Abstract
Many eukaryotic genes are acutely regulated by extra-cellular signals. The c-fos serum response element (SRE) mediates transcriptional activation in response to mitogens through serum response factor (SRF)-dependent recruitment of Elk-1, a mitogen-activated protein kinase (MAPK)-responsive transcription factor. How subsequent events at SRE promoters stimulate initiation of transcription has yet to be fully resolved. Here we show that extra-cellular signal-regulated kinase (ERK) and mitogen and stress-activated kinase (MSK) are recruited to SRE promoter complexes in vitro and in vivo. Their recruitment in vitro correlates with Elk-1 binding and for ERK the D domain/KIM of Elk-1 is specifically involved. In vivo, recruitment of ERK and MSK is stimulated by mitogens, correlates with histone H3 phosphorylation and is impaired by Elk-1 knockdown. Immunocytochemistry and confocal microscopy reveal that ERK appears to associate to some extent with initiating rather than elongating RNA polymerase II. Taken together, our data add to the body of evidence implying that ERK and related MAPKs may fulfil a generic role at the promoters of acutely regulated genes.
INTRODUCTION
Initiation of transcription in eukaryotes is a complex process involving many proteins. Although their access to gene promoters requires permissive chromatin structures controlled by histone-modifying enzymes and chromatin-remodelling activities (1–3), the expression of numerous genes is acutely regulated in response to extra-cellular signals, temporal cues or by factors intrinsic to the cell, for example mitogens, actin dynamics or circadian rhythms (4–6). In such instances, transcription factors and components of the basal transcription machinery can interact with gene promoters without detectable gene expression (7). Formation of active pre-initiation complexes, hallmarks of which are phosphorylation of serine 5 (S5) within the carboxy-terminal domain (CTD) of Rpb1, the largest subunit of RNA polymerase II (RNAPII), local modifications to histone H3, including methylation of lysine 4 (H3K4), phosphorylation of S10 (H3S10) and acetylation of lysine 14 (H3K14) and RNA synthesis are detected only in response to upstream stimuli (1,2,8,9).
The expression of the immediate early (IE) genes c-fos and egr-1 (also known as NGFI-A, zif/268 and Krox24) is triggered by mitogen-activated protein kinase (MAPK) signalling (10,11). Such genes are characterised by serum response elements (SREs) in their promoters, which bind serum response factor (SRF) and recruit ternary complex factors such as Elk-1 (12–14). Elk-1 is phosphorylated by ERKs (also JNK/SAPKs and p38MAPK isoforms) and recruited to the c-fos SRE, but during mitogen-induced c-fos expression, events following Elk-1 phosphorylation are less well understood. It has been proposed that upon phosphorylation Elk-1 adopts an active conformation (15), in which it participates in transcriptional activation through co-activators including MED23 (TRAP150beta/CRSP130/Sur2) and p300/CBP (16–19). More recently, it has been shown that inactive Elk-1 is sumoylated and that upon phosphorylation of Elk-1 the Sumo E3 ligase PIASxα, by desumoylating Elk-1 and disengaging associated histone deacetylases (HDACs), serves as an Elk-1 co-activator (20,21). In these scenarios, the role of ERK is restricted to Elk-1 phosphorylation.
Several reports have described the association of yeast MAPKs with specific gene promoters (22–25). Furthermore, human p38 was recently shown to occupy gene promoters during myogenesis (26) and ERK was found in a complex with the progesterone receptor on the MMTV promoter (27). These findings are consistent with the proposal that MAPKs may be frequent occupants of signal-regulated gene promoters (25,28) and imply that they serve additional roles during transcriptional activation besides the phosphorylation of target transcription factors (29–31).
We studied pre-initiation complexes (PICs) on immobilized, mitogen-responsive SRE promoters and found that both ERK and MSK were recruited to SRE-dependent PICs in a mitogen- and Elk-1-dependent manner. Reconstitution experiments with recombinant proteins indicated that the D-domain/kinase interaction motif (KIM) of Elk-1 was essential for ERK recruitment. Chromatin immunoprecipitation (ChIP) assays confirmed the mitogen-dependent phosphorylation of Elk-1 and recruitment of ERKs and MSK1 to the c-fos and egr1 promoters in cells. However, co-localization of phospho-ERK (pERK) with phospho-Elk-1 (pElk-1) in mitogen-stimulated cells was low and exceeded significantly by co-localization of pERK with RNAPII in which the CTD was phosphorylated on S5 (pS5-CTD). This implies that the interaction between Elk-1 and ERK is transient and that ERK may subsequently associate with and phosphorylate other targets within PICs, thereby contributing directly to the transcriptional activation of mitogen-responsive genes.
MATERIALS AND METHODS
Cell culture and nuclear extract preparation
For nuclear extracts, HeLa cells were grown in suspension in Joklik's MEM supplemented with 10% fetal calf serum (FCS), 100 U ml−1 penicillin, 100 μg ml−1 streptomycin, 1% non-essential amino acids, 2 mM l-glutamine. Cells were serum-starved for 42–44 h in medium containing 0.5% FCS and either harvested directly or after stimulation with 20% FCS for 10 min. Nuclear extracts were prepared from 1.5 × 109 cells, essentially as described (32).
For ChIP assays and immunocytochemistry, HeLa cells were grown as monolayers, on plastic or glass, respectively, in DMEM supplemented with 10% FCS, 2 mM l-glutamine, 100 U ml−1 penicillin and 100 μg ml−1 streptomycin. For reporter assays, NIH3T3 cells were grown in DMEM supplemented with 10% FCS, 2 mM l-glutamine, 100 U ml−1 penicillin and 100 μg ml−1 streptomycin.
Luciferase reporter assays were performed as previously described (33), and in vitro transcription assays were performed as described elsewhere (34).
RNAi knockdown was performed with siRNA from Ambion, reverse-transfected into HeLa cells with the siPORT NeoFX transfection agent (Ambion) according to the manufacturer's instructions.
Reagents, plasmids and antibodies
Streptavidin-coated magnetic beads were from Dynal (Dynabeads M-280 Streptavidin). The generic oligonucleotides for promoter synthesis by PCR were:
b-profor (biotinylated) 5′-CTGCAGGTCGACTCTAGC;
g-prorev 5′-AGTATGTGAGAGTGTAAAAAAGGGCCAAGTGC.
The plasmid pE4-38 CAT, containing the basal promoter from the adenovirus 2 E4 promoter was used to generate the TATA promoter (35). The plasmids pSRE-CAT and pSIDE-CAT, containing a single SRE or a mutant thereof that fails to bind Elk-1 (SIDE) inserted upstream from the TATA box in E4-38 CAT, were used to generate SRE and DSE promoters, respectively. For reporter assays, analogous pGL3-based luciferase constructs containing a single SRE or DSE upstream from the adenovirus 2 E4 basal promoter were transfected alone or with expression vectors for active RhoA (L63) or C-Raf (259D).
Plasmids containing G-free cassettes used for in vitro transcription analyses, pML(C2AT), pTATA-B7 and pWT-TATA-(C2AT)19 (SRE) were provided by R. A. Hipskind and have been described elsewhere (34).
Plasmids used to express his-tagged Elk-1 FxFP mutants (FxLA and dbl) were derived from pQE-Elk-his6 and pQE-ElkΔD, respectively (36) by site-directed mutagenesis.
Antibodies used were as follows: the ElkC and JNK/SAPK are rabbit polyclonal antibodies raised against recombinant proteins; the SRF (H-300), ERK1/ERK2 goat (C-14), MSK1 (H-65), RSK2 mouse monoclonal (E-1), TBP (N-12), p300 (C-20), RAP74 (C-18), MED1 (C-19), Elk-1 (I-20) and pElk-1 mouse monoclonal (B-4) were from Santa Cruz; the MSK1 sheep polyclonal (for immunoblots) and HDAC1 (2E10) and HDAC2 (3F3) mouse monoclonals and phospho/acetyl-H3S10K14 rabbit polyclonal were from Upstate; the pERK rabbit monoclonal (20G11) and p38 (9212) were from Cell Signaling; the MED23 mouse monoclonal (D27) was from BD Biosciences; the RNAPII pS5-CTD (H14), pS2-CTD (H5) and RNAPII-CTD (8WG16) mouse monoclonals were from Abcam; the AlexaFluor 488 goat anti-mouse/AlexaFluor 568 goat anti-rabbit were from Invitrogen, Molecular Probes.
Generation of pre-initiation complexes in vitro
Streptavidin-coated magnetic beads (300 μg), pre-washed in binding buffer (1 M NaCl, 10 mM Tris pH 7.4, 0.2 mM EDTA), were incubated with biotinylated promoter fragment (20–25 pmol) in binding buffer for 1 h at RT, then washed twice in binding buffer and twice in transcription buffer (12 mM HEPES pH 8.0, 12% glycerol, 60 mM KCl, 0.12 mM EDTA, 7.5 mM MgCl2, 1 mM DTT, 0.5 mM PMSF). Nuclear extract (300 μg) was pre-incubated with salmon sperm DNA (3 μg), poly(dIC) (3 μg) in transcription buffer (200 μl) on ice for 15 min. Immobilized promoters and nuclear extract were combined and incubated at 30°C for 45 min with gentle shaking. PICs were washed thrice in transcription buffer with 0.05% NP-40 and proteins were subsequently eluted in 1 M NaCl at 30°C for 15 min.
Reconstituted promoter-binding assays with recombinant proteins
Methods used to generate recombinant coreSRF, his-tagged rElk-1, inactive and active rERK2 have all been described (37,38). For in vitro phosphorylation, rElk-1 (1 μg) and mutant derivatives were incubated with active rERK2 (0.5 μg) in PP buffer (25 mM Tris pH 7.2, 10 mM MgCl2, 1 mM DTT, 0.1 mM EGTA, 0.1 mM Na3VO4, 1 μM okadaic acid, 250 μM ATP) at 37°C, after which Elk-1 proteins were examined by SDS–PAGE and immunoblotting. For promoter binding Elk-1 (3 μg) and coreSRF (0.75 μg) were pre-incubated in PP buffer (4 mM HEPES pH7.5, 150 mM NaCl, 5 mM MgCl2, 0.2 mM EDTA, 0.1 mM Na3VO4, 0.1% Triton X-100, 40 mM β-glycerophosphate and 0.5 mM DTT) containing poly(dIC) and sheared herring sperm DNA (60 μg ml−1 each) on ice for 10 min prior to addition of 12.5 μg ml−1 biotinylated SRE promoter template, incubation for 10 min at 22°C, addition of ERK proteins (2 μg) and further incubation for 20 min at 22°C. Streptavidin-coated magnetic beads (100 μg), pre-incubated in BP buffer containing BSA (1 mg ml−1) were incubated with complexes for 1 h, washed three times in BP buffer and eluted in 1 M NaCl for 15 min at 22°C. Proteins were examined by SDS–PAGE and immunoblotting.
Chromatin immunoprecipitation assays
ChIP assays were performed as described (39) with modifications. HeLa cells were incubated with 1% formaldehyde at 37°C for 10 min, washed twice in ice-cold PBS with 125 mM glycine, 1 mM EDTA, 1 mM PMSF and collected in 1 ml of ice-cold PBS. Cell pellets were re-suspended in lysis buffer (50 mM Tris–HCl pH 8.0, 1% SDS, 10 mM EDTA, 1× protease inhibitor cocktail) and sonicated to produce DNA fragments of 200–500 bp. Lysates were diluted 10-fold in 20 mM Tris–HCl, pH 8.0, 1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 1× protease inhibitor cocktail and incubated with antibodies over night at 4°C. Immune complexes were incubated with sheared salmon sperm DNA for 1 h at 4°C before the addition of protein G Sepharose beads, pre-blocked with BSA and further incubation for 1 h. Immunoprecipitates were washed with TSE I (20 mM Tris–HCl pH 8.0, 0.1% SDS, 1% Triton X-100, 2 mM EDTA, 150 mM NaCl); TSE II (20 mM Tris–HCl pH 8.0, 0.1% SDS, 1% Triton X-100, 2 mM EDTA, 500 mM NaCl); Buffer III (10 mM Tris–HCl pH 8.0, 1% NP-40, 1 mM EDTA, 1% deoxycholate, 0.25 M LiCl) and twice with TE buffer. DNA–protein complexes were eluted twice with 1% SDS in 0.1 M NaHCO3. Eluates were pooled and cross-links reversed at 65°C for 6 h. After proteinase K digestion for 1 h at 45°C, DNA fragments were purified with a PCR purification kit (Qiagen). Primers for the human c-fos promoter (−473 to −276) were: 5′-GGGTCCGCATTGAACCAGGTGC (forward) and 5′-GCCGTGGAAACCTGCTGACGCA (reverse); the human c-fos gene (+1711 to +1865) primers were: 5′-CTGGGAACTCGCCCCACCTGTGTC (forward) and 5′-CACTGCAGGTCCGGACTGGTCGAG (reverse). Primers for the human egr1 promoter were as published elsewhere (40); the egr-1 gene (+1066 to +1305) primers were 5′-ATTTGCGTCAGCTGTTGTTG (forward) and 5′-CAGCACCTTCTCGTTGTTCA (reverse). For PCR quantification, Aida software was used to measure band intensities from digital gel images. Values represent averages from three independent experiments (error bars = SD).
Immunocytochemistry, confocal microscopy and image analysis
Cells were fixed directly in 2% formaldehyde in PBS for 15 min at RT, permeabilized for 10 min in 0.5% Triton X-100 in PBS, incubated for 10 min in 100 mM glycine in PBS, 10 min in 10% BSA and 5 min in PBS containing 0.5% BSA (PBS/BSA). Incubations with primary antibodies (mouse monoclonal α-pElk-1 (B-4, Santa Cruz); rabbit monoclonal α-pERK (20G11) were carried out in PBS/BSA for 1 h at RT [for the mouse monoclonal (H14) to RNAPII at 4°C over night]. Cover slips were subsequently washed four times for 5 min in PBS/BSA and incubated with secondary antibodies (AlexaFluor 488 goat anti-mouse, and AlexaFluor 568 goat anti-rabbit) for 1.5 h in PBS/BSA at RT. Cover slips were washed twice for 5 min in PBS/BSA, twice for 5 min in PBS, for 10 min in PBS containing 0.4 μg ml−1 Hoechst 33258 and 10 min in PBS. Cover slips were mounted in Vectashield (Vector Labs).
Images were collected with a Leica SP2 Confocal Laser Scanning Microscope (CLSM), equipped with a Leica IRBE inverted fluorescence microscope (63× objective, oil-immersion, NA 1.3). The green fluorescence of AlexaFluor 488 or GFP was excited at a wavelength of 488 nm and detected at 498–550 nm using a spectrophotometer detection system; AlexaFluor 568 labelled proteins were excited at 561 nm and detected at 580–667 nm. In all cases, the pinhole was equivalent to 1 Airy disk. Images were collected as single optical sections, averaged 20 times, displayed using the associated Leica SP2 software and compiled using Adobe Photoshop. For image analysis, red and green-channel confocal images were combined into a dual-colour image in Leica SP2 software. Lines were drawn through the nuclei and red and green channel line-scan intensity profiles were obtained. The correlation between the red and the green intensities was determined according to Pearson's correlation coefficient (Rp). For each nucleus examined, five independent line-scans were performed and averages were derived from 10 values.
RESULTS
Generation and validation of PICs on SRE promoters
Three simple promoters were used to produce PICs in HeLa cell nuclear extracts (Figure 1a). The SRE promoter consisted of a single copy of the c-fos SRE, which is known to bind SRF and Elk-1 in a ternary complex (7), centred 27-bp upstream of a TATA element. The DSE promoter carried a mutated ETS-binding site; the TATA promoter lacked an activating element altogether. Complexes recruited to these promoters in HeLa nuclear extracts contained transcription factors as anticipated. Thus SRF, Elk-1 and TBP bound to the SRE promoter (Figure 1b, lanes 2 and 9); SRF and TBP but not Elk-1 bound to the DSE promoter (lanes 3 and 10) whereas neither Elk-1 nor SRF bound to the basal TATA promoter (lanes 4 and 11), irrespective of prior mitogen stimulation of the cells, as indicated by ERK and Elk-1 phosphorylation (Figure 1c). Binding of SRF to the SRE, but not the DSE, is enhanced by Elk-1, as reported earlier (41).
Figure 1.
Functional analysis of simple promoters used to derive pre-initiation complexes. (a) Diagram showing DNA elements present in promoter templates. The SRE sequence is derived from the human c-fos promoter with the CArG box centred 27-bp upstream of the TATA box. In the DSE promoter the Ets-binding site is inactivated by a triple point mutation and the TATA promoter lacks an upstream element. (b) Nuclear extracts from unstimulated (lanes 1–7) or serum-stimulated (lanes 8–14) cells were used for PIC assembly on SRE (lanes 2 and 9), DSE (lanes 3 and 10) or TATA (lanes 4 and 11) templates. Complexes were isolated, washed and resolved by SDS–PAGE for analysis by immunoblotting with antibodies against Elk-1 (upper panel), SRF (middle) and TBP (lower). Inputs were resolved in lanes 1 and 8; flow through fractions in lanes 5–7 and 12–14. (c) Time course of Elk-1 and ERK phosphorylation in response to serum stimulation of HeLa cells. (d) NIH3T3 cells were transfected with SRE-Luc3 or DSE-Luc3 alone or with expression vectors for active RhoA or C-Raf, as indicated. Data are from one representative experiment with triplicate points (error bars = SD). (e) Circular and linear templates, as indicated, containing an SRE (lanes 1–4 and 9–12) or basal promoter (TATA) (lanes 5–8 and 13–16), were incubated at two different concentrations along with a control template derived from the adenovirus 2 major late promoter (MLP) in transcription reactions with nuclear extracts from serum-starved (−) or stimulated (+) cells. RNA transcripts from SRE templates are indicated with arrows. (f) Nuclear extracts from unstimulated (lanes 1–7) or serum-stimulated (lanes 8–14) cells were used for PIC assembly on SRE (lanes 2 and 9), DSE (lanes 3 and 10) or TATA (lanes 4 and 11) templates. Complexes were isolated, washed and resolved by SDS–PAGE for analysis by immunoblotting with antibodies against p300 (upper panel), MED1, MED23, RAP74, HDAC1 and HDAC2 (lower panel). Inputs were resolved in lanes 1 and 8; flow through fractions in lanes 5–7 and 12–14.
Functionality of these simple promoters in vivo was confirmed by reporter assays. The SRE was preferentially activated by C-Raf, a component of the ERK cascade, while the DSE promoter responded better to RhoA, which triggers an Elk-1 independent pathway involving the SRF co-activator MAL/MKL-1 (42–44) (Figure 1d). In in vitro transcription assays, RNA was transcribed accurately from both circular (Figure 1e, left panel) and linear (right panel) templates. However, similar levels of transcription were achieved from both SRE and TATA promoters and regardless of mitogen stimulation, as observed previously (45). Importantly, this result confirmed that the promoters recruit transcriptionally competent PICs in vitro.
Next, we examined PIC occupancy by transcriptional co-activator proteins potentially affected by mitogens. The histone acetyl transferase (HAT) p300 was detected only in PICs formed on SRE and DSE promoters in mitogen-stimulated nuclear extracts (Figure 1f, lanes 9 and 10) (16,19). In contrast to HATs, HDACs have been reported to dissociate from activated transcription factors upon stimulation (46,47), but both HDAC1 and HDAC2 were detected in PICs formed on the SRE, DSE and TATA promoters irrespective of mitogen stimulation (lower panel) (47,48). Likewise, the TFIIF subunit RAP74, which interacts with SRF (49), MED1 (also known as TRAP220 or PPAR-binding protein), a component of the DRIP/CRSP/mediator co-activator complex required for transcriptional activation by nuclear hormone receptors (e.g. thyroid hormone receptor) and other transcription factors (50,51), and MED23, another mediator subunit shown to undergo phosphorylation-dependent interactions with Elk-1 (17,18), were on all three promoters and irrespective of stimulation. This recruitment pattern suggests that basal transcription factors and the mediator complex can associate with SRE promoters prior to mitogen stimulation, which is consistent with published data (52) (see below). On the basis of this data, we concluded that the characteristics of the in vitro PICs were sufficient to warrant further characterization of their composition.
Mitogen-dependent recruitment of ERK and MSK to SRE promoter complexes
Transcriptional activation by Elk-1 in response to mitogens is contingent upon its phosphorylation on multiple sites by ERK and the consequent recruitment of co-activators such as p300 to SREs (12–14). Although active ERK is known to accumulate in the nuclei of mitogen-stimulated cells (53,54) the underlying mechanism is incompletely understood. However, several reports of MAPKs associating with gene promoters have appeared recently (22–27).
Among the proteins present in PICs assembled in vitro we identified ERK. As shown in Figure 2a, ERK was absent from PICs formed on SRE, DSE or TATA promoter templates in extracts from unstimulated cells (upper panel, lanes 2–4) but present in PICs formed on SRE promoters in mitogen-stimulated extracts (lane 9). ERK was not detected on DSE or TATA promoter templates (lanes 10 and 11). Immunoblotting for phospho-ERK confirmed bound ERK to be active. Neither JNK/SAPK nor p38MAPK (lower panels) was detected in any of the PICs, regardless of promoter template or mitogen stimulation. These data suggest that mitogens stimulate recruitment of active ERK into PICs assembled on SRE promoter templates.
Figure 2.
Pre-initiation complexes on SRE promoters contain ERKs and MSK1. (a) Nuclear extracts from unstimulated (lanes 1–7) or serum-stimulated (lanes 8–14) cells were used for PIC assembly on SRE (lanes 2 and 9), DSE (lanes 3 and 10) or TATA (lanes 4 and 11) templates. Complexes were isolated, washed and resolved by SDS–PAGE for analysis by immunoblotting with antibodies against ERKs (upper panel), phospho-ERKs, JNKs and p38 (lower). Inputs were resolved in lanes 1 and 8; flow through fractions in lanes 5–7 and 12–14. (b) As in (a) except with antibodies against MSK1 (upper panel) and RSK2 (lower panel).
The related mitogen and stress-responsive kinase (MSK) and ribosomal S6 kinase (RSK) are MAPK-dependent protein kinases. Both have been linked to the promoters of IE genes such as c-fos and implicated in histone H3S10 and H3S28 phosphorylation in response to EGF (55–58). Mutations in the RSK2 gene are linked to Coffin–Lowry syndrome (59). Although neither kinase was detected in PICs assembled in unstimulated extracts, we consistently observed the association of MSK but not RSK with SRE PICs formed in extracts from serum-stimulated cells (Figure 2b).
Recruitment of ERK to SRE promoter requires docking motifs in Elk-1
As the SRE binds Elk-1 whereas the DSE does not, we used recombinant proteins to test whether Elk-1 recruited ERK to the SRE. In the absence of other proteins or in the presence of the DNA-binding domain of SRF (coreSRF) alone, neither inactive nor active ERK2 was recruited to an immobilized SRE promoter (Figure 3a, lanes 2, 3, 7 and 8). When recombinant Elk-1 was included in the binding reactions, active ERK2 but not inactive ERK2 was recruited into SRE complexes (lanes 5 and 6). Analogous experiments performed with recombinant MSK1 failed to yield promoter complexes containing MSK (data not shown).
ERK interactions with their regulators and substrates are governed by docking domains (60,61). Within Elk-1 the D domain/KIM and the FxFP motif are important for efficient phosphorylation of Elk-1 (62,63). Versions of Elk-1 with MAPK docking sites mutated or removed (Figure 3b and c) were used to test their contribution to ERK recruitment into SRE promoter complexes. In a kinase assay with active ERK2, in which phosphorylation of S383 was assessed with a phospho-specific antibody (Figure 3d) deletion of the D-box/KIM (Elk-ΔD) had only a minor effect on S383 phosphorylation whereas mutation of the FxFP site (FxLA) reduced S383 phosphorylation by ERK2 up to 90%. An Elk-1 mutant lacking both docking sites (dbl) was not phosphorylated by ERK2 under these conditions. When the Elk-1 mutants were tested for the ability to recruit ERK into SRE complexes recruitment of active ERK2 was impaired by mutation of the FxFP motif (Figure 3e, compare lanes 4 and 8) and lost with the D box/KIM mutant (lanes 6 and 10). Thus, although the FxFP motif is necessary for efficient phosphorylation of Elk-1 residues critical for activation, only the D box/KIM appears to be essential for ERK recruitment to the SRE.
Figure 3.
Elk-1 recruitment of ERK to SRE in vitro is dependent on D domain. (a) The SRE promoter template was incubated with recombinant coreSRF alone (lanes 1–3), coreSRF and rElk-1 (lanes 4–6) or neither (lanes 7 and 8) along with inactive rERK (lanes 2, 5 and 7) or active rERK (lanes 3, 6 and 8). Complexes were isolated on magnetic beads and analysed by immunoblotting. The arrows indicate ERK2 (upper panel) and Elk-1 (lower). (b) Representation of Elk-1 including DNA-binding domain (ets), SRF interaction domain (B), suppressor domain (S), docking (D) domain, trans-activation domain (C) and FxFP motif (f). Sites of deletion/mutation are indicated in the corresponding mutants. (c) SDS–PAGE analysis of recombinant proteins used in this study, as indicated. Full-length proteins and coreSRF are identified by arrowheads. (d) Recombinant Elk-1 and the deletions represented in (b) were incubated with active rERK2 for the times indicated and phosphorylation of Ser383 was analysed by SDS–PAGE and immunoblotting with a phospho-specific antibody (upper panel). Elk-1 substrates were controlled with an antibody against Elk-1 (lower panel). N.B. phosphorylation of Elk-1 results in electrophoretic separation of isoforms that increases the signal on the membrane. Relative amounts of Elk-1 proteins in the assay are apparent in lanes 1, 4, 7 and 10. (e) The SRE promoter template was incubated with recombinant coreSRF alone (lanes 1 and 2), with coreSRF and rElk-1 (lanes 3 and 4), rElk-ΔD (lanes 5 and 6), rElk-FxLA (lanes 7 and 8), rElk-dbl (lanes 9 and 10) or without (lanes 11 and 12) along with inactive rERK2 (lanes 1, 3, 5, 7, 9 and 11) or active rERK2 (lanes 2, 4, 6, 8, 10 and 12). Complexes were isolated on magnetic beads and analysed by immunoblotting with antibodies against ERK (upper panel) and Elk-1 (lower panel).
Mitogen-dependent recruitment of ERK and MSK to c-fos and egr-1 promoters
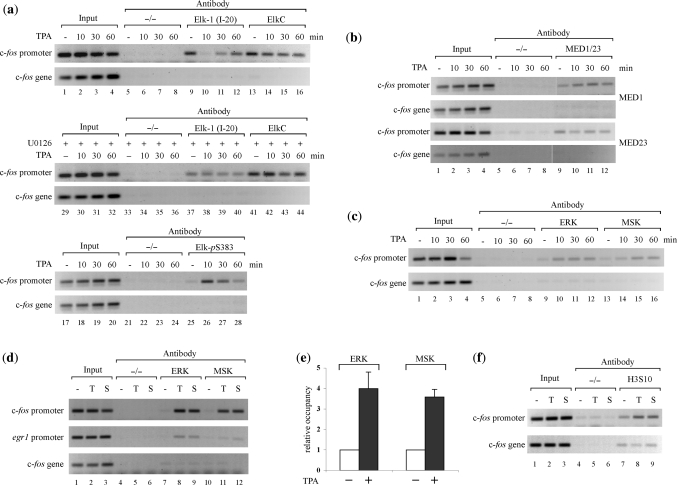
To look for recruitment of kinases to mitogen-responsive gene promoters we examined c-fos promoter complexes by ChIP assay and semi-quantitative PCR (Supplementary Figure S1). First, we used an antibody against Elk-1 (I-20) used previously in such assays (64,65). In serum-starved cells, Elk-1 was detected on the c-fos promoter, but not within the c-fos gene. However, upon TPA stimulation the signal was lost, returning over 1 h (Figure 4a, upper panels, lanes 9–12). In contrast, a second antibody (ElkC) detected Elk-1 on the promoter throughout the time course (lanes 13–16). When cells were treated with the MEK inhibitor U0126, both antibodies detected Elk-1 at the c-fos promoter before and during mitogen stimulation (Figure 4a, middle panels). A third, phospho-specific Elk-1 antibody (ElkpS383) detected phospho-Elk-1 on the promoter transiently after TPA stimulation, which correlated with the time course of Elk-1 phosphorylation in mitogen-stimulated cells (Figure 1c and data not shown). Further experiments confirmed that the I-20 antibody we used detected only unphosphorylated Elk-1 (data not shown). Taken together, these results indicate that unphosphorylated Elk-1 occupies the c-fos SRE prior to stimulation and that phosphorylated Elk-1 is present transiently following mitogen stimulation.
Figure 4.
ERKs and MSK1 are recruited to c-fos promoter in response to mitogen stimulation. (a) HeLa cells were serum starved or starved and stimulated with TPA (100 ng ml−1) for the times indicated, with (+) or without (−) treatment with U0126. Binding of Elk-1 to the c-fos promoter was then determined by chromatin immunoprecipitation (ChIP). PCRs containing primer pairs amplifying a promoter region containing the SRE (upper panel) or a region of the gene (lower panel) were performed after immunoprecipitation of DNA/promoter complexes with the Elk-1 antibodies indicated. (b) As in (a) except binding of mediator components to the c-fos promoter was determined by chromatin immunoprecipitation (ChIP) with antibodies against MED1 and MED23. (c) As in (a) except immunoprecipitations were performed with ERK (lanes 9–12) or MSK1 antibodies (lanes 13–16). (d) As in (c) except cells were treated with 500 ng ml−1 TPA for 15 min (T) or 20% FCS for 30 min (S). (e) Quantification of ERK and MSK binding in three independent experiments similar to those presented in (d). Error bars = SD. (f) As in (d) except immunoprecipitations were performed with a histone H3 pS10 AcK14 antibody (lanes 7–9).
The presence of mediator components in the in vitro PICs (Figure 1f) prompted us to examine their behaviour in similar experiments. Both MED1 and MED23 were detected on the c-fos promoter both before and during mitogen stimulation (Figure 4b, lanes 9–12). An increase in MED23 was not apparent but a modest stimulation of MED1 binding was observed, as reported previously for the egr1 promoter (52).
In parallel analyses, we looked for ERK and MSK at the c-fos promoter. In both cases, we detected increased association upon stimulation with 100 ng ml−1 TPA, whereby recruitment of ERK appeared to precede that of MSK (Figure 4c, lanes 9–12 and 13–16). When cells were stimulated with 500 ng ml−1 TPA for 15 min or 20% FCS for 30 min, recruitment of ERK and MSK to the c-fos promoter was pronounced (Figure 4d, lanes 8, 9, 11 and 12). Under these circumstances, we observed a 3–4-fold increase in ERK and MSK occupancy at the c-fos promoter following stimulation (Figure 4e).
A consequence of MSK activation is H3S10 phosphorylation [often with acetylation of an adjacent lysine (66)], which has been detected in the vicinity of mitogen-responsive gene promoters and correlated with transcriptional activity (57,58,67). We therefore looked for H3S10 phosphorylation in response to TPA and serum at the c-fos promoter. As shown in Figure 4f, H3S10 phosphorylation was significantly greater over the promoter than on the c-fos gene and increased following mitogen stimulation (lanes 8 and 9) in parallel with MSK recruitment.
Next, we looked if these observations extended to other IE gene promoters. TPA stimulation transiently increased the level of phospho-Elk-1 at the egr-1 promoter (Figure 5a, lanes 9–12), which correlated with an increase in MED1 recruitment (lanes 13–16). At the same time, recruitment of ERK and MSK to the egr-1 promoter also increased (Figure 5b) and higher concentrations of TPA or serum were more effective at stimulating ERK recruitment (Figure 5c). TPA also stimulated H3S10 phosphorylation over the egr-1 promoter but not over the gene (Figure 5d). Taken together, these data indicate that ERK and MSK are recruited to the c-fos and egr1 promoters following mitogen stimulation of HeLa cells.
Figure 5.
ERKs and MSK1 are recruited to egr-1 promoter in response to mitogen stimulation. (a) HeLa cells were serum starved or starved and stimulated with TPA (100 ng ml−1) for the times indicated and the presence of pElk-1 and MED1 at the egr-1 promoter was determined by chromatin immunoprecipitation (ChIP). PCRs containing primer pairs amplifying a promoter region containing the promoter proximal SRE (upper panel) or a region of the gene (lower panel) were performed after immunoprecipitation of DNA/promoter complexes with the antibodies indicated. (b) As in (a) except immunoprecipitations were performed with ERK (lanes 9–12), MSK1 (lanes 13–16) or Elk-1 (lanes 17–20) antibodies. (c) As in (b) except cells were treated with 500 ng ml−1 TPA for 15 min (T) or 20% FCS for 30 min (S). (d) As in (c) except immunoprecipitations were performed with a histone H3 pS10 AcK14 antibody (lanes 7–9).
Localization of Elk-1, ERK and RNAPII in HeLa nuclei
Mitogens were first shown to stimulate nuclear accumulation of ERKs by immuno-staining and confocal microscopy (53). We used a similar approach to look for nuclear co-localization of active ERK and Elk-1. Immunoblotting experiments confirmed that two pERK antibodies detected only pERKs (predominantly ERK2 in HeLa cell lysates). A rabbit pElk-1 antibody detected pElk-1 in lysates from mitogen-stimulated cells but also high molecular weight species in both stimulated and unstimulated cells. However, a mouse monoclonal only detected phospho-isoforms of Elk-1 (Supplementary Figure S2a). As a further test of antibody fidelity, quiescent or mitogen-stimulated HeLa cells transfected with a vector for a GFP-Elk-1 fusion protein were fixed and stained for pElk-1. A high coincidence of GFP fluorescence and immunostaining was observed in the mitogen-stimulated cells, confirming the specificity of the pElk-1 antibody (Supplementary Figure S2b and c).
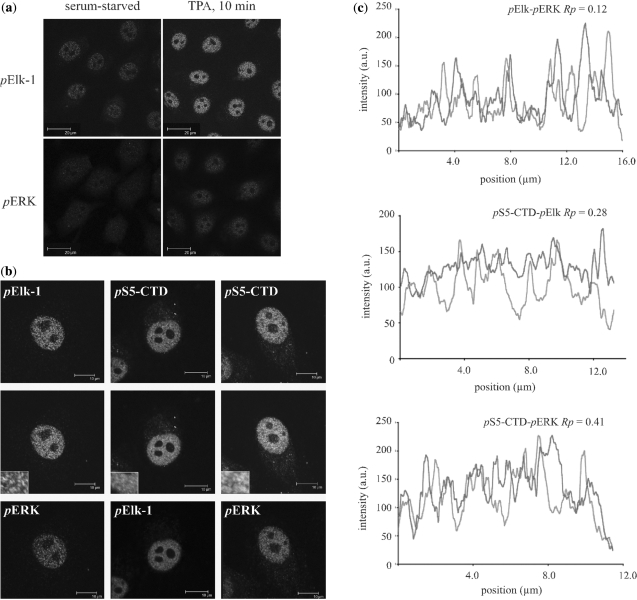
In serum-starved cells weak, punctate pElk-1 staining was seen within the nuclei whereas there was little staining of pERK (Figure 6a, left panels). Following mitogen stimulation, strong, punctate pElk-1 staining was seen, coincident with the appearance of nuclear pERK (Figure 6a, right panels and Figure 6b). No staining was observed with either secondary antibody, either alone or when incubated with unmatched primary antibodies (data not shown). When the pElk-1 and pERK images were merged, coincident staining was apparent although not extensive (Figure 6b left, see inset in middle quadrant), showing that active ERK does not associate predominantly with pElk-1 within the nucleus.
Figure 6.
Nuclear pERK co-localises with RNAPII pS5-CTD. (a) HeLa cells were serum-starved or starved and stimulated with TPA (100 ng ml−1) for 10 min. Cells were fixed, stained with antibodies as indicated and examined by confocal microscopy. (b) Higher magnification images of TPA-stimulated cells stained for pElk-1 and pERK (left column), pS5-CTD and pElk-1 (centre column) and pS5-CTD and pERK (right column). Insets within merged panels represent 3-fold digitally enlarged areas within the respective nuclei. (c) Line-scan analyses of the images shown in (b). Pearson's correlation coefficients (Rp) for the line scans shown are indicated. Mean Rp values (n = 10) for coincident staining were: for pElk-1 and pERK 0.13; for pElk-1 and RNAPII pS5-CTD 0.33; for pERK and RNAPII pS5-CTD 0.41.
To see if the locations of pElk-1 and pERK corresponded to active promoters we looked for co-localization of each with RNAPII pS5-CTD as a marker for sites of transcriptional initiation. The distribution of RNAPII pS5-CTD corresponded with that reported previously (Figure 6b) (68). In the case of pElk-1, we saw co-localization with RNAPII pS5-CTD (Figure 6b, centre panels). For pERK, foci of coincident staining were even more apparent in the nuclei of mitogen-stimulated cells (right-hand panels).
As an objective measure of nuclear co-localization, line-scan analyses were performed (69). For GFP-Elk-1 and pElk-1, we obtained a mean Pearson's correlation coefficient of 0.79, consistent with a high degree of coincident staining (Supplementary Figure S2c). In contrast, the mean value for pERK and pElk-1 was 0.13, which was exceeded by the apparent co-localization of both proteins with RNAPII pS5-CTD (means of 0.33 and 0.42, respectively, see Figure 6c). Analogous experiments performed with antibodies specific for CTD phosphorylated on serine 2 (pS2-CTD) and total RNAPII-CTD showed that Elk-1 co-localized preferentially with RNAPII carrying pS5-CTD, i.e. engaged in transcriptional initiation (Supplementary Figure S2d). Taken together, these observations indicate that pElk-1 and pERK localize more readily with active RNAPII than with each other, suggesting that the interaction between pERK and Elk-1 in mitogen-stimulated cells is transient.
Elk-1 knockdown prevents recruitment of ERK and MSK to c-fos and egr-1 promoters
If Elk-1 is involved in the recruitment of ERK and MSK to SRE promoters, removal of Elk-1 from cells should prevent their access to the c-fos promoter. Thus ChIP assays were performed with cells in which Elk-1 expression was first reduced by RNAi (70). As shown in Figure 7a, a combination of two siRNA species reduced Elk-1 expression in HeLa cells by ∼90%. Parallel analyses were then performed on mock and siRNA-transfected cells that were starved and stimulated with TPA. As anticipated, Elk-1 knockdown caused a significant decrease in Elk-1 at the c-fos promoter (Figure 7b, lanes 9–12). Whereas TPA induced the recruitment of both ERK and MSK to the c-fos promoter in control cells (lanes 14 and 18), Elk-1 knockdown prevented the recruitment of both kinases (lanes 16 and 20). Furthermore, Elk-1 knockdown also prevented ERK and MSK recruitment to the egr-1 promoter (Figure 7c). Taken together, these results demonstrate that Elk-1 is required for the recruitment of ERK and MSK to the promoters of IE genes such as c-fos and egr-1.
Figure 7.
Elk-1 knockdown prevents ERK and MSK recruitment to c-fos and egr-1 promoters. (a) HeLa cells were reverse transfected twice with a combination of two Elk-1 siRNAs or mock transfected. After a further 48 h cells were harvested, nuclear extracts prepared and analysed for Elk-1 expression by SDS–PAGE and immunoblotting. (b) Native (−) or Elk-1 knockdown HeLa cells (+) were serum starved or starved and stimulated with TPA (100 ng ml−1) for 15 min. Binding of Elk-1 (lanes 9–12), ERK (lanes 13–16) and MSK (lanes 17–20) to the c-fos promoter was determined by ChIP assay. (c) As in (b) except binding of Elk-1 (lanes 9–12), ERK (lanes 13–16) and MSK (lanes 17–20) to the egr-1 promoter was determined.
DISCUSSION
In this study, we have examined PICs associated with promoters that contain a mitogen-responsive SRE. We found that ERK and MSK are recruited to these complexes in a mitogen-dependent fashion in vitro, that the ternary complex factor Elk-1 recruits ERK directly via its D-domain/KIM in vitro and is required for recruitment of both ERK and MSK to the c-fos promoter in vivo.
Validation of in vitro PIC formation on SRE promoters
Evidence that the PICs generated reflect the complexes anticipated on SRE promoters included the sequence-specific presence of Elk-1 and SRF, the recruitment of basal factors, co-activators and co-repressors previously reported to interact with SRF and Elk-1 (16–19,47,49). Further evidence for functionality was provided by in vitro transcription reactions. Although the absence of chromatin precluded activator-dependent transcription, these experiments confirmed accurate initiation and transcription by RNAPII on linear templates analogous to those immobilized on beads. In addition, reporter assays confirmed that the single element promoters used for these studies were functional in cells.
Many nuclear proteins have the potential to associate with immobilized DNA molecules in vitro and are potentially present as contaminants rather than valid components of PICs, as earlier analyses of such complexes have indicated (71,72) (our unpublished data; Vougier,S. et al., manuscript in preparation). A specific concern was the presence of basal transcription factors and mediator components in PICs from unstimulated cells. However, ChIP assays confirmed that MED1 and MED23 occupy the c-fos promoter in serum-starved cells, in agreement with our in vitro data and results on the egr1 promoter (52). Set against that, our conclusions that ERK and MSK associated selectively with SRE PICs was based on their absence from DSE and TATA promoters, their failure to associate without mitogen stimulation (although present in unstimulated nuclear extracts), the absence of related MAPKs and RSKs and corroboration by ChIP analyses of the c-fos and egr1 promoters showing mitogen-induced recruitment of both ERK and MSK. The presence of Elk-1 prior to mitogen stimulation is consistent with earlier in vivo footprint analyses (7,73) and appears to support the view that a stable promoter architecture is established over the SRE. However, as with previous work, the present study does not exclude the possibility of protein turnover at SRE promoters.
Although anticipated, this is the first demonstration of ERK recruitment to mitogen-responsive SRE promoters. However, the presence of MSK chimes with earlier reports of its involvement in dynamic histone H3S10 and H3S28 phosphorylation at IE gene promoters (57,58,67). We confirmed that H3S10 phosphorylation at the c-fos promoter increased with mitogen stimulation and, in addition, demonstrated its correlation with MSK recruitment. Furthermore, the detection of MSK is consistent with the interpretation that MSK rather than RSK2 fulfils this role in response to mitogens at IE gene promoters (55,57,66).
Promoter recruitment of ERK through Elk-1 docking domains
Protein interactions by ERKs centre on the common docking (CD) and extended docking (ED) domains (74,75) and complementary kinase interaction motifs have been found on ERK activators, substrates and phosphatases alike (60,76,77). Elk-1 possesses a basic/hydrophobic D domain/KIM and an FxFP motif, each thought to be important for efficient phosphorylation of specific sites (15,63). The FxFP influences predominantly S383/9 phosphorylation (63) while phosphorylation sites in Elk-1 for which the D domain/KIM is critical are less well defined, although reagents that might resolve this issue exist (78). Stable associations between substrates and ERK involving both the D domain/KIM and FxFP motif have been demonstrated (15,63,79) and a dual interaction model has been proposed based on structural analyses of ERK and MKP3/Elk-1-derived peptides whereby docking is initiated by the D domain/KIM and subsequent favourable orientation of phosphorylation sites determined by the FxFP motif (77).
Our data provide the first demonstration of an association between active ERK and Elk-1 bound in a promoter complex. In this scenario, the D domain/KIM was the dominant motif and the FxFP made a lesser contribution to ERK recruitment. The severity of the FxFP mutation was confirmed by the virtual loss of S383 phosphorylation. Extrapolating from the model proposed by Zhang et al. (77) to the context of a PIC, it is conceivable that while ERK is associated with Elk-1 via D domain/KIM-CD interactions, FxFP motifs present on neighbouring proteins might engage ERK to promote trans-phosphorylation events. However, the paucity of Elk-1/ERK co-localization indicates that during active transcription, associations between Elk-1 and ERK are likely to be transient.
Nuclear co-localization of Elk-1 and ERK is limited or transient
The resolution and functional assignment of nuclear structures is a complex undertaking. Sites of ongoing transcription appear to be characterized by granular clusters of RNAPII molecules distributed throughout the nucleus yielding a speckled nuclear staining in proliferating (G1) cells (68,80). A punctate distribution has been observed for a number of other factors and chromatin modifications associated with active transcription, for example H3S10 phosphorylation and H3K4 methylation (67,81). We observed comparable patterns of nuclear distribution for both pElk-1 and pERK in mitogen-stimulated cells. The degree of pERK-pElk-1 co-localization, however, was low, indicating that the bulk of pERK and pElk-1 are not in complexes together (Figure 6b). This may reflect either the limited number of sites at which SRE promoters are transcribed or the transient nature of Elk-1/ERK interactions in the nucleus. In fact, the highest level of coincident staining was observed between pERK and RNAPII pS5-CTD, suggesting that ERK appears to associate to some extent with initiating rather than elongating RNAPII. In light of these and other recent findings, it seems feasible that the association of MAPKs with transcription complexes may be a more general characteristic of acutely regulated promoters (22–27).
The recruitment of ERK by Elk-1 and the progesterone receptor (27) as well as the observation of other MAPKs at promoters (22–28) implies that there may be other ERK targets within promoter complexes. In fact MED1 appears to be phosphorylated by ERK, influencing its stability and transactivation potential (29,31). However, several other protein kinases are associated with PICs, including the CTD kinase CDK7/cyclin H (present with MAT1 in TFIIH), CDK8/cyclin C (present in the negative regulatory ARC/Mediator-like complex with Med230/240) and Cdk9/cyclin T/K, part of the P-TEFb complex (positive transcription elongation factor) responsible for the phosphorylation of the RNAPII CTD on S2, promoting elongation (82). Hence, the identification of bona fide ERK substrates in promoter complexes may not be straightforward. For example, we observed promoter-specific differences in pS5-CTD levels and confirmed that recombinant CTD can be phosphorylated by ERK2 in vitro (82,83), but phosphorylation of S5-CTD in PICs was not blocked by ERK inhibitors (84,85). Further analysis of proteins undergoing modification in SRE PICs is currently underway.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Robert Hipskind for plasmids, Marie Smith and Ian Ward for assistance with confocal microscopy and image analysis. This work was supported by grants to PES from the BBSRC (C17917 and C19734). The Open Access publication charges for this article were waived by Oxford University Press.
Conflict of interest statement. None declared.
REFERENCES
- 1.Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu. Rev. Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- 2.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 3.Saha A, Wittmeyer J, Cairns BR. Chromatin remodelling: the industrial revolution of DNA around histones. Nat. Rev. Mol. Cell Biol. 2006;7:437–447. doi: 10.1038/nrm1945. [DOI] [PubMed] [Google Scholar]
- 4.Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2000;103:211–225. doi: 10.1016/s0092-8674(00)00114-8. [DOI] [PubMed] [Google Scholar]
- 5.Miano JM, Long X, Fujiwara K. Serum response factor: master regulator of the actin cytoskeleton and contractile apparatus. Am. J. Physiol. Cell Physiol. 2007;292:C70–C81. doi: 10.1152/ajpcell.00386.2006. [DOI] [PubMed] [Google Scholar]
- 6.Hunt T, Sassone-Corsi P. Riding tandem: circadian clocks and the cell cycle. Cell. 2007;129:461–464. doi: 10.1016/j.cell.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 7.Herrera RE, Shaw PE, Nordheim A. Occupation of the c-fos serum response element in vivo by a multi-protein complex is unaltered by growth factor induction. Nature. 1989;340:68–70. doi: 10.1038/340068a0. [DOI] [PubMed] [Google Scholar]
- 8.Phatnani HP, Greenleaf AL. Phosphorylation and functions of the RNA polymerase II CTD. Genes Dev. 2006;20:2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- 9.Clayton AL, Mahadevan LC. MAP kinase-mediated phosphoacetylation of histone H3 and inducible gene regulation. FEBS Lett. 2003;546:51–58. doi: 10.1016/s0014-5793(03)00451-4. [DOI] [PubMed] [Google Scholar]
- 10.Pearson G, Robinson F, Beers Gibson T, Xu BE, Karandikar M, Berman K, Cobb MH. Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocrinol. Rev. 2001;22:153–183. doi: 10.1210/edrv.22.2.0428. [DOI] [PubMed] [Google Scholar]
- 11.Turjanski AG, Vaque JP, Gutkind JS. MAP kinases and the control of nuclear events. Oncogene. 2007;26:3240–3253. doi: 10.1038/sj.onc.1210415. [DOI] [PubMed] [Google Scholar]
- 12.Sharrocks AD. The ETS-domain transcription factor family. Nat. Rev. Mol. Cell Biol. 2001;2:827–837. doi: 10.1038/35099076. [DOI] [PubMed] [Google Scholar]
- 13.Shaw PE, Saxton J. Ternary complex factors: prime nuclear targets for mitogen-activated protein kinases. Int. J. Biochem. Cell Biol. 2003;35:1210–1226. doi: 10.1016/s1357-2725(03)00031-1. [DOI] [PubMed] [Google Scholar]
- 14.Buchwalter G, Gross C, Wasylyk B. Ets ternary complex transcription factors. Gene. 2004;324:1–14. doi: 10.1016/j.gene.2003.09.028. [DOI] [PubMed] [Google Scholar]
- 15.Yang S-H, Shore P, Willingham N, Lakey JH, Sharrocks AD. The mechanism of phosphorylation-inducible activation of the ETS-domain transcription factor Elk-1. EMBO J. 1999;18:5666–5674. doi: 10.1093/emboj/18.20.5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Janknecht R, Nordheim A. MAP kinase-dependent transcriptional coactivation by Elk-1 and its cofactor CBP. Biochem. Biophys. Res. Com. 1996;228:831–837. doi: 10.1006/bbrc.1996.1740. [DOI] [PubMed] [Google Scholar]
- 17.Stevens JL, Cantin GT, Wang G, Shevchenko A, Berk AJ. Transcription control by E1A and MAP kinase pathway via Sur2 mediator subunit. Science. 2002;296:755–758. doi: 10.1126/science.1068943. [DOI] [PubMed] [Google Scholar]
- 18.Cantin GT, Stevens JL, Berk AJ. Domain-mediator interactions promote transcription preinitiation complex assembly on promoter DNA. Proc. Natl Acad. Sci. USA. 2003;100:12003–12008. doi: 10.1073/pnas.2035253100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li QJ, Yang SH, Maeda Y, Sladek FM, Sharrocks AD, Martins-Green M. MAP kinase phosphorylation-dependent activation of Elk-1 leads to activation of the co-activator p300. EMBO J. 2003;22:281–291. doi: 10.1093/emboj/cdg028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang SH, Sharrocks AD. PIASx acts as an Elk-1 coactivator by facilitating derepression. EMBO J. 2005;24:2161–2171. doi: 10.1038/sj.emboj.7600690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang SH, Sharrocks AD. PIASxalpha differentially regulates the amplitudes of transcriptional responses following activation of the ERK and p38 MAPK pathways. Mol. Cell. 2006;22:477–487. doi: 10.1016/j.molcel.2006.03.037. [DOI] [PubMed] [Google Scholar]
- 22.Alepuz PM, Jovanovic A, Reiser V, Ammerer G. Stress-induced map kinase Hog1 is part of transcription activation complexes. Mol. Cell. 2001;7:767–777. doi: 10.1016/s1097-2765(01)00221-0. [DOI] [PubMed] [Google Scholar]
- 23.Proft M, Struhl K. Hog1 kinase converts the Sko1-Cyc8-Tup1 repressor complex into an activator that recruits SAGA and SWI/SNF in response to osmotic stress. Mol. Cell. 2002;9:1307–1317. doi: 10.1016/s1097-2765(02)00557-9. [DOI] [PubMed] [Google Scholar]
- 24.Bardwell L, Cook JG, Voora D, Baggott DM, Martinez AR, Thorner J. Repression of yeast Ste12 transcription factor by direct binding of unphosphorylated Kss1 MAPK and its regulation by the Ste7 MEK. Genes Dev. 1998;12:2887–2898. doi: 10.1101/gad.12.18.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pokholok DK, Zeitlinger J, Hannett NM, Reynolds DB, Young RA. Activated signal transduction kinases frequently occupy target genes. Science. 2006;313:533–536. doi: 10.1126/science.1127677. [DOI] [PubMed] [Google Scholar]
- 26.Simone C, Forcales SV, Hill DA, Imbalzano AN, Latella L, Puri PL. p38 pathway targets SWI-SNF chromatin-remodeling complex to muscle-specific loci. Nat. Genet. 2004;36:738–743. doi: 10.1038/ng1378. [DOI] [PubMed] [Google Scholar]
- 27.Vicent GP, Ballare C, Nacht AS, Clausell J, Subtil-Rodriguez A, Quiles I, Jordan A, Beato M. Induction of progesterone target genes requires activation of Erk and Msk kinases and phosphorylation of histone H3. Mol. Cell. 2006;24:367–381. doi: 10.1016/j.molcel.2006.10.011. [DOI] [PubMed] [Google Scholar]
- 28.Edmunds JW, Mahadevan LC. Cell signaling. Protein kinases seek close encounters with active genes. Science. 2006;313:449–451. doi: 10.1126/science.1131158. [DOI] [PubMed] [Google Scholar]
- 29.Misra P, Owuor ED, Li W, Yu S, Qi C, Meyer K, Zhu YJ, Rao MS, Kong AN, Reddy JK. Phosphorylation of transcriptional coactivator peroxisome proliferator-activated receptor (PPAR)-binding protein (PBP). Stimulation of transcriptional regulation by mitogen-activated protein kinase. J. Biol. Chem. 2002;277:48745–48754. doi: 10.1074/jbc.M208829200. [DOI] [PubMed] [Google Scholar]
- 30.De Nadal E, Zapater M, Alepuz PM, Sumoy L, Mas G, Posas F. The MAPK Hog1 recruits Rpd3 histone deacetylase to activate osmoresponsive genes. Nature. 2004;427:370–374. doi: 10.1038/nature02258. [DOI] [PubMed] [Google Scholar]
- 31.Pandey PK, Udayakumar TS, Lin X, Sharma D, Shapiro PS, Fondell JD. Activation of TRAP/Mediator subunit TRAP220/Med1 is regulated by mitogen-activated protein kinase-dependent phosphorylation. Mol. Cell Biol. 2005;25:10695–10710. doi: 10.1128/MCB.25.24.10695-10710.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li L, Shaw PE. Elevated activity of STAT3C due to higher DNA-binding affinity of phosphotyrosine dimer rather than covalent dimer formation. J. Biol. Chem. 2006;281:33172–33181. doi: 10.1074/jbc.M606940200. [DOI] [PubMed] [Google Scholar]
- 34.Hipskind RA, Nordheim A. Functional dissection in vitro of the human c-fos promoter. J. Biol. Chem. 1991;266:19583–19592. [PubMed] [Google Scholar]
- 35.Sadowski I, Ma J, Triezenberg S, Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988;335:563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- 36.Drewett V, Muller S, Goodall J, Shaw PE. Dimer formation by ternary complex factor Elk-1. J. Biol. Chem. 2000;275:1757–1762. doi: 10.1074/jbc.275.3.1757. [DOI] [PubMed] [Google Scholar]
- 37.Gille H, Kortenjann M, Thomae O, Moomaw C, Slaughter C, Cobb MH, Shaw PE. ERK phosphorylation potentiates ELK-1-mediated ternary complex formation and transactivation. EMBO J. 1995;14:951–962. doi: 10.1002/j.1460-2075.1995.tb07076.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khokhlatchev AV, Xu S, English J, Wu P, Schaefer E, Cobb MH. Reconstitution of mitogen-activated protein kinase phosphorylation cascades in bacteria. Efficient synthesis of active protein kinases. J. Biol. Chem. 1997;272:11057–11062. doi: 10.1074/jbc.272.17.11057. [DOI] [PubMed] [Google Scholar]
- 39.Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell. 2000;103:843–852. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- 40.Vickers ER, Kasza A, Kurnaz IA, Seifert A, Zeef LA, O’donnell A, Hayes A, Sharrocks AD. Ternary complex factor-serum response factor complex-regulated gene activity is required for cellular proliferation and inhibition of apoptotic cell death. Mol. Cell Biol. 2004;24:10340–10351. doi: 10.1128/MCB.24.23.10340-10351.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schröter H, Mueller CG, Meese K, Nordheim A. Synergism in ternary complex formation between the dimeric glycoprotein p67SRF, polypeptide p62TCF and the c-fos serum response element. EMBO J. 1990;9:1123–1130. doi: 10.1002/j.1460-2075.1990.tb08218.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Graham R, Gilman M. Distinct protein targets for signals acting at the c-fos serum response element. Science. 1991;251:189–192. doi: 10.1126/science.1898992. [DOI] [PubMed] [Google Scholar]
- 43.Hill CS, Wynne J, Treisman R. The Rho family GTPasses RhoA, Rac1 and CDC42Hs regulate transcriptional activation by SRF. Cell. 1995;81:1159–1170. doi: 10.1016/s0092-8674(05)80020-0. [DOI] [PubMed] [Google Scholar]
- 44.Miralles F, Posern G, Zaromytidou AI, Treisman R. Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell. 2003;113:329–342. doi: 10.1016/s0092-8674(03)00278-2. [DOI] [PubMed] [Google Scholar]
- 45.Hipskind RA, Rao VN, Mueller CGF, Reddy ESP, Nordheim A. Ets-related protein Elk-1 is homologous to the c-fos regulatory factor p62TCF. Nature. 1991;354:531–534. doi: 10.1038/354531a0. [DOI] [PubMed] [Google Scholar]
- 46.Criqui-Filipe P, Ducret C, Maira SM, Wasylyk B. Net, a negative Ras-switchable TCF, contains a second inhibition domain, the CID, that mediates repression through interactions with CtBP and de-acetylation. EMBO J. 1999;18:3392–3403. doi: 10.1093/emboj/18.12.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang SH, Sharrocks AD. SUMO promotes HDAC-mediated transcriptional repression. Mol. Cell. 2004;13:611–617. doi: 10.1016/s1097-2765(04)00060-7. [DOI] [PubMed] [Google Scholar]
- 48.Yang SH, Vickers E, Brehm A, Kouzarides T, Sharrocks AD. Temporal recruitment of the mSin3A-histone deacetylase corepresor complex to the ETS domain transcription factor Elk-1. Mol. Cell Biol. 2001;21:2802–2814. doi: 10.1128/MCB.21.8.2802-2814.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Joliot V, Demma M, Prywes R. Interaction with RAP74 subunit of TFIIF is required for transcriptional activation by serum response factor. Nature. 1995;373:632–635. doi: 10.1038/373632a0. [DOI] [PubMed] [Google Scholar]
- 50.Lewis BA, Reinberg D. The mediator coactivator complex: functional and physical roles in transcriptional regulation. J. Cell Sci. 2003;116:3667–3675. doi: 10.1242/jcs.00734. [DOI] [PubMed] [Google Scholar]
- 51.Malik S, Roeder RG. Dynamic regulation of pol II transcription by the mammalian Mediator complex. Trends Biochem. Sci. 2005;30:256–263. doi: 10.1016/j.tibs.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 52.Wang G, Balamotis MA, Stevens JL, Yamaguchi Y, Handa H, Berk AJ. Mediator requirement for both recruitment and postrecruitment steps in transcription initiation. Mol. Cell. 2005;17:683–694. doi: 10.1016/j.molcel.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 53.Chen RH, Sarnecki C, Blenis J. Nuclear localization and regulation of erk- and rsk-encoded protein kinases. Mol. Cell Biol. 1992;12:915–927. doi: 10.1128/mcb.12.3.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pouyssegur J, Lenormand P. Fidelity and spatio-temporal control in MAP kinase (ERKs) signalling. Eur. J. Biochem. 2003;270:3291–3299. doi: 10.1046/j.1432-1033.2003.03707.x. [DOI] [PubMed] [Google Scholar]
- 55.Sassone-Corsi P, Mizzen CA, Cheung P, Crosio C, Monaco L, Jacquot S, Hanauer A, Allis CD. Requirement of Rsk-2 for epidermal growth factor-activated phosphorylation of histone H3. Science. 1999;285:886–891. doi: 10.1126/science.285.5429.886. [DOI] [PubMed] [Google Scholar]
- 56.Bruning JC, Gillette JA, Zhao Y, Bjorbaeck C, Kotzka J, Knebel B, Avci H, Hanstein B, Lingohr P, Moller DE, et al. Ribosomal subunit kinase-2 is required for growth factor-stimulated transcription of the c-Fos gene. Proc. Natl Acad. Sci. USA. 2000;97:2462–2467. doi: 10.1073/pnas.97.6.2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Soloaga A, Thomson S, Wiggin GR, Rampersaud N, Dyson MH, Hazzalin CA, Mahadevan LC, Arthur JS. MSK2 and MSK1 mediate the mitogen- and stress-induced phosphorylation of histone H3 and HMG-14. EMBO J. 2003;22:2788–2797. doi: 10.1093/emboj/cdg273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Duncan EA, Anest V, Cogswell P, Baldwin AS. The kinases MSK1 and MSK2 are required for epidermal growth factor-induced, but not tumor necrosis factor-induced, histone H3 Ser10 phosphorylation. J. Biol. Chem. 2006;281:12521–12525. doi: 10.1074/jbc.M513333200. [DOI] [PubMed] [Google Scholar]
- 59.Trivier E, De Cesare D, Jacquot S, Pannetier S, Zackai E, Young I, Mandel JL, Sassone-Corsi P, Hanauer A. Mutations in the kinase Rsk-2 associated with Coffin-Lowry syndrome. Nature. 1996;384:567–570. doi: 10.1038/384567a0. [DOI] [PubMed] [Google Scholar]
- 60.Sharrocks AD, Yang SH, Galanis A. Docking domains and substrate-specificity determination for MAP kinases. Trends Biochem. Sci. 2000;25:448–453. doi: 10.1016/s0968-0004(00)01627-3. [DOI] [PubMed] [Google Scholar]
- 61.Tanoue T, Nishida E. Docking interactions in the mitogen-activated protein kinase cascades. Pharmacol. Ther. 2002;93:193–202. doi: 10.1016/s0163-7258(02)00188-2. [DOI] [PubMed] [Google Scholar]
- 62.Yang SH, Yates PR, Whitmarsh AJ, Davis RJ, Sharrocks AD. The Elk-1 ETS-domain transcription factor contains a mitogen-activated protein kinase targeting motif. Mol. Cell Biol. 1998;18:710–720. doi: 10.1128/mcb.18.2.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jacobs D, Glossip D, Xing H, Muslin AJ, Kornfeld K. Multiple docking sites on substrate proteins form a modular system that mediates recognition by ERK MAP kinase. Genes Dev. 1999;13:163–175. [PMC free article] [PubMed] [Google Scholar]
- 64.Aprelikova O, Wood M, Tackett S, Chandramouli GV, Barrett JC. Role of ETS transcription factors in the hypoxia-inducible factor-2 target gene selection. Cancer Res. 2006;66:5641–5647. doi: 10.1158/0008-5472.CAN-05-3345. [DOI] [PubMed] [Google Scholar]
- 65.Zhou J, Hu G, Herring BP. Smooth muscle-specific genes are differentially sensitive to inhibition by Elk-1. Mol. Cell Biol. 2005;25:9874–9885. doi: 10.1128/MCB.25.22.9874-9885.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cheung P, Tanner KG, Cheung WL, Sassone-Corsi P, Denu JM, Allis CD. Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol. Cell. 2000;5:905–915. doi: 10.1016/s1097-2765(00)80256-7. [DOI] [PubMed] [Google Scholar]
- 67.Dyson MH, Thomson S, Inagaki M, Goto H, Arthur SJ, Nightingale K, Iborra FJ, Mahadevan LC. MAP kinase-mediated phosphorylation of distinct pools of histone H3 at S10 or S28 via mitogen- and stress-activated kinase 1/2. J. Cell Sci. 2005;118:2247–2259. doi: 10.1242/jcs.02373. [DOI] [PubMed] [Google Scholar]
- 68.Grande MA, van der Kraan I, de Jong L, van Driel R. Nuclear distribution of transcription factors in relation to sites of transcription and RNA polymerase II. J. Cell Sci. 1997;110:1781–1791. doi: 10.1242/jcs.110.15.1781. [DOI] [PubMed] [Google Scholar]
- 69.van Steensel B, van Binnendijk EP, Hornsby CD, van der Voort HT, Krozowski ZS, de Kloet ER, van Driel R. Partial colocalization of glucocorticoid and mineralocorticoid receptors in discrete compartments in nuclei of rat hippocampus neurons. J. Cell Sci. 1996;109:787–792. doi: 10.1242/jcs.109.4.787. [DOI] [PubMed] [Google Scholar]
- 70.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 71.Drewett V, Molina H, Millar A, Muller S, von Hesler F, Shaw PE. DNA-bound transcription factor complexes analysed by mass-spectrometry: binding of novel proteins to the human c-fos SRE and related sequences. Nucleic Acids Res. 2001;29:479–487. doi: 10.1093/nar/29.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ranish JA, Yi EC, Leslie DM, Purvine SO, Goodlett DR, Eng J, Aebersold R. The study of macromolecular complexes by quantitative proteomics. 2003;33:349–355. doi: 10.1038/ng1101. [DOI] [PubMed] [Google Scholar]
- 73.König H. Cell-type specific multiprotein complex formation over the c-fos serum response element in vivo: ternary complex formation is not required for the induction of c-fos. Nucleic Acids Res. 1991;19:3607–3611. doi: 10.1093/nar/19.13.3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tanoue T, Adachi M, Moriguchi T, Nishida E. A conserved docking motif in MAP kinases common to substrates, activators and regulators. Nat. Cell Biol. 2000;2:110–116. doi: 10.1038/35000065. [DOI] [PubMed] [Google Scholar]
- 75.Tanoue T, Maeda R, Adachi M, Nishida E. Identification of a docking groove on ERK and p38 MAP kinases that regulates the specificity of docking interactions. EMBO J. 2001;20:466–479. doi: 10.1093/emboj/20.3.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tanoue T, Nishida E. Molecular recognitions in the MAP kinase cascades. Cell Signal. 2003;15:455–462. doi: 10.1016/s0898-6568(02)00112-2. [DOI] [PubMed] [Google Scholar]
- 77.Zhang J, Zhou B, Zheng CF, Zhang ZY. A bipartite mechanism for ERK2 recognition by its cognate regulators and substrates. J. Biol. Chem. 2003;278:29901–29912. doi: 10.1074/jbc.M303909200. [DOI] [PubMed] [Google Scholar]
- 78.Cruzalegui FH, Cano E, Treisman R. ERK activation induces phosphorylation of Elk-1 at multiple S/T-P motifs to high stoichiometry. Oncogene. 1999;18:7948–7957. doi: 10.1038/sj.onc.1203362. [DOI] [PubMed] [Google Scholar]
- 79.Ducret C, Maira SM, Lutz Y, Wasylyk B. The ternary complex factor Net contains two distinct elements that mediate different responses to MAP kinase signalling cascades. Oncogene. 2000;19:5063–5072. doi: 10.1038/sj.onc.1203892. [DOI] [PubMed] [Google Scholar]
- 80.Wei X, Somanathan S, Samarabandu J, Berezney R. Three-dimensional visualization of transcription sites and their association with splicing factor-rich nuclear speckles. J. Cell Biol. 1999;146:543–558. doi: 10.1083/jcb.146.3.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 82.Loyer P, Trembley JH, Katona R, Kidd VJ, Lahti JM. Role of CDK/cyclin complexes in transcription and RNA splicing. Cell Signal. 2005;17:1033–1051. doi: 10.1016/j.cellsig.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 83.Bonnet F, Vigneron M, Bensaude O, Dubois MF. Transcription-independent phosphorylation of the RNA polymerase II C-terminal domain (CTD) involves ERK kinases (MEK1/2) Nucleic Acids Res. 1999;27:4399–4404. doi: 10.1093/nar/27.22.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hancock CN, Macias A, Lee EK, Yu SY, Mackerell ADJ, Shapiro P. Identification of novel extracellular signal-regulated kinase docking domain inhibitors. J. Med. Chem. 2005;48:4586–4595. doi: 10.1021/jm0501174. [DOI] [PubMed] [Google Scholar]
- 85.Chen F, Hancock CN, Macias AT, Joh J, Still K, Zhong S, MacKerell AD, Jr, Shapiro P. Characterization of ATP-independent ERK inhibitors identified through in silico analysis of the active ERK2 structure. Bioorg. Med. Chem. Lett. 2006;16:6281–6287. doi: 10.1016/j.bmcl.2006.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.