RhaR NTDs also function in both dimerization and effector (L-rhamnose) binding (Wickstrum et al., 2007) (Kolin, Hunjan and Egan, unpublished results). Our work also indicates that, similar to AraC, RhaS and RhaR may have flexible linker regions that connect their two domains (Carra and Schleif, 1993; Kolin et al., 2007). Given that the effector-binding site is physically separated from the DNA-binding and RNAP-contacting domain in these proteins, an allosteric mechanism must communicate the effector-binding status of each NTD to the respective CTD.
The molecular mechanism of the effector response has been well defined for AraC and its effector L-arabinose, and is referred to as the “light-switch” mechanism (Harmer et al., 2001; Reed and Schleif, 1999; Saviola et al., 1998; Wu and Schleif, 2001a, b). The key to this mechanism is a small group of residues at the very N-terminal end of AraC known as the “N-terminal arm”. The N-terminal arm binds over the L-arabinose binding pocket in the presence of L-arabinose (Soisson et al., 1997b), leaving the two AraC domains in each monomer flexibly connected, and allowing transcription activation from the araBAD promoter-proximal half-sites (Harmer et al., 2001; Seabold and Schleif, 1998; Wu and Schleif, 2001a). In the absence of arabinose, the arms instead bind to the C-terminal domain, rigidly connecting the domains so that a DNA loop forms that prevents transcription activation (Ghosh and Schleif, 2001; Ross et al., 2003; Saviola et al., 1998; Wu and Schleif, 2001a, b), (Lobell and Schleif, 1990; Seabold and Schleif, 1998).
In this study we sought to identify the mechanisms used by RhaS and RhaR to mediate their allosteric L-rhamnose responses. We tested N-terminal deletions of each protein and found that the L-rhamnose responses of RhaS and RhaR are most likely not mediated by N-terminal arms, and therefore may not be similar to the AraC mechanism. RhaS and RhaR variants were then identified that conferred increased activation in the absence of L-rhamnose compared to wild type. Our results suggest that the RhaS and RhaR NTDs likely undergo similar L-rhamnose-mediated structural changes. Substitutions conferring increased activation in the absence of L-rhamnose in the CTDs, however, were found in different regions of RhaS versus RhaR. We propose that in the absence of L-rhamnose, RhaS is limited in its ability to bind to DNA, whereas RhaR is limited in its ability to contact RNAP. Upon L-rhamnose binding, there are allosteric structural changes transmitted from the NTDs to the CTDs in RhaS and RhaR that overcome their respective limitations and thereby allow them to activate transcription.
RESULTS
Our first hypothesis was that the mechanisms of the RhaS and RhaR allosteric effector responses might be similar to that of AraC. In the AraC mechanism, the very N-terminal residues (the arm) of AraC bind to the CTD in the absence of arabinose to prevent transcription activation and cause formation of a DNA loop. Consistent with the arm inhibiting activity in the absence of effector, N-terminal deletions of seven to 20 residues (within the arm) led to increased basal activation by AraC (were constitutive) (Saviola et al., 1998; Soisson et al., 1997b). In spite of the lack of evidence for DNA looping by RhaS or RhaR, binding of an N-terminal arm to their CTDs could, in principle, prevent binding to the adjacent DNA half-sites from which they activate transcription without requiring binding to alternative distant sites. If residues at the very N-terminus of RhaS or RhaR performed functions similar to the AraC N-terminal arm, then one or more N-terminal truncations would be predicted to result in activation of transcription in the absence of L-rhamnose to higher basal levels than wild-type RhaS or RhaR.
N-terminal truncations of RhaS and RhaR
Truncated variants of RhaS were assayed for in vivo transcription activation of a rhaBAD promoter that included the RhaS binding site but not the CRP binding site. In the absence of L-rhamnose, we found that the variants activated transcription to levels comparable to or lower than wild-type RhaS, indicating that none of the truncations conferred a constitutive phenotype (Table 1). In the presence of L-rhamnose, all of the truncations resulted in large defects in the ability of RhaS to activate transcription. Western blots (data not shown) as well as the ability of the variants to activate to nearly wild-type levels in the absence of L-rhamnose support the conclusion that the altered activity was not the result of low protein levels. We also constructed three additional RhaS deletions with intermediate endpoints and found that none of these conferred constitutive phenotypes (data not shown). Overall, these results suggest that the mechanism of the RhaS L-rhamnose response does not involve an N-terminal arm inhibiting activity in the absence of L-rhamnose, and thereby differs from the AraC L-arabinose response.
TABLE 1.
Transcription activation by RhaS and RhaR N-terminal deletion variants
| RhaS or RhaR | β-Galactosidase Activity* | |
|---|---|---|
| Variant | (−) L-rhamnose | (+) L-rhamnose |
| A. RhaS variants | ||
| Wild type | 20 | 1070 |
| Δ2-7 | 14 | 107 |
| Δ2-13 | 23 | 35 |
| Δ2-19 | 12 | 16 |
| B. RhaR variants | ||
| Wild type | 220 | 528 |
| Δ2-6 | 188 | 514 |
| Δ2-12 | 177 | 427 |
| Δ2-18 | 190 | 494 |
| Δ2-24 | 144 | 277 |
| Δ2-29 | 210 | 484 |
| Δ2-34 | 108 | 394 |
| Δ2-40 | 33 | 64 |
| Δ2-46 | 2.6 | 10 |
| Δ2-52 | 0.4 | 0.5 |
β-Galactosidase activity (in Miller Units) was measured from a single-copy lacZ fusions in strain SME1087 (λΦ(rhaB-lacZ)Δ226 rhaS) (A) or strain SME1076 (λΦ(rhaS-lacZ)Δ216 Δ(rhaSR)::Km) (B). Strains were transformed with plasmids (in vector pHG165) encoding either wild-type or N-terminal deletion variants of RhaS or RhaR, as indicated. Cultures were grown in MOPS growth medium containing glycerol and ampicillin, with or without L-rhamnose as indicated. Standard errors were less than 30% of the average units. The activity from each fusion in the absence of plasmid-encoded RhaS or RhaR was less than one Miller Unit.
RhaR has 33 additional N-terminal residues compared with RhaS, so we constructed a larger number of N-terminal deletions and tested their ability to activate transcription from the rhaSR promoter. None resulted in increased activation in the absence of L-rhamnose (Table 1). In addition, deletion of the entire 33 residue N-terminal RhaR extension relative to RhaS had very little affect on RhaR activity. The apparent dispensability of the RhaR N-terminal extension is consistent with the observation that most RhaR orthologs outside of E. coli and Shigella spp. do not have this extension. Deletion of the first 40 or more RhaR residues resulted in increasingly large losses of activity both in the presence and absence of L-rhamnose, and western blots indicated that these same truncations were detected at substantially lower levels than wild-type RhaR (data not shown). Therefore, while the instability of the longest truncations introduces some uncertainty, we conclude that RhaR most likely does not use an N-terminal arm to mediate its response to L-rhamnose.
Isolation of partially constitutive RhaS and RhaR variants
The AraC allosteric response to effector is the only mechanism that has been well characterized in the AraC/XylS family. In addition, our previous results suggested that the RhaS and RhaR linkers are not central to the signal transmission from the NTDs to the CTDs (Kolin et al., 2007). We were therefore unable to hypothesize which residues might be important for the RhaS and RhaR allosteric effector responses, and turned to random mutagenesis. To avoid the problem of distinguishing interesting mutations from the large background of those that reduced activity for reasons unrelated to the effector response, we chose to screen for RhaS and RhaR variants with elevated transcription activation in the absence of L-rhamnose. We used PCR amplification to introduce random mutations, and then transformed the cloned genes into strains carrying single-copy translational fusions to lacZ of either a rhaBAD promoter (for RhaS) or a rhaSR promoter (for RhaR). For simplicity, all promoters lacked the upstream CRP binding site. The level of lacZ expression from these fusions was such that, when plated on media containing X-gal, wild-type transformants yielded white colonies in the absence of L-rhamnose and blue colonies in the presence of L-rhamnose. We therefore screened for blue colonies in the absence of L-rhamnose.
We screened approximately 240,000 clones obtained from 90 independent PCR amplifications of rhaS, and identified blue colonies in 69 of the amplifications. Early in the screening process we began to isolate independent mutants (from separate PCR amplifications) with identical substitutions. Subsequently, we used β-galactosidase assays to identify candidates whose activation levels appeared to differ from those already isolated, in an attempt to isolate as many unique substitutions as possible. Based on the assay results, we sequenced a total of 32 candidates, each from an independent PCR pool. We isolated nine unique RhaS variants, five of which were isolated at least twice (accounting for 19 of the 32 candidates) (Tables 2 and 3). The 13 remaining isolates each had one substitution that was identical to one of the nine unique substitutions as well as a second substitution. In each case, the β-galactosidase activity indicated that the second substitution did not contribute to the phenotype, so we did not further analyze these isolates. The nine unique variants had substitutions that were approximately evenly divided between RhaS-NTD and RhaS-CTD (Tables 2 and 3). These results indicate that, at least within the mutation spectrum of the PCR mutagenesis, a limited number of substitutions could confer a phenotype of increased activity in the absence of L-rhamnose upon RhaS.
TABLE 2.
Transcription activation by partially constitutive variants of RhaS with substitutions that map to RhaS-NTD
| β-Galactosidase Activityc | ||||
|---|---|---|---|---|
| Expt | RhaS varianta | No. of isolatesb | (−) L-rhamnose | (+) L-rhamnose |
| A | Wild type | 0.55 | 218 | |
| F28L/F50L | 1 | 57 | 61 | |
| E37K | 2 | 5.6 | 3.9 | |
| T56I | 2 | 6.1 | 360 | |
| N83H | 1 | 2.9 | 2.7 | |
| P141L | 3 | 12 | 304 | |
| B. | Wild type | 0.34 | 206 | |
| F28L | 0.30 | 1.3 | ||
| F50L | 0.38 | 3.6 | ||
Wild type and RhaS variants were encoded on pHG165 transformed into SME3000 (λΦ(rhaB-lacZ)Δ84 Δ(rhaSR)::Km).
Number of times this substitution was independently isolated in random mutagenesis screen.
Cultures were grown in MOPS growth medium containing glycerol plus ampicillin with or without L-rhamnose as indicated. β-Galactosidase activity (in Miller Units) was assayed. Standard errors were less than 20% of the average units, except for a few of the samples with activities below 3 Miller units. These had errors up to 31% of the average units.
TABLE 3.
Transcription activation by partially constitutive variants of RhaS with substitutions that map to RhaS-CTD
| β-Galactosidase Activityc | ||||
|---|---|---|---|---|
| Expt | RhaS varianta | No. of isolatesb | (−) L-rhamnose | (+) L-rhamnose |
| A | Wild type | 0.55 | 218 | |
| F184Y/Q207R | 1 | 8.6 | 8.9 | |
| L201R | 6 | 54 | 329 | |
| L208F | 2 | 5.6 | 285 | |
| Q210R | 1 | 2.6 | 326 | |
| B | Wild type | 0.34 | 206 | |
| F184Y | 0.69 | 218 | ||
| Q207R | 0.73 | 243 | ||
Wild type and RhaS variants were encoded on pHG165 transformed into SME3000 (λΦ(rhaB-lacZ)Δ84 Δ(rhaSR)::Km).
Number of times this substitution was independently isolated in random mutagenesis screen.
Cultures were grown in MOPS growth medium containing glycerol plus ampicillin with or without L-rhamnose as indicated. β-Galactosidase activity (in Miller Units) was assayed. Standard errors were less than 20% of the average units, except for a few of the samples with activities below 3 Miller units. These had errors up to 31% of the average units.
To identify RhaR variants, we screened approximately 170,000 clones derived from 70 independent rhaR PCR amplifications. Of the twenty-nine colonies that were more blue than wild type in the absence of L-rhamnose (each from an independent PCR amplification), 15 had two-fold or higher activity than wild-type RhaR in the absence of L-rhamnose (data not shown). Full-gene sequencing showed that 13 of the isolates encoded a substitution at residue T279, in RhaR-CTD, changing the wild-type Thr to either Ala (10 times) or Ser (3 times). Some of these had one or more additional substitutions; however, all of them activated transcription to levels that were the same (within error) as RhaR T279A and T279S alone, so those with multiple substitutions were not further analyzed (data not shown). We also found that RhaR T279A and T279S activated to the same levels as each other (data not shown), therefore we proceeded only with RhaR T279A. Only two of the 15 independent RhaR variants did not have a substitution at RhaR T279. Both of these had two substitutions, with all four located outside of RhaR-CTD (in either RhaR-NTD or the unstructured linker between the two domains). Therefore, among the partially constitutive RhaR variants there were only four unique variants, and we further analyzed three of them: F61L/W75R; H165R/D197G; and T279A.
Partially constitutive RhaS variants with substitutions in RhaS-NTD
Among the nine RhaS variants with increased activity in the absence of L-rhamnose, five had substitutions within the L-rhamnose-binding and dimerization domain, RhaS-NTD. Table 2 shows the ability of these RhaS variants to activate transcription. In the absence of L-rhamnose, they activated transcription to levels from six- to more than 100-fold higher than wild-type RhaS. Two of the variants, RhaS T56I and P141L, were able to respond to L-rhamnose, and activated transcription in the presence of L-rhamnose to levels that were somewhat higher than wild-type RhaS. In contrast, the other three variants, RhaS E37K, N83H and the variant with two substitutions, F28L/F50L, were unable to activate transcription to higher levels in the presence of L-rhamnose compared with their respective activities in the absence of L-rhamnose, and therefore we refer to them as uninducible.
We used the structure of AraC-NTD in the presence of L-arabinose (Soisson et al., 1997a) as a model to identify the positions of these RhaS residues (Fig. 1A). All of the substitutions were located within the predicted RhaS β-barrel, relatively near the sugar-binding pocket, with the exception of RhaS P141L. The RhaS substitutions do not appear to form a surface-exposed cluster of residues that might define a site of contact between RhaS-NTD and RhaS-CTD. However, note that the RhaS-CTD would connect to this model of the RhaS-NTD at the upper right corner (as drawn). Given that a second monomer of RhaS is expected to dimerize along the α-helical face of the structure, the RhaS-CTD might be expected to sit above the RhaS-NTD in this orientation, and the RhaS substitutions might provide clues to structural changes transmitted from the L-rhamnose binding pocket toward RhaS-CTD. Western blot analysis (data not shown) indicated that the protein levels of RhaS P141L were at least several fold higher than wild-type RhaS, perhaps explaining the increased activity of this variant, whereas none of the remaining variants were detected at higher levels than wild-type RhaS. We separated the two substitutions in RhaS F28L/F50L and found that neither alone conferred increased activation in the absence of L-rhamnose (Table 2B), indicating that both of these substitutions were necessary for the constitutive phenotype.
Figure 1.
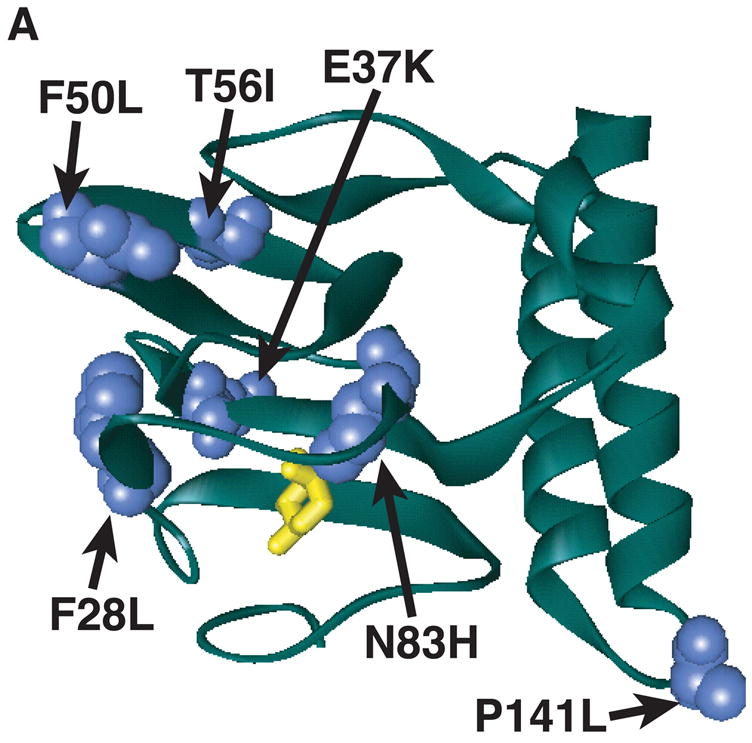
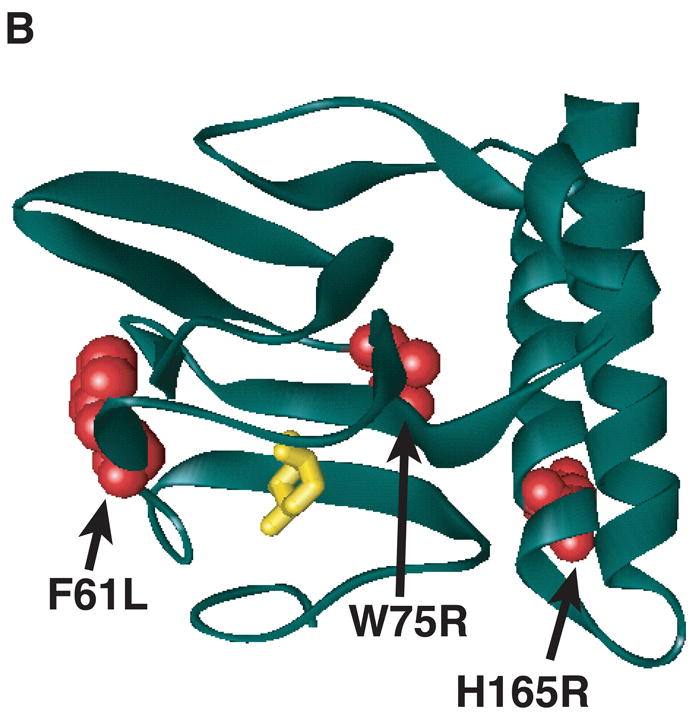
Model of substitutions leading to a partially constitutive phenotype within RhaS- or RhaR-NTD. The model for the structure of RhaS- or RhaR-NTD is the structure of one monomer of AraC-NTD in a ribbon representation (green) in the presence of L-arabinose (yellow stick model) (Soisson et al., 1997b). (A) The positions of RhaS-NTD substitutions are shown in blue as space filling representations. (B) The positions of RhaR-NTD substitutions are shown in red as space filling representations. RhaS F28L/F50L, RhaR F61L/W75R, and RhaR H165R/D197G substitutions were isolated as double mutants. RhaR D197G is not shown, since it is located just beyond the structured residues.
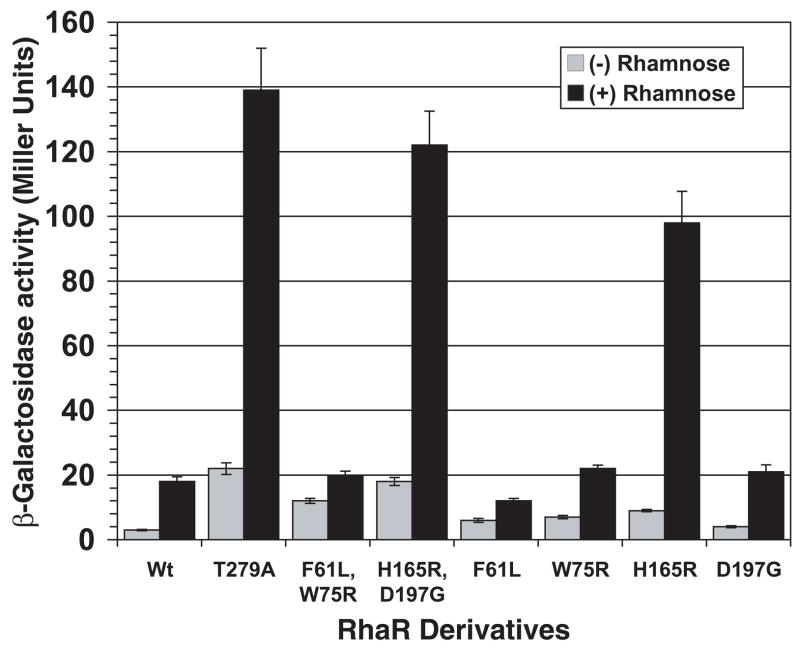
Partially constitutive RhaR variants with substitutions in or near RhaR-NTD
The RhaR variants F61L/W75R and H165R/D197G activated transcription of rhaS-lacZ to levels that were approximately 4- and 6-fold higher, respectively, than wild-type RhaR in the absence of L-rhamnose (Fig. 2). In the presence of L-rhamnose, RhaR F61L/W75R activated to approximately the same level as wild-type RhaR, while H165R/D197G activated to a level 7-fold higher than wild-type RhaR. Using the structure of AraC-NTD in the presence of L-arabinose (Soisson et al., 1997a) as a model we found that RhaR F61L and W75R both mapped within the β-barrel sugar-binding subdomain of AraC-NTD (Fig. 1B). The H165R substitution of RhaR H165R/D197G mapped to the first of the two predicted α-helices of RhaR-NTD (Fig. 1B). While this residue does not map to the β-barrel, it is in position to potentially interact with the β-barrel. The other substitution in this variant, D197G, was located three residues beyond the structured residues of the AraC-NTD model (Soisson et al., 1997b), but within the region defined biochemically as the minimal AraC-NTD (Eustance et al., 1994). This residue is therefore predicted to lie either at the very end of RhaR-NTD or at the beginning of the linker connecting the RhaR domains. Western blots showed that both of the RhaR variants were expressed at levels similar to or slightly lower than wild-type RhaR (data not shown); therefore, increased protein levels did not explain their increased basal activity relative to wild-type RhaR.
Figure 2.
Transcription activation by partially constitutive RhaR variants. Plasmids expressing wild-type RhaR or variants were transformed into strain SME3160 (Φ(rhaS-lacZ)Δ85 Δ(rhaSR)::Km), the cells were grown with or without L-rhamnose, as indicated, and β-galactosidase activity was measured.
We separated the two double mutants into single mutants in order to determine whether both substitutions in the RhaR variants were required for their increased activity in the absence of L-rhamnose. RhaR F61L and W75R each had somewhat higher activation than wild-type RhaR in the absence of L-rhamnose, but both substitutions were required for the full activity of RhaR F61L/W75R in the absence of L-rhamnose (Fig. 2). RhaR H165R activated to higher levels than wild-type RhaR in the absence of L-rhamnose, whereas RhaR D197G did not have significantly higher activation than wild-type RhaR (Fig. 2); but may have a subtle effect on transcription activation in combination with RhaR H165R. RhaR F61L was not fully inducible to the wild-type level in the presence of L-rhamnose, while all of the other single variants were L-rhamnose inducible to at least wild-type levels (Fig. 2).
Partially constitutive RhaS variants with substitutions in RhaS-CTD
Four RhaS variants with increased activity in the absence of L-rhamnose had substitutions in the DNA-binding and transcription activation domain (RhaS-CTD). Table 3 shows the amino acid substitutions found in each of these RhaS variants as well as the level of transcription activation by each. These variants activated transcription in the absence of L-rhamnose to levels ranging from five- to nearly 100-fold higher than wild-type RhaS. Of these variants, only RhaS F184Y/Q207R was uninducible by L-rhamnose. Each of the other variants was able to respond to L-rhamnose, and all activated to levels that were somewhat higher than that of wild-type RhaS. We separated the two substitutions in RhaS F184Y/Q270R, and found that neither alone conferred the phenotype of the parent (Table 3B). Each of them conferred only a very small increase in activity in the absence of L-rhamnose, and the uninducible phenotype of the parent was also lost, indicating that both substitutions were required for the phenotype.
Substitutions in RhaS-CTD could, in principal, result in increased activity in the absence of L-rhamnose due to loss-of-function changes, such as the loss of inhibitory domain-domain contacts. Alternatively, they could result in increased activity due to gain-of-function changes, such as improved DNA binding, interactions with RNAP, or protein folding/stability. Improved DNA binding appeared likely given that all of the RhaS-CTD variants include a substitution in the recognition helix of the first HTH motif (Fig. 3A); three out of four of these residues were changed to positively charged Arg residues; and we have previously shown that nearby residues R202 and R206 make base-specific contacts with the DNA (Bhende and Egan, 1999). The gain-of-function possibilities in RhaS-CTD are predicted to exhibit an altered phenotype (relative to the appropriate wild type) both in full-length RhaS and in RhaS(163-278) (which is RhaS-CTD alone, residues 163-278). In contrast, the loss-of-function possibilities are predicted to exhibit an altered phenotype (relative to the appropriate wild type) in full-length RhaS, but not in RhaS(163-278). Therefore, we could distinguish these possibilities by subcloning the substitutions into truncated rhaS genes that encode RhaS(163-278).
Figure 3.
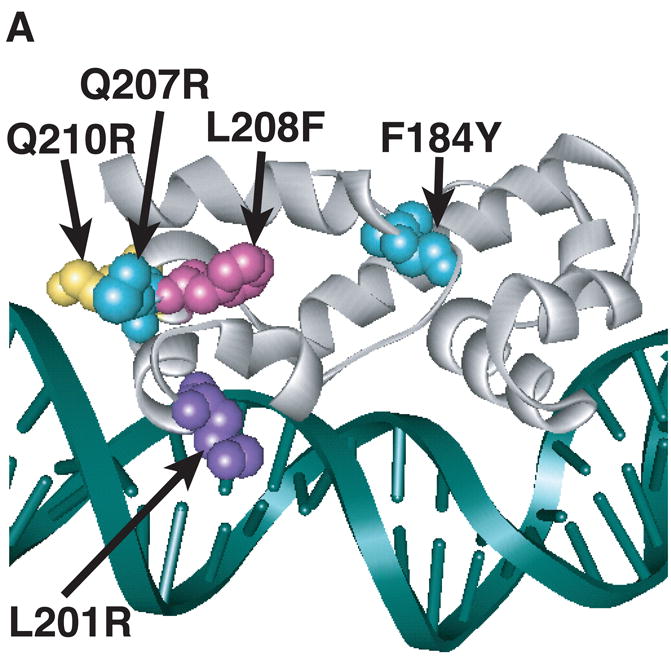
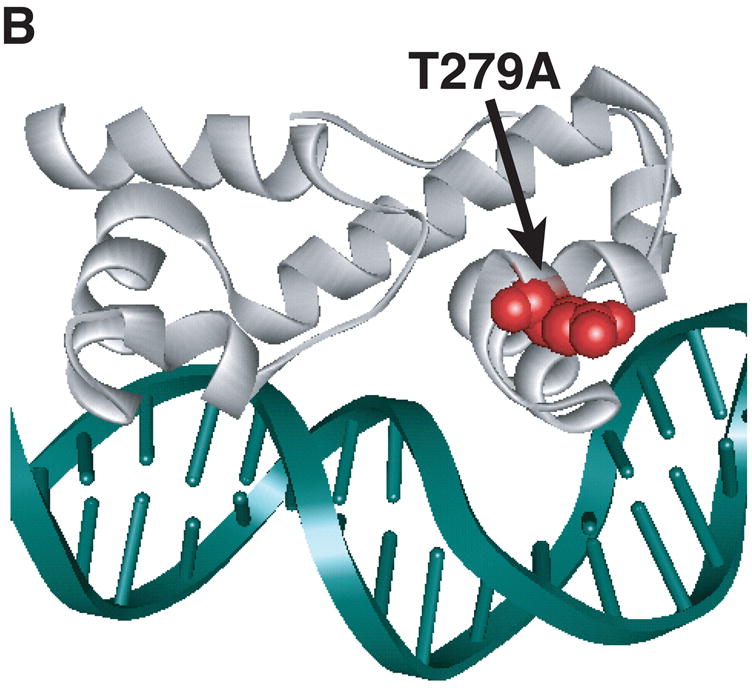
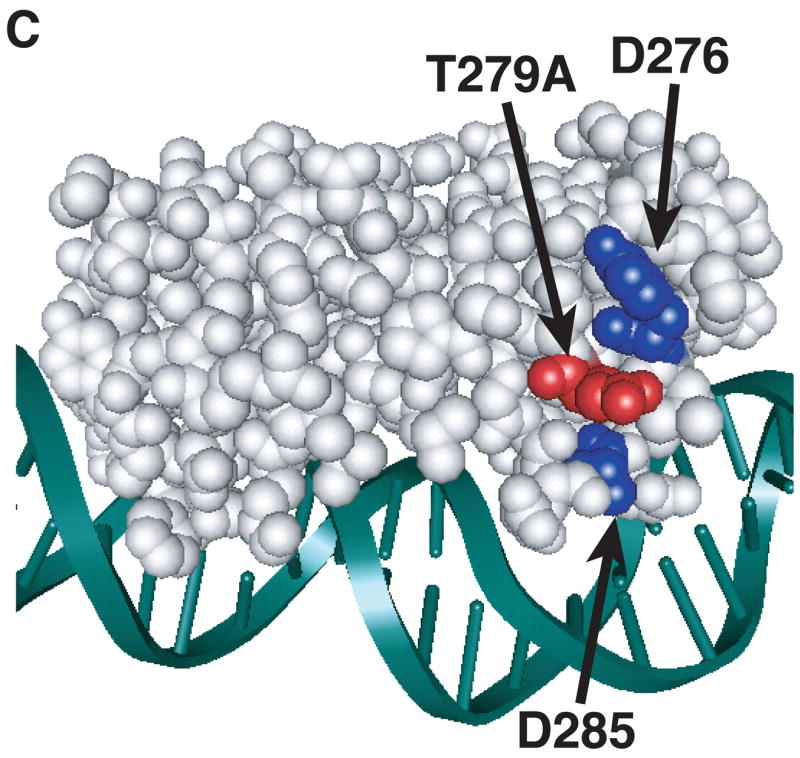
Model of substitutions leading to a constitutive phenotype within RhaS-CTD and RhaR-CTD. The structure of MarA in complex with DNA is used as a model for RhaS-CTD and RhaR-CTD (Rhee et al., 1998), with the DNA in green and MarA in light gray. The overlap with the -35 element is on the right in all cases. (A) The positions of the RhaS-CTD substitutions are shown in various colors as space filling representations on a ribbon model of MarA. The F184Y and Q207R substitutions were isolated as a double mutant. (B) The position of the RhaR-CTD substitution is shown in red as a space-filling representation on a ribbon model of MarA. (C) The position of residue RhaR T279 is shown relative to the positions of residues shown to contact RNAP in RhaS and RhaR. RhaR D276/RhaS D241 and RhaS D250 (that aligns with RhaR D285) have been shown to contact RNAP and are shown in blue (Bhende and Egan, 2000; Wickstrum and Egan, 2004). RhaR T279 is shown in red, all on a space-filling representation of MarA.
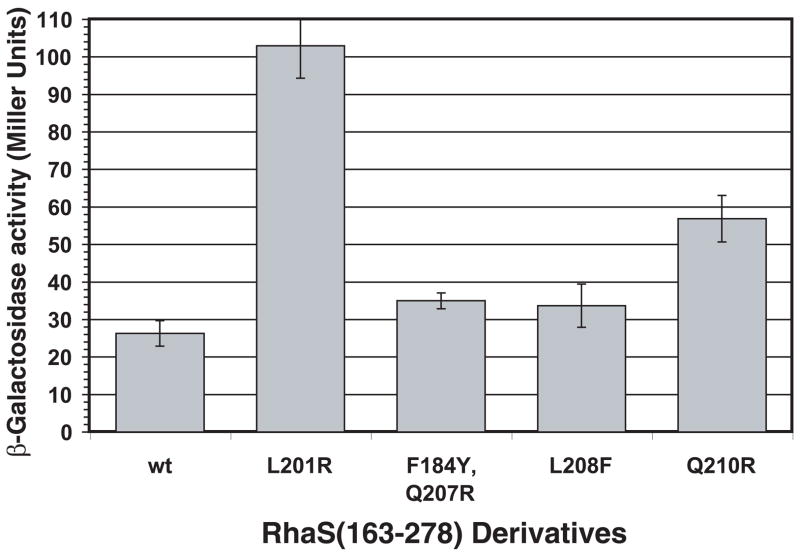
Figure 4 shows the levels of transcription activation by RhaS(163-278) variants carrying each of the substitutions. Three of the RhaS(163-278) variants, L201R, Q210R and F184Y/Q207R, showed significantly higher (1.7- to 4-fold) levels of transcription activation than wild-type RhaS(163-278). The fourth variant (L208F) activated transcription to a slightly higher level than wild-type RhaS(163-278), but this difference was not statistically significant. These results suggest that the substitutions within RhaS-CTD most likely all represent domain autonomous gain-of-function changes in RhaS, and based on their positions, likely confer increased DNA-binding upon RhaS. These results suggest a model in which RhaS is unable to effectively bind DNA in the absence of rhamnose, which is entirely consistent with the previous finding that RhaS DNA binding was only detectable in the presence of L-rhamnose (Egan and Schleif, 1994).
Figure 4.
Transcription activation by variants of the RhaS-CTD expressed alone (RhaS(163-278)). Wild-type RhaS(163-278) or variants (plasmid-encoded) in strain SME 3000 (Φ(rhaB-lacZ)Δ84, Δ(rhaSR)::Km) were assayed for transcription activation of the single copy lacZ fusion. Cultures were grown in minimal glycerol medium with ampicillin and without L-rhamnose.
Partially constitutive RhaR variants with substitutions within RhaR-CTD
All 13 RhaR-CTD variants with increased activity in the absence of L-rhamnose had substitutions at residue T279, and activated transcription of rhaS-lacZ to a level approximately 7-fold higher than wild-type RhaR in the absence of L-rhamnose (Fig. 2). This residue mapped to the stabilizing helix of the second RhaR HTH motif (Fig. 3B). The alignment of this residue with AraC and its position on the MarA structure suggest two different hypotheses to explain the phenotype of RhaR T279A. RhaR T279 aligns with AraC R251, which has been proposed to interact with residues in the N-terminal arm of AraC (Wu and Schleif, 2001b). Although our results suggest that the very N-terminal residues of RhaR are not involved in its L-rhamnose response (Table 1), one hypothesis is that RhaR T279 could interact with a different region of RhaR-NTD as part of its L-rhamnose response. A second hypothesis was based on the location of RhaR T279 on the surface of RhaR-CTD between residues D276 and D285, which we’ve previously identified as sites of contact between RhaS and RhaR and the σ70 subunit of RNAP (Bhende and Egan, 2000; Wickstrum and Egan, 2004) (Fig. 3C). Therefore, our second hypothesis was that the L-rhamnose-independent phenotype of RhaR T279A might be due to increased interaction with the σ70 subunit of RNAP, perhaps by decreasing steric inhibition imposed by T279 in the absence of L-rhamnose.
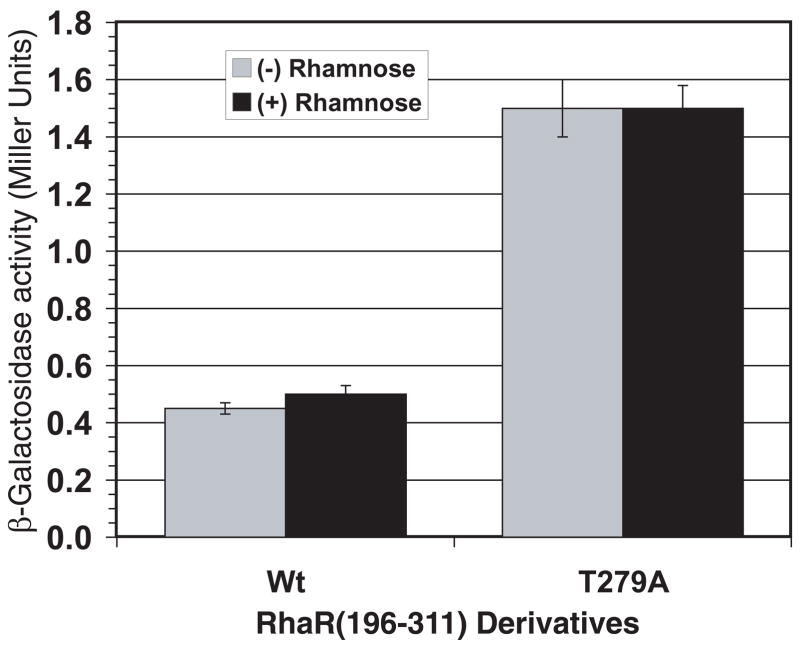
In order to distinguish these two hypotheses for the phenotype of RhaR T279, we subcloned the genes encoding wild-type RhaR and RhaR T279A such that only the CTD of RhaR was expressed (RhaR(196-311), residues 196-311). Similar to the RhaS case above, gain-of-function substitutions that resulted in improved DNA binding, interactions with RNAP, or protein folding/stability are predicted to have an altered phenotype relative to the appropriate wild type in both full-length RhaR and RhaR(196-311). On the other hand, loss-of-function changes that result in the loss of inhibitory domain-domain contacts that altered inter-domain contacts are predicted to have an altered phenotype relative to the appropriate wild type in full-length RhaR but not in RhaR(196-311). Figure 5 shows the ability of RhaR(196-311) T279A to activate rhaS-lacZ expression compared with wild-type RhaR(196-311). As expected, neither wild-type RhaR(196-311) nor RhaR(196-311) T279A was able to respond to L-rhamnose. We found that RhaR(196-311) T279A activated transcription to significantly higher levels (3-fold) than wild-type RhaR(196-311) in the absence of L-rhamnose. This suggests that the increased activity of RhaR T279A (in full-length RhaR) is unlikely to be due to inter-domain interactions. Our RhaR results, therefore, suggest a model in which RhaR is unable to effectively contact RNAP in the absence of L-rhamnose. This model is consistent with the previous finding that RhaR is able to bind to DNA but is not able to activate transcription in the absence of L-rhamnose (Tobin and Schleif, 1990a, b).
Figure 5.
Transcription activation by RhaR T279A in RhaR-CTD expressed alone (RhaR(196-311)). Wild-type RhaR(196-311) or RhaR(196-311) T279A (plasmid-encoded) in strain SME3160 (Φ(rhaS-lacZ)Δ85, Δ(rhaSR)::Km) were assayed for transcription activation of the single copy lacZ fusion. Cultures were grown in minimal glycerol medium with ampicillin and with or without L-rhamnose, as indicated.
DISCUSSION
RhaS and RhaR do not use “light-switch” allosteric effector mechanisms
The goal of this study was to identify the allosteric mechanisms used by the RhaS and RhaR proteins to respond to their common effector, L-rhamnose. Our first hypothesis was that the RhaS and RhaR mechanism might involve inhibition of activity by residues at their very N-termini in the absence of effector. This would be similar to the N-terminal arm that mediates the L-arabinose “light-switch” mechanism in AraC (Harmer et al., 2001; Reed and Schleif, 1999; Saviola et al., 1998). However, unlike AraC, deletion of N-terminal residues of RhaS and RhaR did not result in effector-independent phenotypes (Table 1), suggesting that the mechanism of their effector responses differ from that of AraC.
Alternative allosteric effector hypotheses
Given that AraC’s is the only well-characterized allosteric effector mechanism among AraC/XylS family proteins, we considered other mechanisms that could explain the allosteric effect of L-rhamnose on the activity of RhaS and RhaR. One hypothesis was that the linker could transmit a signal from the RhaS- and RhaR-NTD to their respective CTDs; however, our previous results suggested that the linker was not central to the L-rhamnose signal transmission (Kolin et al., 2007). We therefore hypothesized that the L-rhamnose-binding (NTD) and DNA-binding (CTD) domains might make effector-dependent contacts that could occur, in principle, in the absence of L-rhamnose to inhibit activity, or in the presence of L-rhamnose to stimulate activity. Our N-terminal deletion results indicated that these residues did not inhibit activity in the absence of L-rhamnose. However, these results did not rule-out models in which the very N-terminal residues made stimulatory contacts in the presence of L-rhamnose, or those in which other residues in the NTDs made inhibitory (or stimulatory) contacts in the absence (or presence) of L-rhamnose.
Somewhat surprisingly, we had previous evidence that the mechanism of allosteric activation might differ at some level between RhaS and RhaR. We’ve found that the RhaS C-terminal domain expressed alone, RhaS(163-278), was similar to L-rhamnose-activated full-length RhaS in that both could bind specifically to DNA and activate transcription (2,000- and 7,000-fold compared with ΔrhaS, respectively); whereas L-rhamnose-free full-length RhaS was unable to bind to DNA (Egan and Schleif, 1994; Wickstrum et al., 2007). In contrast, the activity of the RhaR C-terminal domain expressed alone, RhaR(196-311), more closely mimicked L-rhamnose-free full-length RhaR in that both were capable of binding to DNA, but barely if at all (two-fold or less compared with ΔrhaR), able to activate transcription. Only L-rhamnose-activated full-length RhaR was capable of robust transcription activation (Tobin and Schleif, 1990a, b; Wickstrum et al., 2007).
One model to explain the above findings is that the RhaS allosteric effector mechanism involves inhibition in the absence of L-rhamnose and the RhaR mechanism involves stimulation in the presence of L-rhamnose. However, we conclude that a model in which RhaS requires L-rhamnose to effectively bind DNA while RhaR requires L-rhamnose to effectively contact RNAP better fits the available data (further explained below). We also previously noticed that RhaS and RhaR were not alone in this difference in the activities of their DNA-binding domains. Expression of the DNA-binding domains alone of several AraC/XylS family proteins resulted in high levels of transcription activation (Gendlina et al., 2002; Kaldalu et al., 2000; Miyazaki et al., 2003; Poore et al., 2001; Wickstrum et al., 2007), while expression of the DNA-binding domains of others resulted in little or no transcription activation (Howard et al., 2002; Prouty et al., 2005), suggesting that the RhaS and RhaR mechanisms may apply to other AraC/XylS family proteins as well.
L-Rhamnose likely induces allosteric changes in RhaS- and RhaR-NTD
Our screen for RhaS and RhaR substitutions that conferred increased activity in the absence of L-rhamnose resulted in the isolation of a number of substitutions within the NTDs. The positions of these residues within the β-barrel of the AraC-NTD model of RhaS- and RhaR-NTD along with their phenotypes suggests that they may mimic structural changes that normally occur upon binding L-rhamnose, and that may be transmitted from the sugar-binding pocket to the CTDs (Fig. 1). RhaR H165R was instead located in the α-helical region of RhaR-NTD and, unlike RhaS P141L, high protein levels did not explain its elevated activity in the absence of L-rhamnose. The positively charged H165R substitution is positioned, however, across from a negatively charged aspartic acid residue in the RhaR β-barrel (data not shown), suggesting a potential influence of this substitution on the β-barrel structure.
L-rhamnose induces an allosteric change in RhaS-CTD HTH#1
Each of the four RhaS-CTD substitutions that conferred increased activity in the absence of L-rhamnose map to the predicted recognition helix of the first HTH motif (Fig. 3A). Three of these residues were changed to Arg (L201R, Q207R and Q210R) which – combined with our previous identification of two nearby Arg residues in RhaS HTH#1 (R202 and R206) that make base-specific DNA contacts (Bhende and Egan, 1999) – strongly supports the hypothesis that these RhaS variants bind to DNA more tightly than wild-type RhaS. These substitutions also conferred increased activity upon RhaS(163-278), again supporting the hypothesis that DNA-binding is increased (Fig. 4). Although purified RhaS(163-278) is significantly less prone to aggregation than full-length RhaS (Wickstrum et al., 2007), quantitative assays to directly compare the relative DNA binding strength of these variants have not been possible due to residual aggregation. The previous finding that DNA binding by RhaS has not been detected in the absence of L-rhamnose (Egan and Schleif, 1994) supports the model that DNA binding is central to the RhaS L-rhamnose response.
L-rhamnose induces an allosteric change in RhaR-CTD HTH#2
All of the RhaR variants within RhaR-CTD that conferred increased activity in the absence of L-rhamnose substituted small residues (Ala or Ser) for the Thr at position 279. This residue is predicted to lie within the stabilizing helix of the second HTH motif of RhaR-CTD (Fig. 3B), and aligns with AraC R251. Although AraC R251 (in the DNA binding domain of AraC) has been proposed to interact with residues in the N-terminal arm of AraC in the absence of L-arabinose (Wu and Schleif, 2001b), suggesting that RhaR might use a similar mechanism, our results suggest that the RhaR L-rhamnose response is not similar to the AraC light-switch mechanism (Saviola et al., 1998; Wu and Schleif, 2001b). The position of residue T279 on the structural model of RhaR-CTD between two residues that participate in protein-protein interactions with residues in the σ70 subunit of RNAP (Bhende and Egan, 1999; Wickstrum and Egan, 2004) (Fig. 3C), suggests that the substitutions to the smaller Ala or Ser residues may allow RhaR to more effectively contact RNAP in the absence of L-rhamnose. The previous finding that RhaR is capable of binding to DNA both in the absence and presence of L-rhamnose, but cannot activate transcription without L-rhamnose (Tobin and Schleif, 1990a, b) supports the hypothesis that RhaR contacts with σ70 are central to the RhaR L-rhamnose response.
Model for the RhaS and RhaR allosteric effector responses
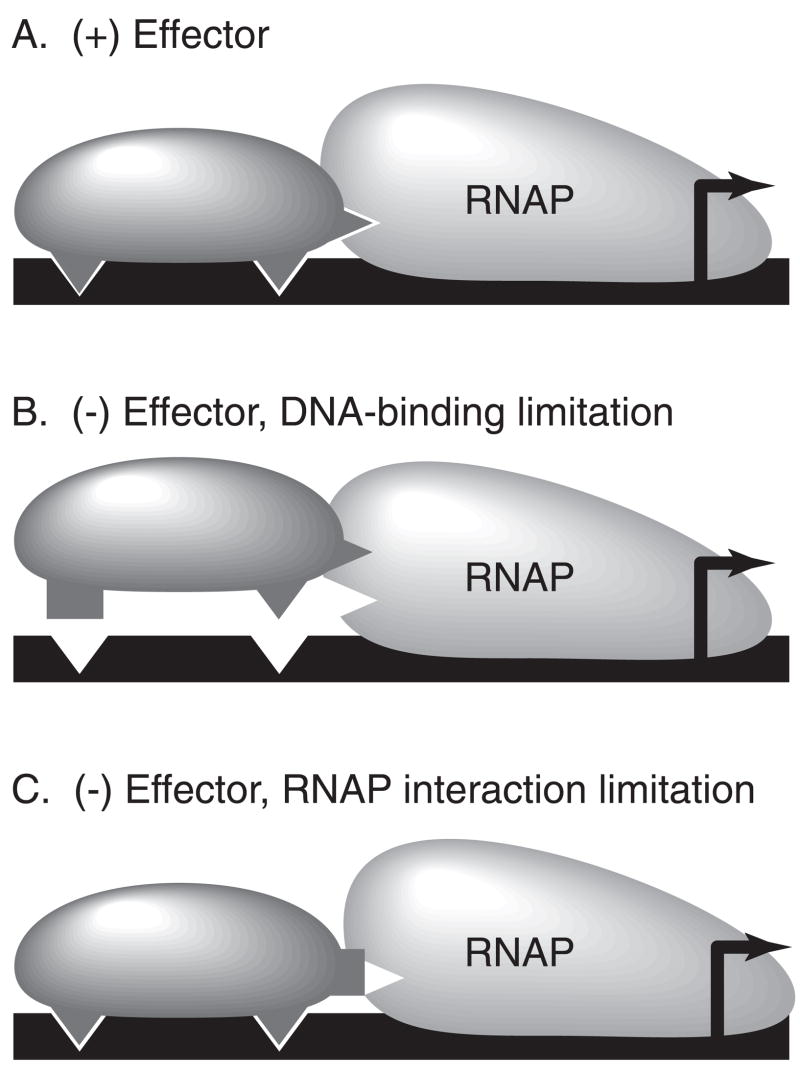
Overall, we propose that both the RhaS and the RhaR L-rhamnose responses involve the stimulation of the activity of the CTDs by the NTDs in the presence of L-rhamnose. We further hypothesize that in the absence of L-rhamnose, RhaS is limited in its ability to bind to DNA, and that L-rhamnose binding induces a structural change that alters HTH#1 and increases DNA binding (Fig. 6B). In contrast, we propose that in the absence of L-rhamnose, RhaR is limited in its ability to contact RNAP, and that a structural change is required in HTH#2 upon L-rhamnose binding to allow RhaR to effectively contact RNAP (Fig. 6C). The previous results that RhaS is unable to bind to DNA in the absence of L-rhamnose, and that RhaR is capable of binding to DNA in the absence of L-rhamnose, but is not capable of activating transcription (Egan and Schleif, 1994; Tobin and Schleif, 1990a, b), are entirely consistent with our proposed mechanisms. The finding that MarA consists of two structurally similar subdomains (Rhee et al., 1998), one including HTH#1 (involved in the RhaS mechanism), and one including HTH#2 (involved in the RhaR mechanism), suggests the possibility that the underlying mechanisms of the RhaS and RhaR allosteric responses may be more similar than they appear. This proposal is also supported by the similarities in our findings in the RhaS- and RhaR-NTDs.
Figure 6.
Model of the effector responses of RhaS and RhaR. (A) In the presence of effector, both RhaS and RhaR can efficiently bind to DNA and contact RNAP to activate transcription. (B) In the absence of an effector, RhaS is limited in its ability to bind to DNA. (C) In the absence of effector, RhaR is limited in its ability to effectively contact RNAP. Black line, DNA; dark gray, the DNA binding domain of the promoter-proximal monomer of either RhaS or RhaR; light gray, RNAP. Triangular protrusions or indentations represent sites of contact between the DNA-binding domain of RhaS or RhaR and DNA or RNAP, whereas rectangular protrusions represent sites that are not in the correct conformation to make contact.
The model that the RhaS- and RhaR-NTD’s must stimulate their respective CTDs in the presence of L-rhamnose in order to activate transcription easily explains why RhaR(196-311) alone could not activate transcription (Wickstrum et al., 2007). However, reconciling the finding that RhaS(163-278) alone activated transcription to high levels (Wickstrum et al., 2007) with this model is more difficult. This apparent contradiction might be explained by the specific nature of the step limiting activation by RhaS-CTD. We propose that the DNA-binding defect of RhaS in the absence of L-rhamnose (and similarly of RhaS(163-278)) may be relatively small, but nonetheless sufficient to prevent the low concentrations of RhaS expressed in the absence of L-rhamnose from binding to DNA. We further propose that the expression of RhaS(163-278) from even relatively low copy number plasmids is sufficiently high (compared with chromosomally expressed full-length RhaS) to overcome this DNA binding defect and to drive RhaS(163-278) onto the DNA. In contrast, similar overexpression of RhaR(196-311) is not expected to overcome limitations in the ability to contact RNAP, since the mass action advantage of overexpression no longer has an influence once a single protein binds to the DNA. Although this aspect of our RhaS model does not incorporate the simplest hypothesis to explain the activity of RhaS(163-278), we find it to be the best model to explain the available data.
Implications for other AraC/XylS family proteins
As mentioned above, the DNA-binding domains of many multi-domain AraC/XylS family proteins appear to fall into two classes: (1) Those that are capable of high levels of transcription activation when expressed alone, including RhaS; and (2) Those that show little or no activation when expressed alone, including RhaR. Our models suggest a possible explanation for these two classes.
The AraC/XylS family proteins whose DNA binding domains activate transcription well –including XylS, UreR and XynR (Gendlina et al., 2002; Kaldalu et al., 2000; Miyazaki et al., 2003; Poore et al., 2001; Wickstrum et al., 2007) – may be similar to RhaS and unable to bind DNA effectively in the absence of effector (Fig. 6B). The DNA-binding domains in each of these studies were overexpressed to some extent, in line with the expected mass action effect on DNA binding. Also consistent with this hypothesis are the findings that: two UreR mutations that confer a constitutive phenotype have been shown to increase DNA binding by the full-length protein (Gendlina et al., 2002); and that XylS expression from the stronger of its two natural promoters causes it to become effector-independent (Gallegos et al., 1996; Gonzalez-Perez et al., 2004; Ramos et al., 1987).
The AraC/XylS family proteins whose DNA binding domains do not activate transcription well – including MelR and ToxT (Howard et al., 2002; Prouty et al., 2005) – may be similar to RhaR and unable to effectively contact RNAP (or otherwise activate transcription at a step after DNA binding) in the absence of effector (Fig. 6C). MelR-CTD binds to DNA but does not activate transcription (Howard et al., 2002). MelR is somewhat of a special case, however, given that in the absence of melibiose, MelR has been proposed to form a DNA loop that actively represses melR transcription (Wade et al., 2000). It remains to be seen whether the MelR mechanism involves the transmission of structural changes from its NTD to its CTD in the presence of melibiose in addition to releasing its DNA loop. Similarities in the mechanisms are suggested by the observation that several melibiose-independent variants of MelR have substitutions that align or are adjacent to substitutions we identified in RhaS and RhaR (Kahramanoglou et al., 2006).
Although the available evidence is consistent with additional AraC/XylS family employing allosteric effector mechanisms similar to those we’ve proposed for RhaS and RhaR, there is no evidence that these mechanisms apply to AraC itself. AraC-NTD does not undergo conformational changes in its β-barrel upon arabinose binding (Soisson et al., 1997b; Weldon et al., 2007), arguing against the transmission of any structural changes from the NTD to the CTD in the presence of L-arabinose. The importance of aligned residues AraC R251 and RhaR T279 in their respective mechanisms, however, suggests that the AraC and RhaR mechanisms may share the common principle of preventing contacts with σ70 in the absence of effector.
EXPERIMENTAL PROCEDURES
Culture media and conditions
Cultures for β-galactosidase assays were grown in morpholinepropanesulfonic acid-buffered minimal medium (Neidhardt et al., 1974), with 0.4% glycerol as the carbon source and in the absence or presence of 0.4% L-rhamnose and appropriate antibiotic (Bhende and Egan, 2000). All other cultures were grown in tryptone-yeast extract (TY) medium (0.8% tryptone, 0.8% yeast extract, 0.5% NaCl, pH 7.0) (Maloy et al., 1996; Miller, 1972). For solid media, cells were grown on nutrient agar plates (2.3% Difco Nutrient agar, 0.5% NaCl) with Xgal (5-bromo-4-chloro-3-indolyl-β-D-galactoside, 40 μg/ml) for testing lacZ expression. Ampicillin (200 μg/ml), chloramphenicol (25 μg/ml) and L-rhamnose (0.2%) were added as indicated. All cultures were grown at 37°C.
General methods and strains
Oligonucleotide primers used in this study were synthesized by MWG-Biotech, Inc. (High Point, NC), and their DNA sequences are available upon request. Restriction endonucleases and T4 DNA ligase were purchased from New England Biolabs (Beverly, MA). Restriction endonuclease digestions, ligations and transformations were performed using standard procedures. All strains used in the study are derived from E. coli strain ECL116 (F− lacU169 endA hsdR thi) (Backman et al., 1981), and genotypes list additional alleles in this genetic background.
Most β-galactosidase assays of activation by RhaS or its variants were performed in strain SME3000 (λΦ(rhaB-lacZ)Δ84 Δ(rhaSR)::Km). The promoter driving lacZ expression in this strain extends to −84 (in all cases, relative to the transcription start site) and does not include the CRP binding site upstream of rhaBAD. The exception was the assays in Table 1 which were performed in SME1087 (λΦ(rhaB-lacZ)Δ226 rhaS). The promoter driving lacZ expression in SME1087 extends to −226 and includes all of the rhaBAD promoter elements. Most β-galactosidase assays of activation by RhaR or its variants were performed in strain SME3160 (λΦ(rhaS-lacZ)Δ85 Δ(rhaSR)::Km recA::cat). The promoter driving lacZ expression in this strain extends to −85 and does not include the CRP binding site upstream of rhaSR. The exception was the assays in Table 1, which were performed in strain SME1076 (λΦ(rhaS-lacZ)Δ216 Δ(rhaSR)::Km). The promoter driving lacZ expression in this strain extends to −216 and includes all of the rhaSR promoter elements.
Mutagenesis of rhaS and rhaR
All full-length wild type and RhaS and RhaR variants (including the N-terminal deletions) were expressed from the plasmid pHG165 (ApR, essentially pUC8 with the copy number of pBR322) (Stewart et al., 1986). Longway-around (inverse) PCR was used to construct 5′ deletions of rhaS and rhaR, using the Expand High Fidelity PCR System (Roche, Indianapolis, IN), with the wild-type rhaS or rhaR in pHG165 (Stewart et al., 1986) as the template. PCR products were designed to carry EarI restriction sites at each end, thereby allowing seamless reconstruction of truncated rhaS or rhaR genes (LaPointe and Taylor, 2000; Wickstrum and Egan, 2004). Expression was under the control of the lac promoter located on the vector. These variants all retained the RhaS or RhaR start codon, but had progressively longer deletions ranging from codons 2 through 4 (in RhaS) up to deletion of codons 2 through 52 (in RhaR) (Table 1).
Random mutagenesis of rhaS or rhaR was performed by PCR amplification of the entire open reading frame using Taq DNA polymerase (Promega, Madison, WI) under standard reaction conditions (Zhou et al., 1991). PCR products of randomly mutagenized rhaS or rhaR were ligated into the EcoRI and HindIII restriction sites of pGH165 (Stewart et al., 1986) such that expression was driven by a vector-encoded lac promoter. Separation of the rhaS or rhaR L-rhamnose-independent double mutants into single mutants was performed by PCR amplification using the Expand High Fidelity PCR system (Roche; Indianapolis, IN). Fragments of rhaS or rhaR carrying a single mutation were seamlessly pieced together with fragments lacking mutations (LaPointe and Taylor, 2000; Wickstrum and Egan, 2004) upon cloning into pHG165.
To test the phenotypes of the L-rhamnose-independent rhaS mutants when expressed in RhaS-CTD alone (residues 163-278, referred to as RhaS(163-278)), the portion of rhaS that encodes RhaS(163-278) was PCR amplified using the Expand High Fidelity PCR system (Roche; Indianapolis, IN) and ligated into pSE257 using BamHI and Hind III restriction sites. pSE257 was derived from pSU18 (CmR, essentially the pUC18 multiple cloning site/lacZα region on a plasmid with the P15A ori) (Bartolome et al., 1991) by the addition of a promoter (driving downstream gene expression) that combines a near-consensus σ70 −35 element (TTGACT) with the −10 element from the rhaSR promoter (TACTAT), to construct a moderately strong, constitutive promoter (Wickstrum and Egan, unpublished).
The genes encoding the wild type and the L-rhamnose-independent variant T279A of RhaR-CTD alone (residues 196-311, referred to as RhaR(196-311)) were cloned into pSE262. pSE262 was derived from pHG165 (in a manner analogous to pSE257) by the addition of a promoter to drive downstream gene expression that combines a consensus σ70 −35 element (TTGACA) with the −10 element from the rhaSR promoter (TACTAT), to construct a relatively strong, constitutive promoter (Wickstrum and Egan, unpublished). PCR amplification was performed using the Expand High Fidelity PCR system (Roche; Indianapolis, IN). For these constructs, the insert contained the translation start codon, whereas the Shine-Dalgarno sequence was plasmid-encoded, with a BamHI site between them to provide proper spacing (8 nucleotides) for ribosome recognition. Plasmids expressing either the wild-type RhaR(196-311) or RhaR(196-311) T279A were transformed into a strain that carried a rhaSR deletion and a single-copy fusion of lacZ with a rhaSR promoter that did not include the CRP binding site.
DNA sequencing
DNA sequencing was performed at the Molecular Research Core Facility of Idaho State University (Pocatello, ID) using an ABI3100 automated sequencer or the Northwestern University Biotechnology Laboratory (Chicago, IL) using an ABI3100 or an ABI3730 automated sequencer. The DNA sequences of both strands were determined for the entire cloned region of all cloned and mutagenized DNA fragments.
β-galactosidase assays
β-galactosidase assays were performed as previously described (Bhende and Egan, 1999) using the method of Miller (Miller, 1972). Briefly, all strains for β-galactosidase assays were grown in three serial steps: tryptone-yeast extract culture with ampicillin; overnight culture (MOPS-buffered minimal medium containing 0.04% glycerol as limiting carbon source and ampicillin); and growth culture (MOPS-buffered mimimal medium containing 0.4% glycerol as the carbon source, with or without 0.4% L-rhamnose and with ampicillin), based on the method of Neidhardt (Neidhardt et al., 1974). Specific activities were averaged from at least two, usually three, independent assays with two replicates in each assay. β-galactosidase activity is expressed in Miller units (Miller, 1972).
Acknowledgments
We would like to thank members of the lab of William Picking, especially Marianela Espina and Andrew Olive, for help with Western blots and image analysis. Western blot images were obtained on an Odyssey Infrared Imager through collaboration with LI-COR Inc. We also thank Liskin Swint-Kruse (KU Med Cntr) and Shivakumara Siddaramappa for comments on the manuscript, Matthew Buechner for use of a gradient thermocycler, and Brian Easley, Remealle How and Christian Stopp for technical assistance.
This work was supported by NIH grant GM55099 from the National Institute of General Medical Sciences, NIH Grant P20 RR17708 from the Institutional Development Award (IDeA) Program of the National Center for Research Resources, and the Department of Molecular Biosciences, University of Kansas, all to S.M.E.
References
- Backman K, Chen Y-M, Magasanik B. Physical and genetic characterization of the gln A-glnG region of the Escherichia coli chromosome. Proc Nat Acad Sci, USA. 1981;78:3743–3747. doi: 10.1073/pnas.78.6.3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolome B, Jubee YME, de la Cruz F. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene. 1991;102:75–78. doi: 10.1016/0378-1119(91)90541-i. [DOI] [PubMed] [Google Scholar]
- Bhende PM, Egan SM. Amino acid-DNA contacts by RhaS: an AraC family transcription activator. J Bacteriol. 1999;181:5185–5192. doi: 10.1128/jb.181.17.5185-5192.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhende PM, Egan SM. Genetic evidence that transcription activation by RhaS involves specific amino acid contacts with sigma 70. J Bacteriol. 2000;182:4959–4969. doi: 10.1128/jb.182.17.4959-4969.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carra JH, Schleif RF. Variation of half-site organization and DNA looping by AraC protein. EMBO J. 1993;12:35–44. doi: 10.1002/j.1460-2075.1993.tb05629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangi B, Gronenborn AM, Rosner JL, Martin RG. Versatility of the carboxy-terminal domain of the alpha subunit of RNA polymerase in transcriptional activation: use of the DNA contact site as a protein contact site for MarA. Mol Microbiol. 2004;54:45–59. doi: 10.1111/j.1365-2958.2004.04250.x. [DOI] [PubMed] [Google Scholar]
- Egan SM, Schleif RF. A regulatory cascade in the induction of rhaBAD. J Mol Biol. 1993;234:87–98. doi: 10.1006/jmbi.1993.1565. [DOI] [PubMed] [Google Scholar]
- Egan SM, Schleif RF. DNA-dependent renaturation of an insoluble DNA binding protein. Identification of the RhaS binding site at rhaBAD. J Mol Biol. 1994;243:821–829. doi: 10.1006/jmbi.1994.1684. [DOI] [PubMed] [Google Scholar]
- Egan SM. Growing repertoire of AraC/XylS activators. J Bacteriol. 2002;184:5529–5532. doi: 10.1128/JB.184.20.5529-5532.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eustance RJ, Bustos SA, Schleif RE. Reaching out: Locating and lengthening the interdomain linker in AraC protein. J Mol Biol. 1994;242:330–338. doi: 10.1006/jmbi.1994.1584. [DOI] [PubMed] [Google Scholar]
- Gallegos M-T, Michan C, Ramos JL. The XylS/AraC family of regulators. Nucl Acids Res. 1993;21:807–810. doi: 10.1093/nar/21.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallegos M-T, Marques S, Ramos JL. Expression of the TOL plasmid xylS gene in Pseudomonas putida occurs fom a σ70-dependent promoter or from σ70- and σ54-dependent tandem promoters according to the compound used for growth. J Bacteriol. 1996;178:2356–2361. doi: 10.1128/jb.178.8.2356-2361.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallegos M-T, Schleif R, Bairoch A, Hofmann K, Ramos JL. AraC/XylS family of transcriptional regulators. Microbiol Mol Biol Rev. 1997;61:393–410. doi: 10.1128/mmbr.61.4.393-410.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendlina I, Gutman DM, Thomas V, Collins CM. Urea-dependent signal transduction by the virulence regulator UreR. J Biol Chem. 2002;277:37349–37358. doi: 10.1074/jbc.M203462200. [DOI] [PubMed] [Google Scholar]
- Ghosh M, Schleif RF. Biophysical evidence of arm-domain interactions in AraC. Anal Biochem. 2001;295:107–112. doi: 10.1006/abio.2001.5213. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Perez MM, Ramos JL, Marques S. Cellular XylS levels are a function of transcription of xylS from two independent promoters and the differential efficiency of translation of the two mRNAs. J Bacteriol. 2004;186:1898–1901. doi: 10.1128/JB.186.6.1898-1901.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grainger DC, Belyaeva TA, Lee DJ, Hyde EI, Busby SJ. Transcription activation at the Escherichia coli melAB promoter: interactions of MelR with the C-terminal domain of the RNA polymerase alpha subunit. Mol Microbiol. 2004a;51:1311–1320. doi: 10.1111/j.1365-2958.2003.03930.x. [DOI] [PubMed] [Google Scholar]
- Grainger DC, Webster CL, Belyaeva TA, Hyde EI, Busby SJ. Transcription activation at the Escherichia coli melAB promoter: interactions of MelR with its DNA target site and with domain 4 of the RNA polymerase sigma subunit. Mol Microbiol. 2004b;51:1297–1309. doi: 10.1111/j.1365-2958.2003.03929.x. [DOI] [PubMed] [Google Scholar]
- Harmer T, Wu M, Schleif R. The role of rigidity in DNA looping-unlooping by AraC. Proc Natl Acad Sci U S A. 2001;98:427–431. doi: 10.1073/pnas.98.2.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holcroft CC, Egan SM. Interdependence of activation at rhaSR by cyclic AMP receptor protein, the RNA polymerase alpha subunit C-terminal domain and RhaR. J Bacteriol. 2000;182:6774–6782. doi: 10.1128/jb.182.23.6774-6782.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard VJ, Belyaeva TA, Busby SJ, Hyde EI. DNA binding of the transcription activator protein MelR from Escherichia coli and its C-terminal domain. Nucleic Acids Res. 2002;30:2692–2700. doi: 10.1093/nar/gkf370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jair K, Martin RG, Rosner JL, Fujita N, Ishihama A, Wolf RE., Jr Purification and regulatory properties of MarA protein, a transcriptional activator of Escherichia coli multiple antibiotic and superoxide resistance promoters. J Bacteriol. 1995;177:7100–7104. doi: 10.1128/jb.177.24.7100-7104.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jair K-M, Yu X, Skarstad K, Thony B, Fujita N, Ishihama A, Wolf RE., Jr Transcriptional activation of promoters of the superoxide and multiple antibiotic resistance regulons by Rob, a binding protein of the Escherichia coli origin of chromosomal replication. J Bacteriol. 1996a;178:2507–2513. doi: 10.1128/jb.178.9.2507-2513.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jair K-W, Fawcett WP, Fujita N, Ishihama A, Wolf RE., Jr Ambidextrous transcriptional activation by SoxS: requirement for the C-terminal domain of the RNA polymerase alpha subunit in a subset of Escherichia coli superoxide-inducible genes. Molec Microbiol. 1996b;19:307–317. doi: 10.1046/j.1365-2958.1996.368893.x. [DOI] [PubMed] [Google Scholar]
- Kahramanoglou C, Webster CL, El-Robh MS, Belyaeva TA, Busby SJ. Mutational analysis of the Escherichia coli melR gene suggests a two-state concerted model to explain transcriptional activation and repression in the melibiose operon. J Bacteriol. 2006;188:3199–3207. doi: 10.1128/JB.188.9.3199-3207.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaldalu N, Toots U, de Lorenzo V, Ustav M. Functional domains of the TOL plasmid transcription factor XylS. J Bacteriol. 2000;182:1118–1126. doi: 10.1128/jb.182.4.1118-1126.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolin A, Jevtic V, Swint-Kruse L, Egan SM. Linker regions of the RhaS and RhaR proteins. J Bacteriol. 2007;189:269–271. doi: 10.1128/JB.01456-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon HJ, Bennik MHJ, Demple B, Ellenberger T. Crystal structure of the Escherichia coli Rob transcription factor in complex with DNA. Nature Structural Biology. 2000;7:424–430. doi: 10.1038/75213. [DOI] [PubMed] [Google Scholar]
- Landini P, Gaal T, Ross W, Volkert MR. The RNA polymerase α subunit carboxyl-terminal domain is required for both basal and activated transcription from the alkA promoter. J Biol Chem. 1997;272:15914–15919. doi: 10.1074/jbc.272.25.15914. [DOI] [PubMed] [Google Scholar]
- LaPointe CF, Taylor RK. The type 4 prepilin peptidases comprise a novel family of aspartic acid proteases. J Biol Chem. 2000;275:1502–1510. doi: 10.1074/jbc.275.2.1502. [DOI] [PubMed] [Google Scholar]
- Lobell RB, Schleif RF. DNA looping and unlooping by AraC protein. Science. 1990;250:528. doi: 10.1126/science.2237403. [DOI] [PubMed] [Google Scholar]
- Lonetto MA, Rhodius V, Lamberg K, Kiley P, Busby S, Gross C. Identification of a contact site for different transcription activators in region 4 of the Escherichia coli RNA polymerase σ70 subunit. J Mol Biol. 1998;284:1353–1365. doi: 10.1006/jmbi.1998.2268. [DOI] [PubMed] [Google Scholar]
- Maloy SR, Stewart VJ, Taylor RK. Genetic analysis of pathogenic bacteria. Cold Spring Harbor, N. Y: Cold Spring Harbor Laboratory Press; 1996. [Google Scholar]
- Martin RG, Gillette WK, Martin NI, Rosner JL. Complex formation between activator and RNA polymerase as the basis for transcriptional activation by MarA and SoxS in Escherichia coli. Mol Microbiol. 2002;43:355–370. doi: 10.1046/j.1365-2958.2002.02748.x. [DOI] [PubMed] [Google Scholar]
- Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- Miyazaki K, Miyamoto H, Mercer DK, Hirase T, Martin JC, Kojima Y, Flint HJ. Involvement of the multidomain regulatory protein XynR in positive control of xylanase gene expression in the ruminal anaerobe Prevotella bryantii B(1)4. J Bacteriol. 2003;185:2219–2226. doi: 10.1128/JB.185.7.2219-2226.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidhardt FC, Bloch PL, Smith DF. Culture medium for enterobacteria. J Bacteriol. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poore CA, Coker C, Dattelbaum JD, Mobley HL. Identification of the domains of UreR, an AraC-like transcriptional regulator of the urease gene cluster in Proteus mirabilis. J Bacteriol. 2001;183:4526–4535. doi: 10.1128/JB.183.15.4526-4535.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power J. The L-rhamnose genetic system in Escherichia coli K-12. Genetics. 1967;55:557–568. doi: 10.1093/genetics/55.3.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prouty MG, Osorio CR, Klose KE. Characterization of functional domains of the Vibrio cholerae virulence regulator ToxT. Mol Microbiol. 2005;58:1143–1156. doi: 10.1111/j.1365-2958.2005.04897.x. [DOI] [PubMed] [Google Scholar]
- Ramos JL, Mermod N, Timmis KN. Regulatory circuits controlling transcription of TOL plasmid operon encoding meta-cleavage pathway for degradation of alkylbenzoates by Pseudomonas. Mol Microbiol. 1987;1:293–300. doi: 10.1111/j.1365-2958.1987.tb01935.x. [DOI] [PubMed] [Google Scholar]
- Ramos JL, Rojo F, Zhou L, Timmis KN. A family of positive regulators related to the Pseudomonas putida TOL plasmid XylS and the Escherichia coli AraC activators. Nucl Acids Res. 1990;18:2149–2152. doi: 10.1093/nar/18.8.2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed WL, Schleif RF. Hemiplegic mutations in AraC protein. J Mol Biol. 1999;294:417–425. doi: 10.1006/jmbi.1999.3224. [DOI] [PubMed] [Google Scholar]
- Rhee S, Martin RG, Rosner JL, Davies DR. A novel DNA-binding motif in MarA: the first structure for an AraC family transcriptional activator. Proc Natl Acad Sci USA. 1998;95:10413–10418. doi: 10.1073/pnas.95.18.10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross JJ, Gryczynski U, Schleif R. Mutational analysis of residue roles in AraC function. J Mol Biol. 2003;328:85–93. doi: 10.1016/s0022-2836(03)00262-6. [DOI] [PubMed] [Google Scholar]
- Ruiz R, Ramos JL, Egan SM. Interactions of the XylS regulators with the C-terminal domain of the RNA polymerase α subunit influence the expression level from the cognate Pm promoter. FEBS Letters. 2001;491:207–211. doi: 10.1016/s0014-5793(01)02192-5. [DOI] [PubMed] [Google Scholar]
- Saviola B, Seabold R, Schleif RF. Arm-domain interactions in AraC. J Mol Biol. 1998;278:539–548. doi: 10.1006/jmbi.1998.1712. [DOI] [PubMed] [Google Scholar]
- Seabold RR, Schleif RF. Apo-AraC actively seeks to loop. J Mol Biol. 1998;278:529–538. doi: 10.1006/jmbi.1998.1713. [DOI] [PubMed] [Google Scholar]
- Shah IM, Wolf RE., Jr Novel Protein-Protein Interaction Between Escherichia coli SoxS and the DNA Binding Determinant of the RNA Polymerase alpha Subunit: SoxS Functions as a Co-sigma Factor and Redeploys RNA Polymerase from UP-element-containing Promoters to SoxS-dependent Promoters during Oxidative Stress. J Mol Biol. 2004;343:513–532. doi: 10.1016/j.jmb.2004.08.057. [DOI] [PubMed] [Google Scholar]
- Soisson SM, MacDougall-Shackleton B, Schleif R, Wolberger C. The 1.6 A crystal structure of the AraC sugar-binding and dimerization domain complexed with D-fucose. J Mol Biol. 1997a;273:226–237. doi: 10.1006/jmbi.1997.1314. [DOI] [PubMed] [Google Scholar]
- Soisson SM, MacDougall-Shackleton B, Schleif R, Wolberger C. Structural basis for ligand-regulated oligomerization of AraC. Science. 1997b;276:421–425. doi: 10.1126/science.276.5311.421. [DOI] [PubMed] [Google Scholar]
- Stewart GSAB, Lubinsky-Mink S, Jackson CG, Cassel A, Kuhn J. pHG165: a pBR322 copy number derivative of pUC8 for cloning and expression. Plasmid. 1986;15:172–181. doi: 10.1016/0147-619x(86)90035-1. [DOI] [PubMed] [Google Scholar]
- Tate CG, Muiry JAR, Henderson PJF. Mapping, cloning, expression, and sequencing of the rhaT gene which encodes a novel L-Rhamnose-H+ transport protein in Salmonella typhimurium and Escherichia coli. J Biol Chem. 1992;287:6923–6932. [PubMed] [Google Scholar]
- Tobin JF, Schleif RF. Positive regulation of the Escherichia coli L-rhamnose operon is mediated by the products of tandemly repeated regulatory genes. J Mol Biol. 1987;196:789–799. doi: 10.1016/0022-2836(87)90405-0. [DOI] [PubMed] [Google Scholar]
- Tobin JF, Schleif RF. Purification and properties of RhaR, the positive regulator of the L-rhamnose operons of Escherichia coli. J Mol Biol. 1990a;211:75–89. doi: 10.1016/0022-2836(90)90012-B. [DOI] [PubMed] [Google Scholar]
- Tobin JF, Schleif RF. Transcription from the rha operon psr promoter. J Mol Biol. 1990b;211:1–4. doi: 10.1016/0022-2836(90)90003-5. [DOI] [PubMed] [Google Scholar]
- Via P, Badia J, Baldoma L, Obradors N, Aguilar J. Transcriptional regulation of the Escherichia coli rhaT gene. Microbiology. 1996;142:1833–1840. doi: 10.1099/13500872-142-7-1833. [DOI] [PubMed] [Google Scholar]
- Wade JT, Belyaeva TA, Hyde EI, Busby SJ. Repression of the Escherichia coli melR promoter by MelR: evidence that efficient repression requires the formation of a repression loop. Mol Microbiol. 2000;36:223–229. doi: 10.1046/j.1365-2958.2000.01850.x. [DOI] [PubMed] [Google Scholar]
- Weldon JE, Rodgers ME, Larkin C, Schleif RF. Structure and properties of a truely apo form of AraC dimerization domain. Proteins. 2007;66:646–654. doi: 10.1002/prot.21267. [DOI] [PubMed] [Google Scholar]
- Wickstrum JR, Egan SM. Amino acid contacts between sigma 70 domain 4 and the transcription activators RhaS and RhaR. J Bacteriol. 2004;186:6277–6285. doi: 10.1128/JB.186.18.6277-6285.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickstrum JR, Skredenske JM, Kolin A, Jin DJ, Fang J, Egan SM. Transcription Activation by the DNA-Binding Domain of the AraC Family Protein RhaS in the Absence of Its Effector-Binding Domain. J Bacteriol. 2007;189:4984–4993. doi: 10.1128/JB.00530-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Schleif R. Strengthened arm-dimerization domain interactions in AraC. J Biol Chem. 2001a;276:2562–2564. doi: 10.1074/jbc.M008705200. [DOI] [PubMed] [Google Scholar]
- Wu M, Schleif R. Mapping arm-DNA-binding domain interactions in AraC. J Mol Biol. 2001b;307:1001–1009. doi: 10.1006/jmbi.2001.4531. [DOI] [PubMed] [Google Scholar]
- Zhou YH, Zhang XP, Ebright RH. Random mutagenesis of gene-sized DNA molecules by use of PCR with Taq DNA polymerase. Nucleic Acids Res. 1991;19:6052. doi: 10.1093/nar/19.21.6052. [DOI] [PMC free article] [PubMed] [Google Scholar]