Abstract
Aminoacyl-tRNA synthetases (AARS) are an essential family of enzymes that catalyze the attachment of amino acids to specific tRNAs during translation. Previously, we showed that base-specific recognition of the tRNAPro acceptor stem is critical for recognition by Escherichia coli prolyl-tRNA synthetase (ProRS), but not for human ProRS. To further delineate species-specific differences in acceptor stem recognition, atomic group mutagenesis was used to probe the role of sugar–phosphate backbone interactions in recognition of human tRNAPro. Incorporation of site-specific 2′-deoxynucleotides, as well as phosphorothioate and methylphosphonate modifications within the tRNA acceptor stem revealed an extensive network of interactions with specific functional groups proximal to the first base pair and the discriminator base. Backbone functional groups located at the base of the acceptor stem, especially the 2′-hydroxyl of A66, are also critical for aminoacylation catalytic efficiency by human ProRS. Therefore, in contrast to the bacterial system, backbone-specific interactions contribute significantly more to tRNA recognition by the human enzyme than base-specific interactions. Taken together with previous studies, these data show that ProRS-tRNA acceptor stem interactions have co-adapted through evolution from a mechanism involving ‘direct readout’ of nucleotide bases to one relying primarily on backbone-specific ‘indirect readout’.
INTRODUCTION
The accuracy of translation of the genetic code depends on the recognition of specific tRNAs by cognate aminoacyl-tRNA synthetases (AARS) (1). Based on known co-crystal structures of synthetases with their cognate tRNAs, both base-specific and backbone interactions contribute to the RNA–protein-binding interface (2). Nucleotide base contacts are believed to be critical for specific recognition of both the anticodon loop and the acceptor stem sequences, and are key elements in defining tRNA identity (3). Although there is also evidence for backbone interactions in these domains, the functional relevance of these interactions and their contribution to aminoacylation catalytic efficiency has not been as extensively investigated as base-specific contacts.
Prolyl-tRNA synthetase (ProRS) sequences can be divided into two evolutionarily distant groups (4, 5). These groups are characterized not only by the presence or absence of a distinct prokaryotic-specific insertion domain that functions in post-transfer editing of mischarged Ala-tRNAPro (6, 7), but also by species-specific differences in tRNA recognition (4, 8, 9). Whereas prokaryotic-like ProRS recognizes both the tRNA acceptor stem and anticodon domain in a base-specific manner (9, 10), members of the eukaryotic-like group, including human ProRS, have evolved to possess only weak acceptor stem recognition and appear to rely predominantly on anticodon interactions for tRNA discrimination (4, 8, 9).
Although a co-crystal structure of Thermus thermophilus ProRS complexed to tRNAPro has been reported, the tRNA acceptor stem is not bound (11), and therefore, the structural basis for acceptor stem recognition by ProRS is unknown. However, biochemical data has revealed the significance of base-specific acceptor stem interactions with the major groove of G72 and A73 (9). In contrast to the bacterial tRNAPro species, which contain a unique C1:G72 base pair, the eukaryotic proline tRNAs contain a more typical G1:C72 pair and a C73 ‘discriminator’ base. Mutagenesis at either C72 or C73 resulted in only minor (≤2-fold) effects on the overall kcat/KM for aminoacylation by human ProRS (4). Domain swap experiments, wherein 13 of 15 acceptor stem nucleotides of human tRNAPro were changed, also strongly support the conclusion that base-specific interactions in the acceptor stem do not make an important contribution to recognition by the human enzyme (9). Therefore, the human tRNA acceptor stem has changed significantly through evolution, and the homologous synthetase has not adapted to recognize it in a base-specific fashion. Based on these results, we hypothesize that eukaryotic ProRS systems may have co-adapted to interact with the tRNA acceptor stem in a manner that is distinct from the prokaryotic system and that involves exclusively backbone functional group interactions.
To probe this hypothesis, in this work, the 3′-strand of the acceptor stem of human tRNAPro was modified by incorporation of specific 2′-deoxynucleotides, phosphorothioates and 2′-deoxy-methylphosphonates. The 3′-strand was chosen as the focus of this study based on both class II AARS–tRNA co-crystal structures and footprinting experiments, which suggest that the majority of backbone interactions are localized to this strand (12–16). Exceptions include the findings that the 2′-hydroxyl of C3 in Escherichia coli tRNAThr weakly interacts with E. coli threonyl-tRNA synthetase (ThrRS) (16), and that the phosphate of U1 in yeast tRNAAsp interacts with yeast aspartyl-tRNA synthetase (AspRS) (14). Here, we show that there are extensive interactions between human ProRS and tRNAPro throughout the acceptor stem, including specific functional groups near the first base pair, the discriminator base, and A66. These results support the conclusion that acceptor stem backbone interactions by human ProRS are more significant than base-specific interactions, and provide new insights into how a specific protein–RNA interaction can co-adapt through evolution from a mechanism relying on ‘direct readout’ to one primarily involving ‘indirect readout’.
MATERIALS AND METHODS
Enzyme purification
Purification of human ProRS was performed as described previously using plasmid pKS-509 (17), which express the ProRS portion (codons 926–1440) of the human glutamyl-prolyl-tRNA synthetase fusion protein containing a histidine-tag at the N-terminus. The concentration of human ProRS used in all kinetic experiments was determined by the adenylate burst assay (18).
tRNA preparation and site-specific atomic group modification
Semi-synthetic human tRNAPro was prepared by annealing a 5′-57-mer fragment to a 3′-16-mer. The 5′-57-mer was prepared by in vitro transcription as described (19). Briefly, the gene for a human tRNAPro A57G mutant (20) was assembled by cloning a set of six overlapping synthetic DNA oligonucleotides into the EcoRI and BamHI sites of pUC119 (21). The upstream consensus promoter sequence of T7 RNA polymerase was also introduced into the plasmid. The A57G mutation introduced a BstBI restriction site within the gene, but had no effect on catalytic activity. BstBI-linearized DNA was in vitro transcribed using an established procedure (22, 23), resulting in a 57-nt long RNA corresponding to the 5′-fragment of human tRNAPro. Transcripts were purified on denaturing 12% polyacrylamide gels. The 3′-fragment was chemically synthesized using the phosphoramidite method on an Expedite™ 8909 Nucleic Acid Synthesis System (PerSeptive Biosystems). All phosphoramidites were purchased from Glen Research Corp. (Sterling, VA) and the modified nucleotide analogues were incorporated into synthetic 3′-16-mer oligonucleotides during automated chemical synthesis. In the case of phosphorothioate incorporation, the sulfurizing agent, 3-ethoxy-1,2,4-dithiazoline-5-one (EDITH), was used in the chemical synthesis as described (24, 25). The 3′-16-mer oligonucleotides were purified by electrophoresis on denaturing 16% polyacrylamide gels.
To investigate chiral phosphorothioate effects, anion exchange chromatography (DNAPac™ PA-100, 4 × 250 mm, DIONEX Corp.) or reversed phase chromatography (Nucleosil-100 C-18, 5 µm, 4.6 × 250 mm, Alltech) was used to purify the desired chiral phosphorothioates by high performance liquid chromatography (HPLC) (26). The anion exchange column was employed at 60°C with a flow rate of 1.0 ml/min in buffer containing 0.1 M ammonium acetate (pH 8.0) with 2% CH3CN and increasing KCl concentrations (from 300 to 500 mM over the course of 65 min). RNA purity and Rp or Sp configuration were determined by re-injecting each purified isomer onto a reversed phase C-18 column. Chromatography was performed at room temperature in this case, with running buffer 0.1 M triethylamine acetate (pH 7.0) and an increasing CH3CN ratio (up to 40% over 60 min). Based on literature precedent, faster eluting isomers were assigned as Rp and slower eluting isomers as Sp (26–28). To confirm this assignment, selected purified oligonucleotides were subjected to enzymatic digestion followed by reversed phase HPLC analysis (27). As expected, the Rp isomer is more resistant to cleavage by nuclease P1 (Amersham) while the Sp isomer is more resistant to cleavage by snake venom phosphodiesterase (Amersham). Purification of chiral methylphosphonate-containing oligonucleotides was also successfully carried out using the anion exchange HPLC system under similar conditions at 45°C. Product purity was confirmed in a similar manner by re-injecting purified samples onto a reversed phase HPLC column. The concentration of RNA was determined by measuring the optical density at 260 nm (ε260 = 51.8 × 104 M−1 for 57-mer and 12.5 × 104 M−1 for 16-mers).
Aminoacylation assays
Semi-synthetic tRNA molecules were annealed immediately prior to use in aminoacylation assays by heating the 5′ and 3′ fragments together at 60°C for three min at a 57-mer:16-mer ratio of 1:1.5, followed by addition of MgCl2 to 10 mM, cooling at room temperature for 5 min, and placement on ice. Human ProRS stored at −80°C was diluted immediately before use and stored on ice. l-proline and [3H]-labeled l-proline (100 Ci/mmol) were purchased from Sigma and Amersham Biosciences, respectively. The assays were carried out at room temperature using conditions described previously (29). Relative kcat/KM values of semi-synthetic tRNA molecules were determined by Michaelis–Menten analysis by measuring initial rates of aminoacylation obtained by using 0.04 µM ProRS and tRNA concentrations ranging from 1 to 6 µM. To compare each mutant's aminoacylation activity with wild-type tRNAPro, relative kcat/KM values were converted to the difference in the transition state free energy, −ΔΔG‡, defined as RT·ln[(kcat/KM)variant/(kcat/KM)wild-type].
RESULTS
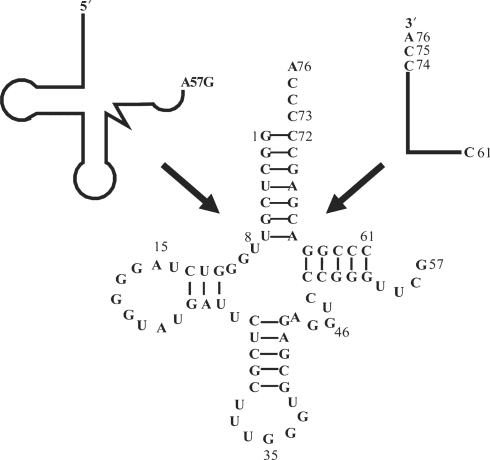
We previously demonstrated that E. coli ProRS can efficiently aminoacylate semi-synthetic E. coli tRNAs prepared by annealing two RNA fragments (19, 29). In the present work, semi-synthetic human tRNAPro was constructed by annealing an in vitro transcribed 5′-57-mer with a chemically synthesized 3′-16-mer (Figure 1). The wild-type semi-synthetic construct is a good substrate for human ProRS, despite the fact that it contains a break in the phosphodiester backbone in the TΨC loop. The efficiency of aminoacylation of semi-synthetic human tRNAPro was similar to that of in vitro transcribed full-length human tRNAPro and aminoacylation assays were performed as previously described (29). To investigate the contribution of specific acceptor stem backbone functional groups in aminoacylation by human ProRS, three different modifications were incorporated into semi-synthetic tRNAs using automated chemical synthesis: 2′-deoxy, phosphorothioate and methylphosphonate. Substitutions were incorporated into positions 63–75 of the 3′-strand of the tRNA, which can be arbitrarily divided into three local regions: a single-strand top region, positions 73–76; a double-stranded acceptor stem region, positions 66–72; and a double-stranded TΨC stem region, position 63–65.
Figure 1.
Sequence of the semi-synthetic A57G human tRNAPro variant used in these studies. The tRNA was constructed by annealing an in vitro transcribed 5′-57-mer with a chemically synthesized 3′-16-mer containing site-specific backbone modifications.
2′-Deoxynucleotide modification
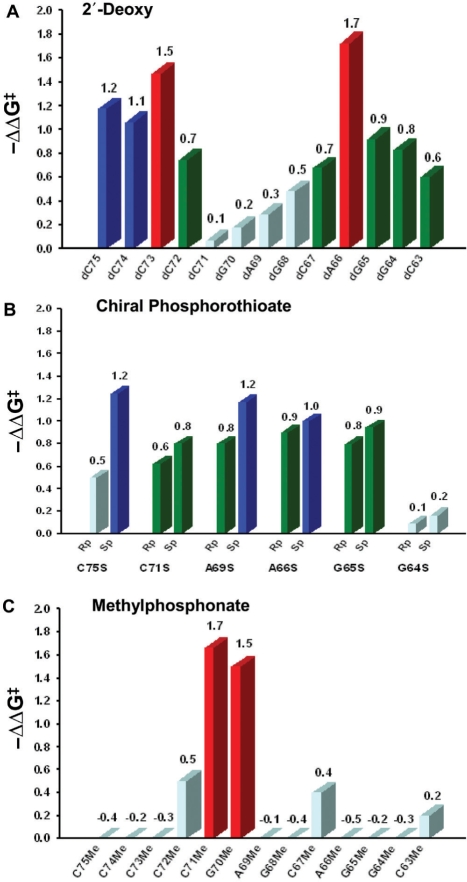
The 2′-deoxy substitution data are summarized in Table 1 and Figure 2A. The results show that each 2′-hydroxyl group in the top single-stranded region of the acceptor stem contributes more than 1.0 kcal/mol to recognition by ProRS. Interestingly, the largest effect was observed upon deletion of the 2′-hydroxyl of A66 located at the bottom of the acceptor stem domain (−ΔΔG‡ = 1.7 kcal/mol). Significant, but smaller effects were observed at positions 63–65, 67 and 72 (−ΔΔG‡ = 0.6–0.9 kcal/mol). Because 2′-hydroxyls can be both hydrogen-bond donors and acceptors, nucleotides 66 and 73–75, which had the largest effects (−ΔΔG‡ = 1.1–1.7 kcal/mol), may interact with human ProRS through a hydrogen-bonding network. In particular, nucleotides in the single-stranded region are likely to penetrate into the catalytic active site, resulting in direct contacts with protein functional groups. The large effect observed upon A66 substitution is consistent with interactions observed in the known crystal structures of class II E. coli ThrRS-tRNAThr, T. thermophilus seryl-tRNA synthetase (SerRS)-tRNASer and T. thermophilus phenylalanyl-tRNA synthetase (PheRS)-tRNAPhe (12,15, 16, 30).
Table 1.
The effect of 2′-deoxy substitutions in the 3′-strand of human tRNAPro on aminoacylation efficiency
| Variant | Relative kcat/KM (±SD) | Activity change (x-fold) | −ΔΔG‡ (kcal/mol) |
|---|---|---|---|
| WT | 1.00 | 1.0 | 0.0 |
| dC75 | 0.14 (±0.05) | −7.3 | 1.2 |
| dC74 | 0.17 (±0.03) | −6.0 | 1.1 |
| dC73 | 0.08 (±0.02) | −12.0 | 1.5 |
| dC72 | 0.29 (±0.09) | −3.5 | 0.7 |
| dC71 | 0.89 (±0.21) | −1.1 | 0.1 |
| dG70 | 0.74 (±0.13) | −1.3 | 0.2 |
| dA69 | 0.61 (±0.10) | −1.6 | 0.3 |
| dG68 | 0.44 (±0.08) | −2.3 | 0.5 |
| dC67 | 0.32 (±0.08) | −3.1 | 0.7 |
| dA66 | 0.05 (±0.01) | −18.5 | 1.7 |
| dG65 | 0.21 (±0.03) | −4.7 | 0.9 |
| dG64 | 0.25 (±0.05) | −4.1 | 0.8 |
| dC63 | 0.36 (±0.18) | −2.7 | 0.6 |
The kcat/KM of the wide-type semi-synthetic tRNA was assigned a value of 1.0, and all other values are reported relative to this. Under the experimental conditions, initial rates of aminoacylation were proportional to substrate concentrations. The values reported here are averages of at least three determinations with the standard deviation (±SD) indicated. The −ΔΔG‡ value was calculated as described in the Experimental Procedures section.
Figure 2.
Bar graphs summarizing the effects of 2′-deoxy (A) chiral phosphorothioate (B) and methylphosphonate (C) substitutions on aminoacylation kinetic efficiency. The values reported here are averages of at least three determinations. Red corresponds to −ΔΔG‡ ≥ 1.5 kcal/mol, blue corresponds to −ΔΔG‡ = 1.0–1.5 kcal/mol, green corresponds to −ΔΔG‡ = 0.6–1.0 kcal/mol and cyan corresponds to −ΔΔG‡ ≤ 0.5 kcal/mol.
Phosphorothioate modification
Chemically synthesized oligonucleotides containing phosphorothioate substitutions were initially obtained as a mixture of Rp and Sp diastereomers (24, 25). Before separating each component, we first screened the racemic mixtures to identify potential candidates for interaction with human ProRS. Incorporation of a phosphorothioate linkage between A76 and C75 (which is called C75S in this work) has among the largest effects on aminoacylation efficiency (Table 2). The phosphates located at the bottom of the acceptor stem (A66S) and in the TΨC stem (G65S and G64S) also appear to be important for binding and/or catalysis (Table 2). Although modifications at positions 64 and 66 contribute only ∼1 kcal/mol to transition state stabilization, these results overlap with the results of 2′-deoxy substitution (Table 1), consistent with the presence of an important hydrogen-bonding network in this region of the tRNA.
Table 2.
The effect of phosphorothioate substitutions in the 3′-strand of human tRNAPro on aminoacylation efficiency
| Variant | Relative kcat/KM (±SD) | Activity change (x-fold) | −ΔΔG‡ (kcal/mol) |
|---|---|---|---|
| WT | 1.00 | 1.0 | 0.0 |
| C75S | 0.14 (±0.06) | −7.3 | 1.17 |
| C74S | 0.59 (±0.21) | −1.7 | 0.32 |
| C73S | 0.99 (±0.14) | −1.0 | 0.01 |
| C72S | 0.96 (±0.25) | −1.0 | 0.02 |
| C71S | 0.25 (±0.08) | −4.0 | 0.82 |
| G70S | 0.80 (±0.16) | −1.2 | 0.13 |
| A69S | 0.25 (±0.09) | −4.1 | 0.83 |
| G68S | 0.38 (±0.09) | −2.7 | 0.58 |
| C67S | 0.57 (±0.19) | −1.8 | 0.34 |
| A66S | 0.13 (±0.02) | −7.8 | 1.22 |
| G65S | 0.19 (±0.05) | −5.2 | 0.98 |
| G64S | 0.19 (±0.04) | −5.2 | 0.98 |
| C63S | 0.34 (±0.11) | −2.9 | 0.63 |
The reported kcat/KM and −ΔΔG‡ values were obtained as described in the legend to Table 1. Racemic mixtures of the phosphorothioate-containing oligonucleotides were used in this study.
Based on the assay results with racemic mixtures, six phosphorothioate-containing oligonucleotides (−ΔΔG‡ > 0.7 kcal/mol) were further purified by anion exchange or reversed phase HPLC (Table 3). Assays carried out with chiral phosphorothioate-containing substrates revealed that the Sp isomers at positions C75 and A69 made larger contributions to aminoacylation than the corresponding Rp isomers (Table 3, Figure 2B). The difference between the Sp and the Rp isomer at these positions was 0.75 and 0.4 kcal/mol, respectively.
Table 3.
The effect of chiral phosphorothioate substitutions in the 3′-strand of human tRNAPro on aminoacylation efficiency
| Variant | Relative kcat/KM (±SD) | Activity change (x-fold) | −ΔΔG‡ (kcal/mol) |
|---|---|---|---|
| WT | 1.00 | 1.0 | 0.0 |
| C75S-Rp | 0.43 (±0.11) | −2.3 | 0.50 |
| C75S-Sp | 0.12 (±0.04) | −8.2 | 1.25 |
| C71S-Rp | 0.35 (±0.03) | −2.9 | 0.62 |
| C71S-Sp | 0.28 (±0.04) | −3.6 | 0.76 |
| A69S-Rp | 0.27 (±0.03) | −3.7 | 0.77 |
| A69S-Sp | 0.14 (±0.02) | −7.3 | 1.17 |
| A66S-Rp | 0.23 (±0.04) | −4.4 | 0.88 |
| A66S-Sp | 0.19 (±0.04) | −5.2 | 0.98 |
| G65S-Rp | 0.26 (±0.03) | −3.8 | 0.80 |
| G65S-Sp | 0.20 (±0.06) | −4.9 | 0.94 |
| G64S-Rp | 0.86 (±0.37) | −1.2 | 0.09 |
| G64S-Sp | 0.76 (±0.31) | −1.3 | 0.16 |
The reported kcat/KM and −ΔΔG‡ values were obtained as described in the legend to Table 1. Purification and stereochemistry assignment methods are described in the Experimental procedures section.
Methylphosphonate modification
Methylphosphonate substitution, which replaces one of two non-bridging oxygens with a methyl group, has been used to identify phosphate contacts in protein–DNA (31–33) and to a lesser extent, protein–RNA interactions (34–36). Unlike a phosphorothioate substitution, the methylphosphonate linkage is uncharged, which neutralizes a small section of the RNA backbone. This change in electrostatic potential is likely to reduce the local water organization, as well as weaken the ionic attraction between proteins and RNAs. Thus, it is possible that methylphosphonate substitutions at certain positions may allow detection of charge–charge interactions not apparent by phosphorothioate modification (36).
The methylphosphonate oligonucleotides used here contained an additional 2′-deoxy modification. Therefore, to isolate the effect of the methylphosphonate linkage, the kinetic data generated using 2′-deoxy-methylphosphonate-containing variants was corrected by subtracting the contribution of 2′-deoxy substitution (Table 1). Whereas most of the single methylphosphonate substitutions tested did not result in a significant decrease in aminoacylation efficiency (Table 4, Figure 2C), methylphosphonate substitutions at positions G70 and C71 have substantial effects on catalysis, resulting in −ΔΔG‡ values of 1.5 and 1.7 kcal/mol, respectively. In this region, salt bridges and water-mediated hydrogen bonds are commonly observed in AARS–tRNA co-crystal structures. Since we did not observe major effects upon 2′-deoxy or phosphorothioate substitution at these positions (Figure 3), an indirect electrostatic interaction is more likely. We conclude that the phosphate oxygens of G70 and C71 are likely to participate in interactions with human ProRS, either directly or indirectly through salt bridges. This analysis is based on the results obtained with racemic mixtures. Although we did successfully separate the diastereomers of methylphosphonate-containing oligonucleotides by anion exchange chromatography, under all assay conditions tested, the tRNAs containing chiral methylphosphonate substitutions at positions G70 and C71 reached low extents of plateau-level charging in 20 s, and thus, were not amenable to kinetic analysis.
Table 4.
The effect of 2′-deoxy-methylphosphonate substitutions in the 3'-strand of human tRNAPro on aminoacylation efficiency
| Variant | Relative kcat/KM (±SD) | Activity change (x-fold) | −ΔΔG‡ (kcal/mol) | (−ΔΔG‡)Me a (kcal/mol) |
|---|---|---|---|---|
| WT | 1.00 | 1.0 | 0.0 | 0.0 |
| dC75Me | 0.28 (±0.05) | −3.6 | 0.76 | −0.41 |
| dC74Me | 0.23 (±0.05) | −4.3 | 0.87 | −0.19 |
| dC73Me | 0.14 (±0.07) | −7.3 | 1.17 | −0.30 |
| dC72Me | 0.11 (±0.03) | −8.8 | 1.29 | 0.55 |
| dC71Me | 0.05 (±0.02) | −18.5 | 1.73 | 1.66 |
| dG70Me | 0.06 (±0.03) | −17.0 | 1.68 | 1.50 |
| dA69Me | 0.77 (±0.05) | −1.3 | 0.16 | −0.13 |
| dG68Me | 0.80 (±0.04) | −1.2 | 0.13 | −0.35 |
| dC67Me | 0.16 (±0.03) | −6.2 | 1.08 | 0.40 |
| dA66Me | 0.13 (±0.08) | −7.8 | 1.22 | −0.51 |
| dG65Me | 0.29 (±0.15) | −3.5 | 0.74 | −0.18 |
| dG64Me | 0.38 (±0.12) | −2.6 | 0.57 | −0.26 |
| dC63Me | 0.26 (±0.07) | −3.8 | 0.80 | 0.20 |
Figure 3.
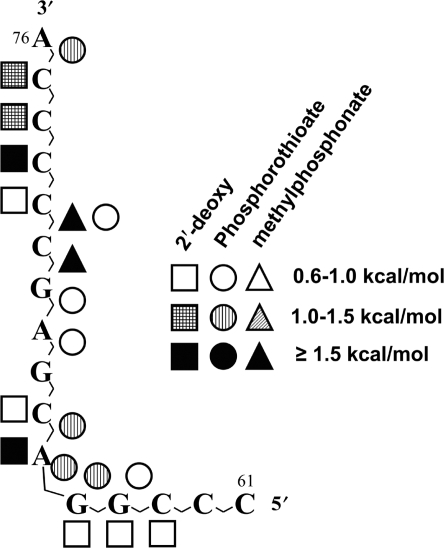
Summary of sugar–phosphate backbone substitution effects mapped onto the sequence of the 3′-16 nucleotides of human tRNAPro. Symbols correspond to effects of 2′-deoxy (squares), phosphorothioate (circles) or methylphosphonate (triangles) substitutions. Solid symbols correspond to −ΔΔG‡ ≥ 1.5 kcal/mol, striped symbols correspond to −ΔΔG‡ = 1.0–1.5 kcal/mol and open symbols correspond to −ΔΔG‡ = 0.6–1.0 kcal/mol.
DISCUSSION
Although the importance of nucleotide base interactions for sequence-specific recognition of nucleic acids by proteins is well-documented (37–39), the more exposed phosphodiester backbone also contains the potential for functionally important interactions. These may occur either through direct hydrogen-bonding interactions to protein functional groups, or indirectly through salt bridges, water molecules or metal ions. In fact, there are numerous examples of backbone interactions among the solved AARS–tRNA co-crystal structures. For example, the class I E. coli glutaminyl-tRNA synthetase (GlnRS)-tRNAGln complex revealed that sugar–phosphate backbone functional groups in positions 69 and 72–76 of the acceptor stem directly interact with GlnRS (40). In class II T. thermophilus SerRS-tRNASer and E. coli ThrRS-tRNAThr co-crystal structures, direct backbone-specific interactions are observed between positions 66 and 71 in the tRNA acceptor stem, while base-specific contacts are dominant at positions 74–76 (12,16,30). Interestingly, E. coli AspRS and T. thermophilus PheRS contain extensive backbone-specific contacts throughout the tRNA acceptor stem (14,15). Thus, it is clear that backbone-specific interactions with tRNA by AARS appear to play an important role in recognition.
To elucidate critical sugar–phosphate backbone contacts, site-directed atomic group substitutions of 2′-deoxynucleotide and phosphorothioate functionalities have been applied to study protein–nucleic acid interactions, including limited studies involving AARS–tRNA contacts. In particular, the functional role of specific 2′-OH groups in aminoacylation has been probed by specific 2′-deoxynucleotide substitutions in the E. coli ProRS (41), E. coli cysteinyl-tRNA synthetase (CysRS) (42,43), E. coli alanyl-tRNA synthetase (AlaRS) (44) and E. coli AspRS (45) systems. Additionally, phosphorothioate modification has been used to map the contacts of human and E. coli CysRS, E. coli SerRS, T. thermophilus PheRS and yeast AspRS with their cognate tRNAs (43,46–48). Unlike 2′-deoxynucleotides and phosphorothioates, 2′-deoxy-methylphosphonate substitution has not been widely applied to investigate tRNA contacts with synthetases, although the modification has been used in mapping protein–nucleic acid interactions in other systems (36,49–51). Collectively, site-directed atomic group substitutions have proven to be valuable in the characterization of AARS–tRNA contacts when co-crystal structures are not available and also in understanding the functional significance of contacts observed in high-resolution structures. Nevertheless, despite their importance, relatively few quantitative studies have been performed to understand how sugar–phosphate backbone contacts provide specificity in AARS–tRNA recognition (2).
Since previous studies failed to identify critical nucleotide recognition elements in the acceptor stem of human tRNAPro, we hypothesized that the human synthetase may have evolved to interact with RNA backbone elements (4,9). In this work, we evaluated the energetic contributions of individual backbone functional groups in catalysis by human ProRS. By performing aminoacylation assays using semi-synthetic tRNAs containing site-specific modifications, we provide quantitative information supporting a critical role for sugar–phosphate backbone interactions in recognition by human ProRS.
Since the top of the tRNA acceptor stem is single-stranded, the flexibility of this region may facilitate direct functional interactions with protein residues, resulting in the relatively severe effects observed upon deletion of even one 2′-hydroxyl (dC75, dC74 and dC73) or upon substitution of one phosphate oxygen (C75S-Sp) (Figure 3). The 2′-hydroxyl group of the discriminator base (C73) is the most critical within this single-stranded region, making the largest contribution to catalysis (−ΔΔG‡ = 1.5 kcal/mol). The Sp phosphate oxygen between C75 and A76, which is near the site of amino acid attachment, also makes a notable contribution to catalysis (−ΔΔG‡ = 1.25 kcal/mol), and is likely to make a functionally important contact with active site residues. In this top region of the tRNA acceptor stem, therefore, direct interactions through a hydrogen-bonding network appear to dominate and contribute to the enzyme's catalytic efficiency.
The first G1:C72 base pair in eukaryotic proline tRNAs, including human tRNAPro, play only a minor role in aminoacylation (4). In contrast, the unique C1:G72 base pair found in all bacterial proline tRNAs, is critical for ProRS recognition (10,52). Although human ProRS does not recognize the first base pair via base-specific contacts, extensive direct and/or indirect interactions with atomic groups near the first base pair are likely to occur. As shown here, the total energetic contribution of backbone functional groups located in the vicinity of the first base pair of human tRNAPro is quite large. For instance, the 2′-hydroxyl of C72 contributes 0.70 kcal/mol and the combined results of phosphorothioate and methylphosphonate substitutions suggest that the non-bridging phosphate oxygens of C71 are also critical (−ΔΔG‡ = 0.6–1.66 kcal/mol). The double-helical region between positions 68 and 72 of the tRNA acceptor stem showed phosphorothioate and also strong methylphosphonate effects (Figure 3). In accordance with the co-crystal structures of AspRS-tRNAAsp and GlnRS-tRNAGln (40,53), this region may participate in interactions with human ProRS through salt bridges and/or a water-mediated hydrogen-bond network.
Functional groups located near the bottom of the tRNA acceptor stem (i.e. A66 and C67) also appear to interact with the enzyme. Cross-subunit interactions between the second monomer in dimeric structures and nucleotides at the bottom of the tRNA acceptor stem have been observed in class II synthetase co-crystal structures (12,16,30). For example, in the E. coli ThrRS-tRNAThr structure, strong cross-subunit interactions involved the backbone of U66 and A67, but did not use base-specific contacts (16). Known SerRS-tRNASer co-crystal structures also revealed that both phosphate oxygens and the 3′-hydroxyl at position 66 were in contact with the second monomer of the complexes (12,30).
In summary, previous work has shown that bacterial ProRS relies heavily on base-specific functional group interactions for specific tRNAPro acceptor stem recognition. In contrast, the human enzyme appeared to lack base-specific recognition. We show here that the human enzyme has evolved to recognize an extensive network of backbone functional groups throughout the acceptor stem and extending into the TΨC arm. Although the entire panel of backbone substitutions tested here for human ProRS has not been studied in the bacterial system, previous 2′-deoxynucleotide substitutions were incorporated into the 3′-strand of E. coli tRNAPro (41). Only minimal effects on aminoacylation efficiency (<2-fold) were observed at positions 63–72 of the acceptor stem. Notably, deoxynucleotide substitution at position 66, which had the largest effect on catalysis by human ProRS (−ΔΔG‡ = 1.7 kcal/mol) resulted in a 2-fold increase in catalytic efficiency by E. coli ProRS. Taken together, this work reveals how a specific AARS–tRNA system has changed through evolution from a mode of acceptor stem recognition in bacteria that relies heavily on direct readout of atomic groups on the nucleotide bases to use of a functionally relevant indirect readout mechanism in higher eukaryotes.
ACKNOWLEDGEMENTS
Funding was provided by National Institutes of Health (GM049928 to K.M.-F.). We thank Dr Brian Burke for preparing the plasmid of human A57G tRNAPro used in this work and Dr Abbey Rosen for chemical synthesis of all 3′-16-mer oligonucleotides. We also thank Drs Olke C. Uhlenbeck and Dagmar Dertinger for helpful discussions about purification of chiral phosphorothioate-containing oligonucleotides. Funding to pay the Open Access publication charges for this article was provided by NIH GM049928.
Conflict of interest statement. None declared.
REFERENCES
- 1.Ibba M, Söll D. Quality control mechanisms during translation. Science. 1999;286:1893–1897. doi: 10.1126/science.286.5446.1893. [DOI] [PubMed] [Google Scholar]
- 2.Perona JJ, Hou YM. Indirect readout of tRNA for aminoacylation. Biochemistry. 2007;46:10419–10432. doi: 10.1021/bi7014647. [DOI] [PubMed] [Google Scholar]
- 3.Giegé R, Sissler M, Florentz C. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res. 1998;26:5017–5035. doi: 10.1093/nar/26.22.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stehlin C, Burke B, Yang F, Liu H, Shiba K, Musier-Forsyth K. Species-specific differences in the operational RNA code for aminoacylation of tRNA(Pro) Biochemistry. 1998;37:8605–8613. doi: 10.1021/bi980364s. [DOI] [PubMed] [Google Scholar]
- 5.Cusack S, Yaremchuk A, Krikliviy I, Tukalo M. tRNA(Pro) anticodon recognition by Thermus thermophilus prolyl-tRNA synthetase. Structure. 1998;6:101–108. doi: 10.1016/s0969-2126(98)00011-2. [DOI] [PubMed] [Google Scholar]
- 6.Wong FC, Beuning PJ, Nagan M, Shiba K, Musier-Forsyth K. Functional role of the prokaryotic proline-tRNA synthetase insertion domain in amino acid editing. Biochemistry. 2002;41:7108–7115. doi: 10.1021/bi012178j. [DOI] [PubMed] [Google Scholar]
- 7.Wong FC, Beuning PJ, Silvers C, Musier-Forsyth K. An isolated class II aminoacyl-tRNA synthetase insertion domain is functional in amino acid editing. J. Biol. Chem. 2003;278:52857–52864. doi: 10.1074/jbc.M309627200. [DOI] [PubMed] [Google Scholar]
- 8.Burke B, Lipman RS, Shiba K, Musier-Forsyth K, Hou YM. Divergent adaptation of tRNA recognition by Methanococcus jannaschii prolyl-tRNA synthetase. J. Biol. Chem. 2001;276:20286–20291. doi: 10.1074/jbc.m100456200. [DOI] [PubMed] [Google Scholar]
- 9.Burke B, Yang F, Chen F, Stehlin C, Chan B, Musier-Forsyth K. Evolutionary coadaptation of the motif 2–acceptor stem interaction in the class II prolyl-tRNA synthetase system. Biochemistry. 2000;39:15540–15547. doi: 10.1021/bi001835p. [DOI] [PubMed] [Google Scholar]
- 10.Liu H, Peterson R, Kessler J, Musier-Forsyth K. Molecular recognition of tRNA(Pro) by Escherichia coli proline tRNA synthetase in vitro. Nucleic Acids Res. 1995;23:165–169. doi: 10.1093/nar/23.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yaremchuk A, Cusack S, Tukalo M. Crystal structure of a eukaryote/archaeon-like protyl-tRNA synthetase and its complex with tRNA(Pro)(CGG) EMBO J. 2000;19:4745–4758. doi: 10.1093/emboj/19.17.4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Biou V, Yaremchuk A, Tukalo M, Cusack S. The 2.9 A crystal structure of T. thermophilus seryl-tRNA synthetase complexed with tRNA(Ser) Science. 1994;263:1404–1410. doi: 10.1126/science.8128220. [DOI] [PubMed] [Google Scholar]
- 13.Cusack S, Yaremchuk A, Tukalo M. The crystal structures of T. thermophilus lysyl-tRNA synthetase complexed with E. coli tRNA(Lys) and a T. thermophilus tRNA(Lys) transcript: anticodon recognition and conformational changes upon binding of a lysyl-adenylate analogue. EMBO J. 1996;15:6321–6334. [PMC free article] [PubMed] [Google Scholar]
- 14.Cavarelli J, Rees B, Ruff M, Thierry JC, Moras D. Yeast tRNA(Asp) recognition by its cognate class II aminoacyl-tRNA synthetase. Nature. 1993;362:181–184. doi: 10.1038/362181a0. [DOI] [PubMed] [Google Scholar]
- 15.Goldgur Y, Mosyak L, Reshetnikova L, Ankilova V, Lavrik O, Khodyreva S, Safro M. The crystal structure of phenylalanyl-tRNA synthetase from thermus thermophilus complexed with cognate tRNAPhe. Structure. 1997;5:59–68. doi: 10.1016/s0969-2126(97)00166-4. [DOI] [PubMed] [Google Scholar]
- 16.Sankaranarayanan R, Dock-Bregeon AC, Romby P, Caillet J, Springer M, Rees B, Ehresmann C, Ehresmann B, Moras D. The structure of threonyl-tRNA synthetase-tRNA(Thr) complex enlightens its repressor activity and reveals an essential zinc ion in the active site. Cell. 1999;97:371–381. doi: 10.1016/s0092-8674(00)80746-1. [DOI] [PubMed] [Google Scholar]
- 17.Heacock D, Forsyth CJ, Shiba K, Musier-Forsyth K. Synthesis and aminoacyl-tRNA synthetase inhibitory activity of prolyl adenylate analogs. Bioorg. Chem. 1996;24:273–289. [Google Scholar]
- 18.Fersht AR, Ashford JS, Bruton CJ, Jakes R, Koch GL, Hartley BS. Active site titration and aminoacyl adenylate binding stoichiometry of aminoacyl-tRNA synthetases. Biochemistry. 1975;14:1–4. doi: 10.1021/bi00672a001. [DOI] [PubMed] [Google Scholar]
- 19.Liu H, Musier-Forsyth K. Escherichia coli proline tRNA synthetase is sensitive to changes in the core region of tRNA(Pro) Biochemistry. 1994;33:12708–12714. doi: 10.1021/bi00208a023. [DOI] [PubMed] [Google Scholar]
- 20.Steinberg S, Misch A, Sprinzl M. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1993;21:3011–3015. doi: 10.1093/nar/21.13.3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sampson JR, Uhlenbeck OC. Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc. Natl Acad. Sci. USA. 1988;85:1033–1037. doi: 10.1073/pnas.85.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi JP, Francklyn C, Hill K, Schimmel P. A nucleotide that enhances the charging of RNA minihelix sequence variants with alanine. Biochemistry. 1990;29:3621–3626. doi: 10.1021/bi00467a005. [DOI] [PubMed] [Google Scholar]
- 24.Xu Q, Musier-Forsyth K, Hammer RP, Barany G. Use of 1,2,4-dithiazolidine-3,5-dione (DtsNH) and 3-ethoxy-1,2,4-dithiazoline-5-one (EDITH) for synthesis of phosphorothioate-containing oligodeoxyribonucleotides. Nucleic Acids Res. 1996;24:1602–1607. doi: 10.1093/nar/24.9.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu Q, Barany G, Hammer RP, Musier-Forsyth K. Efficient introduction of phosphorothioates into RNA oligonucleotides by 3-ethoxy-1,2,4-dithiazoline-5-one (EDITH) Nucleic Acids Res. 1996;24:3643–3644. doi: 10.1093/nar/24.18.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dertinger D, Behlen LS, Uhlenbeck OC. Using phosphorothioate-substituted RNA to investigate the thermodynamic role of phosphates in a sequence specific RNA-protein complex. Biochemistry. 2000;39:55–63. doi: 10.1021/bi991769v. [DOI] [PubMed] [Google Scholar]
- 27.Slim G, Gait MJ. Configurationally defined phosphorothioate-containing oligoribonucleotides in the study of the mechanism of cleavage of hammerhead ribozymes. Nucleic Acids Res. 1991;19:1183–1188. doi: 10.1093/nar/19.6.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thorogood H, Grasby JA, Connolly BA. Influence of the phosphate backbone on the recognition and hydrolysis of DNA by the EcoRV restriction endonuclease. A study using oligodeoxynucleotide phosphorothioates. J. Biol. Chem. 1996;271:8855–8862. doi: 10.1074/jbc.271.15.8855. [DOI] [PubMed] [Google Scholar]
- 29.Yap LP, Stehlin C, Musier-Forsyth K. Use of semi-synthetic transfer RNAs to probe molecular recognition by Escherichia coli proline-tRNA synthetase. Chem. Biol. 1995;2:661–666. doi: 10.1016/1074-5521(95)90029-2. [DOI] [PubMed] [Google Scholar]
- 30.Cusack S, Yaremchuk A, Tukalo M. The crystal structure of the ternary complex of T.thermophilus seryl-tRNA synthetase with tRNA(Ser) and a seryl-adenylate analogue reveals a conformational switch in the active site. EMBO J. 1996;15:2834–2842. [PMC free article] [PubMed] [Google Scholar]
- 31.Noble SA, Fisher EF, Caruthers MH. Methylphosphonates as probes of protein-nucleic acid interactions. Nucleic Acids Res. 1984;12:3387–3404. doi: 10.1093/nar/12.7.3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Botfield MC, Weiss MA. Bipartite DNA recognition by the human Oct-2 POU domain: POUs-specific phosphate contacts are analogous to those of bacteriophage lambda repressor. Biochemistry. 1994;33:2349–2355. doi: 10.1021/bi00175a001. [DOI] [PubMed] [Google Scholar]
- 33.Smith SA, McLaughlin LW. Probing contacts to the DNA backbone in the trp repressor-operator sequence-specific protein-nucleic acid complex using diastereomeric methylphosphonate analogues. Biochemistry. 1997;36:6046–6058. doi: 10.1021/bi9700781. [DOI] [PubMed] [Google Scholar]
- 34.Hamy F, Asseline U, Grasby J, Iwai S, Pritchard C, Slim G, Butler PJ, Karn J, Gait MJ. Hydrogen-bonding contacts in the major groove are required for human immunodeficiency virus type-1 tat protein recognition of TAR RNA. J. Mol. Biol. 1993;230:111–123. doi: 10.1006/jmbi.1993.1129. [DOI] [PubMed] [Google Scholar]
- 35.Pritchard CE, Grasby JA, Hamy F, Zacharek AM, Singh M, Karn J, Gait MJ. Methylphosphonate mapping of phosphate contacts critical for RNA recognition by the human immunodeficiency virus tat and rev proteins. Nucleic Acids Res. 1994;22:2592–2600. doi: 10.1093/nar/22.13.2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dertinger D, Uhlenbeck OC. Evaluation of methylphosphonates as analogs for detecting phosphate contacts in RNA-protein complexes. RNA. 2001;7:622–631. doi: 10.1017/s1355838201002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sarai A, Kono H. Protein-DNA recognition patterns and predictions. Annu. Rev. Biophys. Biomol. Struct. 2005;34:379–398. doi: 10.1146/annurev.biophys.34.040204.144537. [DOI] [PubMed] [Google Scholar]
- 38.Hoffman MM, Khrapov MA, Cox JC, Yao J, Tong L, Ellington AD. AANT: the amino acid-nucleotide interaction database. Nucleic Acids Res. 2004;32:D174–181. doi: 10.1093/nar/gkh128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ibba M, Francklyn C, Cusack S. The Aminoacyl-tRNA Synthetases. Georgetown, TX: Landes Bioscience; 2005. [Google Scholar]
- 40.Rould MA, Perona JJ, Söll D, Steitz TA. Structure of E. coli glutaminyl-tRNA synthetase complexed with tRNA(Gln) and ATP at 2.8 A resolution. Science. 1989;246:1135–1142. doi: 10.1126/science.2479982. [DOI] [PubMed] [Google Scholar]
- 41.Yap LP, Musier-Forsyth K. Transfer RNA aminoacylation: identification of a critical ribose 2'-hydroxyl-base interaction. RNA. 1995;1:418–424. [PMC free article] [PubMed] [Google Scholar]
- 42.Hou YM, Zhang X, Holland JA, Davis DR. An important 2'-OH group for an RNA-protein interaction. Nucleic Acids Res. 2001;29:976–985. doi: 10.1093/nar/29.4.976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ming X, Smith K, Suga H, Hou YM. Recognition of tRNA backbone for aminoacylation with cysteine: evolution from Escherichia coli to human. J. Mol. Biol. 2002;318:1207–1220. doi: 10.1016/s0022-2836(02)00232-2. [DOI] [PubMed] [Google Scholar]
- 44.Musier-Forsyth K, Schimmel P. Functional contacts of a transfer RNA synthetase with 2'-hydroxyl groups in the RNA minor groove. Nature. 1992;357:513–515. doi: 10.1038/357513a0. [DOI] [PubMed] [Google Scholar]
- 45.Vortler CS, Fedorova O, Persson T, Kutzke U, Eckstein F. Determination of 2'-hydroxyl and phosphate groups important for aminoacylation of Escherichia coli tRNA(Asp): a nucleotide analogue interference study. RNA. 1998;4:1444–1454. doi: 10.1017/s1355838298980967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schatz D, Leberman R, Eckstein F. Interaction of Escherichia coli tRNA(Ser) with its cognate aminoacyl-tRNA synthetase as determined by footprinting with phosphorothioate-containing tRNA transcripts. Proc. Natl Acad. Sci. USA. 1991;88:6132–6136. doi: 10.1073/pnas.88.14.6132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rudinger J, Puglisi JD, Pütz J, Schatz D, Eckstein F, Florentz C, Giegé R. Determinant nucleotides of yeast tRNA(Asp) interact directly with aspartyl-tRNA synthetase. Proc. Natl Acad. Sci. USA. 1992;89:5882–5886. doi: 10.1073/pnas.89.13.5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kreutzer R, Kern D, Giegé R, Rudinger J. Footprinting of tRNA(Phe) transcripts from Thermus thermophilus HB8 with the homologous phenylalanyl-tRNA synthetase reveals a novel mode of interaction. Nucleic Acids Res. 1995;23:4598–4602. doi: 10.1093/nar/23.22.4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Soliva R, Monaco V, Gomez-Pinto I, Meeuwenoord NJ, Marel GA, Boom JH, Gonzalez C, Orozco M. Solution structure of a DNA duplex with a chiral alkyl phosphonate moiety. Nucleic Acids Res. 2001;29:2973–2985. doi: 10.1093/nar/29.14.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thiviyanathan V, Vyazovkina KV, Gozansky EK, Bichenchova E, Abramova TV, Luxon BA, Lebedev AV, Gorenstein DG. Structure of hybrid backbone methylphosphonate DNA heteroduplexes: effect of R and S stereochemistry. Biochemistry. 2002;41:827–838. doi: 10.1021/bi011551k. [DOI] [PubMed] [Google Scholar]
- 51.Okonogi TM, Alley SC, Harwood EA, Hopkins PB, Robinson BH. Phosphate backbone neutralization increases duplex DNA flexibility: a model for protein binding. Proc. Natl Acad. Sci. USA. 2002;99:4156–4160. doi: 10.1073/pnas.072067799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McClain WH, Schneider J, Gabriel K. Distinctive acceptor-end structure and other determinants of Escherichia coli tRNA(Pro) identity. Nucleic Acids Res. 1994;22:522–529. doi: 10.1093/nar/22.3.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moulinier L, Eiler S, Eriani G, Gangloff J, Thierry JC, Gabriel K, McClain WH, Moras D. The structure of an AspRS-tRNA(Asp) complex reveals a tRNA-dependent control mechanism. EMBO J. 2001;20:5290–5301. doi: 10.1093/emboj/20.18.5290. [DOI] [PMC free article] [PubMed] [Google Scholar]