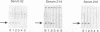Abstract
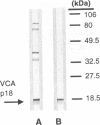
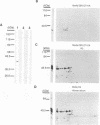
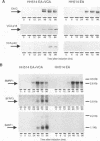
The genomic localization of two immunodominant genes encoding two proteins of the Epstein-Barr virus capsid antigen (VCA) complex, VCA-p18 and VCA-p40, has been identified. For that purpose, lambda gt11-based cDNA libraries were constructed from HH514.c16 cells induced for virus production. The libraries were screened with a monoclonal antibody, EBV.OT41A, directed against VCA-p40 or with affinity-purified human antibodies against VCA-p18. Sequencing of the inserts of positive plaques showed that VCA-p18 and VCA-p40 are encoded within open reading frames (ORFs) BFRF3 and BdRF1, respectively. Peptide scanning analysis of the predicted protein of ORF BdRF1 resulted in defining the epitope of monoclonal antibody EBV.OT41A at the C-terminal region. The dominant VCA-p18 reactivity of human sera can be completely inhibited by preadsorption with Escherichia coli-expressed BFRF3-beta-galactosidase. Serum of a rabbit immunized with BFRF3-beta galactosidase reacts with a VCA-specific protein of 18 kDa. In addition, BFRF3-beta-galactosidase affinity-purified antibodies react with VCA-p18 of virus-producing cells (HH514.c16). Complete inhibition of viral DNA polymerase activity by phosphonoacetic acid is associated with the absence of RNAs and protein products of both ORFs, indicating that VCA-p18 and VCA-p40 are true late antigens.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baer R., Bankier A. T., Biggin M. D., Deininger P. L., Farrell P. J., Gibson T. J., Hatfull G., Hudson G. S., Satchwell S. C., Séguin C. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature. 1984 Jul 19;310(5974):207–211. doi: 10.1038/310207a0. [DOI] [PubMed] [Google Scholar]
- Berbers G. A., Kunnen R., van Bergen en Henegouwen P. M., van Wijk R. Localization and quantitation of hsp84 in mammalian cells. Exp Cell Res. 1988 Aug;177(2):257–271. doi: 10.1016/0014-4827(88)90460-0. [DOI] [PubMed] [Google Scholar]
- Chee M. S., Bankier A. T., Beck S., Bohni R., Brown C. M., Cerny R., Horsnell T., Hutchison C. A., 3rd, Kouzarides T., Martignetti J. A. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr Top Microbiol Immunol. 1990;154:125–169. doi: 10.1007/978-3-642-74980-3_6. [DOI] [PubMed] [Google Scholar]
- Chen M. R., Hsu T. Y., Lin S. W., Chen J. Y., Yang C. S. Cloning and characterization of cDNA clones corresponding to transcripts from the BamHI G region of the Epstein-Barr virus genome and expression of BGLF2. J Gen Virol. 1991 Dec;72(Pt 12):3047–3055. doi: 10.1099/0022-1317-72-12-3047. [DOI] [PubMed] [Google Scholar]
- Cho M. S., Milman G., Hayward S. D. A second Epstein-Barr virus early antigen gene in BamHI fragment M encodes a 48- to 50-kilodalton nuclear protein. J Virol. 1985 Dec;56(3):860–866. doi: 10.1128/jvi.56.3.860-866.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolyniuk M., Wolff E., Kieff E. Proteins of Epstein-Barr Virus. II. Electrophoretic analysis of the polypeptides of the nucleocapsid and the glucosamine- and polysaccharide-containing components of enveloped virus. J Virol. 1976 Apr;18(1):289–297. doi: 10.1128/jvi.18.1.289-297.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dölken G., Hecht T., Röckel D., Hirsch F. W. Characterization of the Epstein-Barr virus-induced early polypeptide complex p50/58 EA-D using rabbit antisera, a monoclonal antibody, and human antibodies. Virology. 1987 Apr;157(2):460–471. doi: 10.1016/0042-6822(87)90288-1. [DOI] [PubMed] [Google Scholar]
- Edson C. M., Cohen L. K., Henle W., Strominger J. L. An unusually high-titer human anti-Epstein Barr virus (EBV) serum and its use in the study of EBV-specific proteins synthesized in vitro and in vivo. J Immunol. 1983 Feb;130(2):919–924. [PubMed] [Google Scholar]
- Geysen H. M., Meloen R. H., Barteling S. J. Use of peptide synthesis to probe viral antigens for epitopes to a resolution of a single amino acid. Proc Natl Acad Sci U S A. 1984 Jul;81(13):3998–4002. doi: 10.1073/pnas.81.13.3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geysen H. M., Rodda S. J., Mason T. J., Tribbick G., Schoofs P. G. Strategies for epitope analysis using peptide synthesis. J Immunol Methods. 1987 Sep 24;102(2):259–274. doi: 10.1016/0022-1759(87)90085-8. [DOI] [PubMed] [Google Scholar]
- Gong M., Kieff E. Intracellular trafficking of two major Epstein-Barr virus glycoproteins, gp350/220 and gp110. J Virol. 1990 Apr;64(4):1507–1516. doi: 10.1128/jvi.64.4.1507-1516.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubler U., Hoffman B. J. A simple and very efficient method for generating cDNA libraries. Gene. 1983 Nov;25(2-3):263–269. doi: 10.1016/0378-1119(83)90230-5. [DOI] [PubMed] [Google Scholar]
- Katz D. A., Baumann R. P., Sun R., Kolman J. L., Taylor N., Miller G. Viral proteins associated with the Epstein-Barr virus transactivator, ZEBRA. Proc Natl Acad Sci U S A. 1992 Jan 1;89(1):378–382. doi: 10.1073/pnas.89.1.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Middeldorp J. M., Herbrink P. Epstein-Barr virus specific marker molecules for early diagnosis of infectious mononucleosis. J Virol Methods. 1988 Sep;21(1-4):133–146. doi: 10.1016/0166-0934(88)90060-2. [DOI] [PubMed] [Google Scholar]
- Middeldorp J. M., Meloen R. H. Epitope-mapping on the Epstein-Barr virus major capsid protein using systematic synthesis of overlapping oligopeptides. J Virol Methods. 1988 Sep;21(1-4):147–159. doi: 10.1016/0166-0934(88)90061-4. [DOI] [PubMed] [Google Scholar]
- Ooka T., de Turenne-Tessier M., Stolzenberg M. C. Relationship between antibody production to Epstein-Barr virus (EBV) early antigens and various EBV-related diseases. Springer Semin Immunopathol. 1991;13(2):233–247. doi: 10.1007/BF00201471. [DOI] [PubMed] [Google Scholar]
- Pearson G. R. ELISA tests and monoclonal antibodies for EBV. J Virol Methods. 1988 Sep;21(1-4):97–104. doi: 10.1016/0166-0934(88)90056-0. [DOI] [PubMed] [Google Scholar]
- Pfitzner A. J., Strominger J. L., Speck S. H. Characterization of a cDNA clone corresponding to a transcript from the Epstein-Barr virus BamHI M fragment: evidence for overlapping mRNAs. J Virol. 1987 Sep;61(9):2943–2946. doi: 10.1128/jvi.61.9.2943-2946.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston V. G., Rixon F. J., McDougall I. M., McGregor M., al Kobaisi M. F. Processing of the herpes simplex virus assembly protein ICP35 near its carboxy terminal end requires the product of the whole of the UL26 reading frame. Virology. 1992 Jan;186(1):87–98. doi: 10.1016/0042-6822(92)90063-u. [DOI] [PubMed] [Google Scholar]
- Robinson P. A., Anderton B. H., Loviny T. L. Nitrocellulose-bound antigen repeatedly used for the affinity purification of specific polyclonal antibodies for screening DNA expression libraries. J Immunol Methods. 1988 Apr 6;108(1-2):115–122. doi: 10.1016/0022-1759(88)90409-7. [DOI] [PubMed] [Google Scholar]
- Sample C., Kieff E. Molecular basis for Epstein-Barr virus induced pathogenesis and disease. Springer Semin Immunopathol. 1991;13(2):133–146. doi: 10.1007/BF00201464. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sculley D. G., Sculley T. B., Pope J. H. Reactions of sera from patients with rheumatoid arthritis, systemic lupus erythematosus and infectious mononucleosis to Epstein-Barr virus-induced polypeptides. J Gen Virol. 1986 Oct;67(Pt 10):2253–2258. doi: 10.1099/0022-1317-67-10-2253. [DOI] [PubMed] [Google Scholar]
- Sculley T. B., Sculley D. G., Pope J. H. Identification of Epstein-Barr virus-induced polypeptides in P3HR-1 cells by protein immunoblot. J Gen Virol. 1985 May;66(Pt 5):1113–1122. doi: 10.1099/0022-1317-66-5-1113. [DOI] [PubMed] [Google Scholar]
- Seibl R., Wolf H. Mapping of Epstein-Barr virus proteins on the genome by translation of hybrid-selected RNA from induced P3HR1 cells and induced Raji cells. Virology. 1985 Feb;141(1):1–13. doi: 10.1016/0042-6822(85)90177-1. [DOI] [PubMed] [Google Scholar]
- Takada K., Fujiwara S., Yano S., Osato T. Monoclonal antibody specific for capsid antigen of Epstein-Barr virus. Med Microbiol Immunol. 1983;171(4):225–231. doi: 10.1007/BF02123496. [DOI] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA transferred or dotted nitrocellulose paper. Methods Enzymol. 1983;100:255–266. doi: 10.1016/0076-6879(83)00060-9. [DOI] [PubMed] [Google Scholar]
- Vroman B., Luka J., Rodriguez M., Pearson G. R. Characterization of a major protein with a molecular weight of 160,000 associated with the viral capsid of Epstein-Barr virus. J Virol. 1985 Jan;53(1):107–113. doi: 10.1128/jvi.53.1.107-113.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wielaard F., Scherders J., Dagelinckx C., Middeldorp J. M., Sabbe L. J., Van Belzen C. Development of an antibody-capture IgM-enzyme-linked immunosorbent assay for diagnosis of acute Epstein-Barr virus infections. J Virol Methods. 1988 Sep;21(1-4):105–115. doi: 10.1016/0166-0934(88)90057-2. [DOI] [PubMed] [Google Scholar]
- Zagursky R. J., Berman M. L. Cloning vectors that yield high levels of single-stranded DNA for rapid DNA sequencing. Gene. 1984 Feb;27(2):183–191. doi: 10.1016/0378-1119(84)90139-2. [DOI] [PubMed] [Google Scholar]