Abstract
Replication protein A (RPA) is required for both DNA replication and nucleotide excision repair. Previous studies have shown that RPA interacts with the tumor suppressor p53. Herein, we have mapped a 20-amino acid region in the N-terminal part of p53 that is essential for its binding to RPA. This region is distinct from the minimal activation domain of p53 previously identified. We also demonstrate that UV radiation of cells greatly reduces the ability of RPA to bind to p53. Interestingly, damage-induced hyperphosphorylated RPA does not associate with p53. Furthermore, down-regulation of the RPA/p53 interaction is dependent upon the capability of cells to perform global genome repair. On the basis of these data, we propose that RPA may participate in the coordination of DNA repair with the p53-dependent checkpoint control by sensing UV damage and releasing p53 to activate its downstream targets.
To prevent permanent changes in the genome, damaged cells activate checkpoint control to arrest cell cycle progression, inhibit DNA replication, and facilitate DNA repair (1, 2). The p53-dependent damage response pathway represents the major checkpoint control in mammalian cells (3, 4). DNA damage leads to elevated levels of p53, which in turn activates transcription of several downstream targets including the gene that encodes the Cdk inhibitor p21. The p53-dependent pathway is also responsible for damage-induced apoptosis in certain cell types. Compared with our knowledge of the downstream events of p53 function, less is known about the upstream events that lead to activation of p53. Several studies have suggested that single-strand DNA (ssDNA) breaks are likely to be the proximal cause for triggering increase in p53 activity and downstream cell cycle arrest (5, 6). ssDNA breaks can be generated either by DNA damage directly inflicted by ionizing radiation (IR) or by repair intermediates such as those present in nucleotide excision repair (NER), the major type of repair in response to UV radiation. In the latter case, elevation of p53 is dependent upon repair efficiency of UV-damaged cells (6). Although it remains unclear how the ssDNA signal is recognized and transduced, it is an attractive model that cellular proteins that recognize DNA damage may act in concert with p53 to sense damage and activate the p53-dependent checkpoint (4).
Replication protein A (RPA) is a multifunctional ssDNA-binding protein complex composed of 70-, 34-, and 11-kDa subunits (7–9). It facilitates DNA unwinding and DNA synthesis in the initiation and elongation stages of DNA replication (10). In addition, RPA is also involved in NER (11). In the latter case, RPA not only participates in the gap-filling stage of NER but also facilitates recognition of pyrimidine dimers in the first step of the repair process by interacting with repair proteins such as XPA and XPG (12–14). This raises an interesting possibility that RPA may coordinate DNA repair with other events in response to DNA damage, such as inhibition of DNA replication and cell cycle arrest. In keeping with this hypothesis, several lines of evidence suggest that RPA activity is regulated after DNA damage. (i) Extracts from UV-irradiated cells lack RPA activity for supporting simian virus 40 DNA replication in vitro (15). (ii) The middle subunit of RPA (RPA-34) is phosphorylated in many irradiated cell types (16, 17). (iii) Furthermore, IR-induced RPA phosphorylation is delayed in Ataxia telangiectasia mutant cells, which are defective in several checkpoints (16). Thus, the damage-dependent change in RPA and its pivotal role in NER support the notion that RPA may be actively involved in damage response.
RPA interacts with the activation domains of several transcription factors from a variety of sources (18–20). In particular, the tumor suppressor p53 has been demonstrated to associate with RPA both in vivo and in vitro. Previous studies also show that the first 73 amino acids of p53, which contain the transcriptional activation domain, are sufficient for RPA binding. In this study, we further characterized this region of p53 for RPA binding. Our results revealed a small RPA-binding region located downstream of the minimal activation domain of p53. To test the hypothesis that the RPA–p53 interaction may play a role in linking p53 to DNA damage, we examined the effects of UV damage on RPA binding to p53. RPA in irradiated cells is precluded from interacting with p53 in a UV dose- and time-dependent manner. Furthermore, this modulation is correlated with the ability of damaged cells to perform global genome repair.
MATERIALS AND METHODS
Cell Culture and Treatment.
RKO cells were obtained from Michael Kastan (Johns Hopkins University). WI38 cells were purchased from American Type Culture Collection. Both cell types were grown in DMEM plus 10% fetal bovine serum. The following mutant human fibroblast cell lines were purchased from Coriell Cell Repositories (Camden, NJ) and grown in MEM plus 20% fetal bovine serum: XPA (GM0710B); XPC (GM03176); XPG (GM03021A); CSA (GM01856B); CSB (GM01098B). For UV irradiation, cells were washed with PBS and irradiated at various doses using either a UV crosslinker (Stratagene) or a 254-nm UV lamp precalibrated by a UV radiometer. For IR, a 137Cs-source γ radiator was used at a dose rate of approximately 1 Gy/min. The cells were then incubated in medium for various periods of time. Cells were harvested and incubated on ice for 15 min in the following lysis buffer: 50 mM Tris⋅HCl (pH 7.4), 125 mM NaCl, 50 mM NaF, 5 mM EDTA, 1 mM Na3VO4, and 0.1% Nonidet P-40. Cell debris was removed by centrifugation at 10,000 rpm for 10 min in an ice-cold microcentrifuge. Supernatant was either used immediately for binding assays or stored in a −70°C freezer.
Protein Expression and Purification.
The deletion mutants were generated by PCR using the appropriate primers corresponding to the 5′ and 3′ sequences of the amplified region. The PCR fragments were subsequently cloned into the plasmid pGEX-2TK between the BamHI and EcoRI sites. The sequences were verified by restriction digestion and dideoxynucleotide sequencing and confirmed by expression of fusion proteins from isopropyl β-dthiogalactoside-induced bacteria cultures. The point mutants were either obtained from Arnold Levine at Princeton University (21) or generated in our laboratory by site-directed mutagenesis. The mutant cDNA fragments encoding the first 73 amino acids of p53 were amplified by PCR and cloned into the glutathione S-transferase (GST) expression vector. The GST fusion proteins were expressed and affinity-purified as described (18). The recombinant RPA was expressed in bacteria and purified by an established purification scheme; the three-subunit RPA expression vector was obtained from Marc Wold (22).
GST Pull-Down Assay.
For the binding assays using purified recombinant RPA, 200 ng RPA was mixed with an excess of GST–p53 wild-type or various mutant fusion proteins bound to glutathione-agarose beads in buffer D containing 0.2 M NaCl (buffer D = 20 mM Hepes, pH 7.5/1 mM EDTA/0.5 mM dithiothreitol/0.1% Nonidet P-40). As a carrier, insulin was also added to a final concentration of 10 ng/μl. The mixture was rotated at 4°C for 4 hr. After extensive washes with the same buffer, the beads were eluted with buffer D containing 1.2 M NaCl. The amount of RPA in the eluates was analyzed by immunoblotting using a polyclonal anti-RPA antiserum (a gift from Marc Wold). For the binding studies using mammalian cell lysates, GST–p53-(1–73) bound to glutathione-agarose beads was mixed with extracts in lysis buffer and incubated with agitation at 4°C for 1 hr. The beads were washed repeatedly with the lysis buffer and the bound protein fractions were eluted in protein sample buffer. The presence of RPA in the bound fractions was detected by SDS/PAGE and immunoblotting using either mono- or polyclonal anti-RPA antibodies (23) (S. Din and B. Stillman, personal communication).
Coimmunoprecipitation.
Cell lysates described above were incubated with a monoclonal anti-p53 antibody (pAb410; a gift from Bruce Stillman, Cold Spring Harbor Laboratory) covalently crosslinked to protein A-Sepharose beads. After extensive washes with lysis buffer, the immunoprecipitates were resolved by SDS/PAGE and immunoblotted with either anti-RPA or anti-p53 antibodies.
RESULTS
Mutational Analysis of the RPA-Interacting Region of p53.
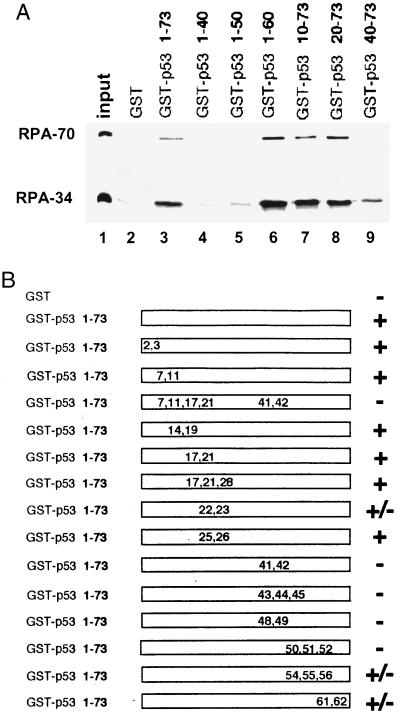
We (18) and others (19, 20) have demonstrated that the first 73 amino acids of p53 are sufficient for interaction with RPA. This region of p53 contains the transcriptional activation domain of the protein and is also responsible for p53 interaction with several other cellular proteins including TAF40, TAF60, mdm2, and TFIIH (4, 24). To identify the minimal region for RPA binding, we generated a series of truncated versions of this region fused to GST. The fusion proteins immobilized on glutathione-agarose beads were mixed with purified recombinant human RPA. The amount of RPA bound to the beads was detected by immunoblotting (Fig. 1A). Mutants with deletions up to the first 40 amino acids were still capable of binding to RPA, although RPA binding of GST–p53-(40–73) was weaker than that of the other positive binding N-terminal deletion constructs. Removal of amino acids 60–73 also did not affect RPA binding. However, further deletion from the C-terminal end completely abolished the interaction with RPA. These results suggest that the region between amino acids 40–60 is most critical for RPA binding. To more closely characterize the interaction domain, we also tested RPA binding of a number of point mutants spanning the 73-amino acid region. In keeping with the results from the deletion analysis, most proteins that carried mutations between the amino acids 40 and 60 completely abrogated RPA binding (Fig. 1B). In contrast, most mutations outside the 20-amino acid region did not affect RPA binding. The only exceptions were the mutations at amino acids 22 and 23 and at amino acids 61 and 62 that reduced RPA binding. Thus, with the data from the deletion analysis, we conclude that amino acids 40–60 play a pivotal role in p53–RPA interaction. Our results are also consistent with a possible minor contribution from residues outside this 20-amino acid region.
Figure 1.
Characterization of the RPA-interacting domain of p53. (A). Deletion analysis of the N-terminal domain of p53. One-fourth of the input purified RPA (50 ng) and 40% of the bound fractions were loaded on the gel. Approximately 25–30% of the input RPA was bound to the wild-type GST–p53 beads. (B) RPA-binding ability of various point mutants of the p53 activation domain. +, Affinity no less than the wild type; −, signal equivalent to GST alone; ±, reduced binding affinity similar to the one shown in Fig. 1A, lane 9. The positions and types of these mutations are E2K, E3K, D7H, E11K, L14Q, E17K, F19S, D21H, L22Q, W23S, L25Q, L26H, E28K, D41H, D42H, L43A, M44A, L45A, D48H, D49H, I50A, E51A, Q52A, F54A, T55A, E56A, D61H, and E62K.
UV Radiation Abolishes RPA Interaction with the Activation Domain of p53.
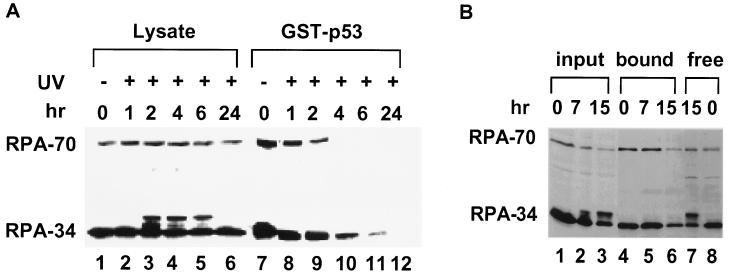
Previous studies suggest that UV radiation causes several changes in the biochemical properties of RPA, including decreased replication activity and increased phosphorylation of the RPA-34 subunit (15–17). To test the effect of UV radiation on RPA interaction with p53, lysates were prepared at different times after UV radiation at 50 J/m2 and were mixed with GST–p53-(1–73) beads. The amount of RPA bound to the beads was analyzed by immunoblotting as shown in Fig. 2A. Consistent with previous studies, the total protein level of RPA was not affected by UV damage. Furthermore, phosphorylation of RPA-34 increased upon UV radiation, as indicated by the appearance of an additional band migrating above the major RPA-34 band (e.g., compare lanes 1 and 3). However, most RPA lost the ability of p53 binding by the 4- and 6-hr time points (lanes 10 and 11). No bound RPA could be detected by 24 hr. The interaction was also abolished after irradiation at 20 J/m2 and partially lost at 10 J/m2 (data not shown). Because the decrease in binding activity occurred gradually after UV damage, loss of RPA activity is unlikely due to nonspecific UV crosslinking. The same effect was observed in a variety of other irradiated cell types, including HeLa and 293 cells (data not shown and see below).
Figure 2.
RPA from UV-irradiated RKO cells loses its ability to bind the activation domain of p53. (A) Cells were irradiated at 50 J/m2 and subsequently incubated in medium at 37°C as indicated before harvesting. RPA from these lysates was tested for binding to GST–p53-(1–73). RPA was detected by immunoblotting using monoclonal antibodies raised against RPA-70 and RPA-34. The species migrating slightly above the major RPA-34 band in the lysates from UV-irradiated cells represents hyperphosphorylated RPA-34. In this and the following experiments, 5% of the input lysates (lanes 1–6) and 50% of the bound fractions (lanes 7–12) were loaded on the gel. The 0-hr point represents the sample without irradiation. (B) RPA binding to p53 is not significantly affected by 20 Gy of γ-radiation. Lysates (lanes 1–3) were harvested at different times and used in the binding assay with GST–p53 (lanes 4–6). The supernatants of the completed binding reactions at 0 and 15 hr were included (lanes 7 and 8).
It is worth noting that no hyperphosphorylated RPA-34 could be detected in the GST–p53-bound fraction, even when most binding activity of the hypophosphorylated RPA was still present (e.g., Fig. 2, compare lanes 1 and 3 with lanes 7 and 9), indicating that hyperphosphorylated RPA in the lysates has much less affinity for the activation domain of p53. However, RPA hyperphosphorylation is not likely to be the only cause for loss of RPA binding activity, because the two events did not correlate in time with each other. The hyperphosphorylated form of RPA-34 appeared 2 hr after irradiation (lane 3), was sustained at a relatively constant level over a period of time, and disappeared at the 24-hr point (lane 6). On the other hand, RPA binding activity decreased steadily to an undetectable level over a 24-hr period after UV radiation.
UV and IR elicit common cellular responses such as elevation of p53 protein levels and cell cycle arrest (1, 2, 25). However, there are important differences between the two types of damage. IR induces DNA breaks, whereas the initial damage caused by UV is photodimers. This may account for the slower kinetics of p53 induction after UV radiation (26), because ssDNA breaks, not the photodimers, appear to be the direct signal for p53 activation (6). To test whether IR could have a similar effect to UV irradiation on the RPA–p53 interaction, RKO cells were irradiated at 20 Gy and lysates were prepared at different times after damage. As expected, RPA-34 was phosphorylated after γ-radiation (Fig. 2B, lanes 2 and 3). Consistent with the findings in the UV treatment, little binding was detected between the GST–p53 beads and the hyperphosphorylated RPA (Fig. 2B, compare lanes 2 and 3 with lanes 5 and 6). This was not due to inadvertent dephosphorylation of RPA during incubation, because the hyperphosphorylated form was present in the unbound fraction at the completion of the binding reaction (Fig. 2B, lane 7). In contrast to the inhibitory effect of UV on the RPA–p53 interaction, the IR effect was much delayed and more subtle; a modest decrease in binding activity of the hypophosphorylated RPA was detected only at much later times (e.g., 15 hr from data in Fig. 2B). The different effects of UV and IR may reflect different pathways of cellular response after the two types of damage. Alternatively, it could simply be due to differences in the number of lesions and repair intermediates.
UV Damage also Disrupts the Interaction Between the Endogenous RPA and p53.
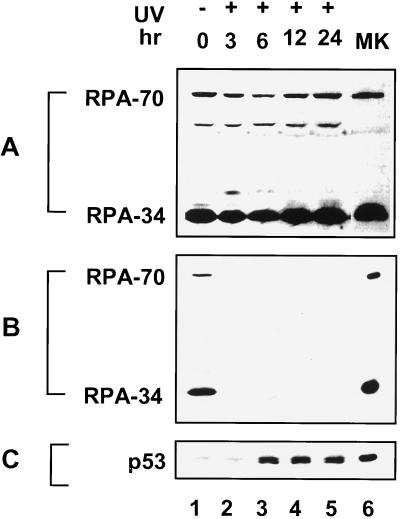
DNA damage including UV radiation is known to trigger elevation of the p53 protein levels (26, 27). Therefore, one possible scenario is that RPA from irradiated cells was sequestered by a saturating amount of damage-induced endogenous p53 and, hence, unavailable for binding to the GST–p53 beads. We did not favor this possibility because (i) RPA is a relatively abundant protein and (ii) RPA from UV-irradiated cells with no endogenous p53 was still prevented from binding to the GST–p53 beads (data not shown). Nevertheless, we sought to directly examine the effect of UV radiation on the interaction of the endogenous RPA and p53. RKO cells are known to contain functional p53 protein and maintain an intact p53-dependent checkpoint control. The total level of detectable RPA did not change through the time course of the experiment (Fig. 3A). Lysates from RKO cells were immunoprecipitated with an anti-p53 antibody and the precipitates were analyzed for the amount of associated RPA by immunoblotting with an anti-RPA antibody (Fig. 3B). In line with previously published results using different cell lines (20), RPA was coprecipitated with p53 from the unirradiated RKO cells (Fig. 3B, lane 1). As a control, RPA was not coprecipitated from lysates of Saos2 cells, which lack endogenous p53 protein (data not shown). When RKO cells were irradiated with UV, the amount of RPA associated with p53 was greatly reduced after damage (lanes 2–5). Concomitantly, an increasing amount of p53 was precipitated by the same anti-p53 antibody as a result of UV induction (Fig. 3C). Thus, the data clearly indicate that the endogenous RPA–p53 complexes are disrupted in UV-irradiated cells.
Figure 3.
UV radiation disrupts the interaction between the endogenous p53 and RPA. (A) Lysates were prepared at different times after UV irradiation at 50 J/m2. Total RPA in extracts was detected by immunoblotting using an anti-RPA polyclonal antiserum. The 50-kDa band is a degradation product of RPA-70. (B) The lysates were immunoprecipitated by an anti-p53 monoclonal antibody crosslinked to protein G-Sepharose beads. The immunoprecipitates were resolved by SDS/PAGE and immunoblotted using an anti-RPA polyclonal antiserum. (C) The same blot shown in B was stripped and reprobed with the anti-p53 antibody. Lanes 6 in A–C contain purified recombinant proteins as markers.
Modulation of RPA Interaction with p53 Is Dependent upon Nucleotide Excision Repair.
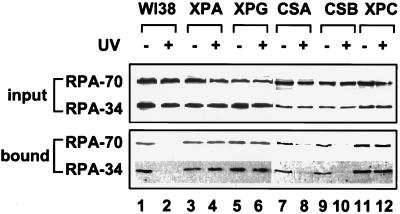
Given the important role of RPA in recognition of the damage site and in gap-filling synthesis of NER, we speculated that down-regulation of the RPA–p53 interaction in UV-irradiated cells might be linked to its function in DNA repair. To probe this hypothesis, we compared the RPA interaction with p53 in normal cells and several mutant cell types that are defective in various aspects of NER (25, 28). As shown in Fig. 4, lysates from normal and mutant cells contained equivalent amounts of RPA before and after UV treatment. Consistent with the results from the RKO cells (Fig. 2), RPA from normal human primary fibroblasts (WI38) also displayed reduced ability to bind GST–p53 after UV radiation (Fig. 4, compare lanes 1 and 2). When different repair-deficient cells were tested, it was found that CSA and CSB (deficient in transcription-coupled repair) behaved in a manner similar to the wild-type cells, whereas the other mutant cells (XPA, XPG, and XPC) that are deficient in global genome repair did not show significant decrease in RPA binding activity after UV damage. Therefore, modulation of the RPA binding activity was correlated well with the capability of the damaged cells to perform global repair. Deficiency in transcription-coupled repair did not appear to affect the cell’s ability to down-regulate the p53–RPA interaction. These results were confirmed by coimmunoprecipitation of the endogenous RPA and p53 from the normal and mutant cell lysates (data not shown). These data also indicate that the presence of UV-inflicted photoproducts alone is not sufficient to down-regulate the interaction between RPA and p53.
Figure 4.
RPA binding to p53 is modulated by UV radiation in a NER-dependent manner. Normal human primary fibroblasts (WI38) and mutant cells with defects in various aspects of NER were irradiated at 50 J/m2. Lysates were prepared 4 hr after irradiation and tested for binding of RPA to the GST–p53 beads. RPA in the input lysates (Upper) and in the bound fractions (Lower) was detected by immunoblotting using anti-RPA-70 and anti-RPA-34 monoclonal antibodies.
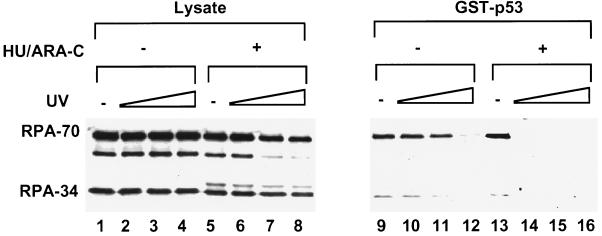
If NER intermediates that contain RPA are indeed required for modulation of the p53–RPA interaction, we surmised that inhibition of the gap-filling step of NER in UV-irradiated cells would give rise to more stalled repair intermediates and, thus, incapacitate more RPA from binding to p53. To test this assumption, WI38 cells were treated with increasing doses of UV and incubated afterwards in the presence or absence of hydroxyurea (HU) and cytosine arabinonucleoside (araC), which inhibit repair synthesis of NER (6, 29, 30). When the lysates were tested for RPA binding to GST–p53, we found that HU/araC exacerbated the UV-induced inhibition of RPA binding to p53, namely, complete inhibition of the interaction could be detected at much lower UV doses (10 vs. 50 J/m2; Fig. 5). The drug treatment alone did not significantly affect RPA–p53 binding in unirradiated cells (Fig. 5, compare lanes 9 and 13). Thus, these results strongly support the notion that the RPA-dependent global genome repair is a pivotal determinant for the dissociation of RPA and p53 after UV damage. Because the loss of the RPA–p53 interaction is UV-dose-dependent, it will be important to determine the numbers and types of repair intermediates that are required for dissociation of the complex.
Figure 5.
RPA binding to p53 is more sensitive to UV radiation when the gap-filling synthesis of NER is inhibited. WI38 cells were irradiated with increasing doses of UV (10, 20, and 50 J/m2) and incubated for 3 hr in the absence (lanes 1–4 and 9–12) or presence of 2 mM HU and 10 μM araC (lanes 5–8 and 13–16). RPA binding to the GST–p53 beads was detected by immunoblotting with an anti-RPA polyclonal antiserum.
DISCUSSION
The first 73 amino acids of p53 contain the transcriptional activation domain of the protein (24). The present study demonstrates that the most critical region for RPA interaction resides in amino acids 41–73. This is consistent with a recently published study that shows that mutations at amino acids 53–54 or 48–49 greatly affect p53’s ability to bind to RPA (31). Because the first 40 amino acids are sufficient for transcriptional activation and binding to the TATA box-binding protein-associated factors of the basal transcription machinery (32, 33), RPA binding is not likely to be required for p53 trans-activation. In support of this notion, mutations in the p53 gene that disrupt RPA binding do not affect p53’s ability to activate transcription or suppress cell growth (31). Furthermore, the fact that UV radiation results in dissociation of RPA and p53 also suggest that RPA is unlikely to be involved in downstream events of p53 functions. Instead, we postulate that the RPA–p53 interaction may play a role in other upstream aspects of the p53-dependent damage response.
A pivotal step in the p53-dependent checkpoint control involves induction of the p53 protein level and increased p53 activity. Although it remains largely unclear exactly how DNA damage triggers elevation of the p53 levels, several studies have shown that DNA strand breaks are sufficient for triggering p53 induction (5, 6). For UV irradiation, induction of p53 appears to be dependent upon the presence of ssDNA intermediates from NER (6). Furthermore, this dependence becomes more prominent when the gap-filling step of NER is inhibited by HU/araC. Interestingly, these findings bear significant analogy to our observation on UV-induced modulation of the RPA–p53 interaction. (i) p53 induction after UV treatment is correlated with the disappearance of RPA–p53 interaction. (ii) The phenomenon is dependent upon the capability of cells to carry out global genome repair. (iii) Treatment of irradiated cells with HU/araC results in elimination of the RPA–p53 interaction at lower UV doses. Recently, Miller et al. (34) reported that RPA inhibited p53 binding to its DNA sites in its target promoters and that ssDNA could overcome the inhibitory effect of RPA, thus providing biochemical hints as to the impact of RPA binding on p53 function. On the basis of these results, the following hypothesis can be put forth: RPA normally sequesters p53 in unirradiated cells. Upon UV irradiation, RPA participates in NER and simultaneously releases the bound p53, thus transducing the damage signal and activating the p53-dependent checkpoint control. This simple idea may be further elaborated by the possibility that other repair proteins may participate in higher-order complexes that contain RPA and p53. Alternatively, p53 released from RPA after UV radiation could directly participate in certain steps of NER. In this regard, p53 has been shown to interact with XPB and XPD, two components of the TFIIH complex required for both transcription and repair (35). In addition, cells that carry homozygous mutations in the p53 loci are deficient in global genome repair but proficient in transcription-coupled repair (36). Again, it coincides with the observation that modulation of the RPA–p53 interaction depends on global genome repair but not transcription-coupled repair. Therefore, it is conceivable that release of p53 by RPA may facilitate p53’s function in NER. Further studies are required to distinguish between these possibilities.
What is the biochemical basis for damage-induced dissociation of RPA–p53? In light of the fact that RPA from irradiated cells fails to bind either the endogenous p53 or the purified GST–p53 fusion protein, we believe that disruption of the interaction is more likely due to changes in RPA rather than p53. Because the protein level of RPA does not change before and after DNA damage, we consider protein modification and/or sequestration as the two most likely causes. The middle subunit of RPA (RPA-34) is phosphorylated in a cell-cycle-dependent manner (23) and also in many UV- or γ-irradiated cell types (refs. 15 and 16; also see Fig. 3). The damage-induced phosphorylation of RPA-34 appears to be dependent on key checkpoint genes in mammalian and yeast cells (16, 17). It is interesting that hyperphosphorylated RPA induced by either UV or IR does not associate with p53. Given the fact that the RPA-70 subunit contains the domain that interacts with p53 (19, 37), conformational change due to phosphorylation of RPA-34 could result in masking the p53-interacting domain on RPA-70. Alternatively, the hyperphosphorylated RPA may preferentially bind to other factors that can preclude RPA binding to p53. Although the functional significance of RPA phosphorylation awaits further investigation, the in vitro study clearly reveals a distinct behavior of phosphorylated RPA.
Hyperphosphorylated RPA does not associate with p53 in our assays; however, this state cannot entirely account for the loss of RPA interaction with p53 after UV damage. This is because (i) the hyperphosphorylated form of RPA from most irradiated cells represents only a minor portion of the total RPA; (ii) UV-induced phosphorylation of RPA is hardly detectable in certain cell types (e.g., WI38), yet the RPA–p53 interaction is inhibited to a similar degree in these cells following UV damage; (iii) the time course of RPA hyperphosphorylation does not correlate well with that of inhibition of the RPA–p53 interaction. Therefore, we suspect that there may exist additional causes for UV-induced modulation of the interaction or that minor yet specific phosphorylation may lead to the disruption of the interaction. Biochemical characterization is underway to determine the underlying mechanisms of RPA inactivation. However, it is tempting to speculate that an active “repairsome” that includes RPA, XPA, and XPG may be responsible for precluding RPA from binding to p53 and this could trigger activation of the p53-dependent checkpoint pathway.
Acknowledgments
We thank Dr. Bruce Stillman for the anti-RPA and anti-p53 antibodies, Dr. Arnold Levine for sharing some of the mutant p53 constructs used in our study, Dr. Michael Kastan for the RKO cells, and Dr. Marc Wold for the anti-RPA antiserum and RPA expression vector. R.L. is also grateful to Drs. Bruce Stillman and Arne Stenlund for helpful discussion and encouragement during the course of the work. This work was supported in part by a Special Fellow Award from Leukemia Society of America, an American Cancer Society institutional research grant, a start-up research fund from University of Virginia, funds from the National Institute on Environmental Health Sciences Center (ES-01896), and National Cancer Institute Grant CA-30490.
ABBREVIATIONS
- RPA
replication protein A
- NER
nucleotide excision repair
- IR
ionizing radiation
- ssDNA
single-stranded DNA
- GST
glutathione S-transferase
- HU
hydroxyurea
- araC
cytosine arabinonucleoside
References
- 1.Elledge S J. Science. 1996;274:1664–1672. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- 2.Paulovich A G, Toczyski D P, Hartwell L H. Cell. 1997;88:315–321. doi: 10.1016/s0092-8674(00)81870-x. [DOI] [PubMed] [Google Scholar]
- 3.Vogelstein B, Kinzler K W. Cell. 1992;70:523–526. doi: 10.1016/0092-8674(92)90421-8. [DOI] [PubMed] [Google Scholar]
- 4.Levine A J. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 5.Huang L-C, Clarkin K C, Wahl G M. Proc Natl Acad Sci USA. 1996;93:4827–4832. doi: 10.1073/pnas.93.10.4827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nelson W G, Kastan M B. Mol Cell Biol. 1994;14:1815–1823. doi: 10.1128/mcb.14.3.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fairman M P, Stillman B. EMBO J. 1988;7:1211–1218. doi: 10.1002/j.1460-2075.1988.tb02933.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wobbe C R, Weissbach L, Borowiec J A, Dean F B, Murakami Y, Bullock P, Hurwitz J. Proc Natl Acad Sci USA. 1987;84:1834–1838. doi: 10.1073/pnas.84.7.1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wold M S, Kelly T. Proc Natl Acad Sci USA. 1988;85:2523–2527. doi: 10.1073/pnas.85.8.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wold M. Annu Rev Biochem. 1997;66:61–91. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 11.Coverley D, Kenny M K, Munn M, Rupp W D, Lane D P, Wood R D. Nature (London) 1991;349:538–541. doi: 10.1038/349538a0. [DOI] [PubMed] [Google Scholar]
- 12.He Z, Henricksen L A, Wold M S, Ingles C J. Nature (London) 1995;374:566–569. doi: 10.1038/374566a0. [DOI] [PubMed] [Google Scholar]
- 13.Matsuda T, Saijo M, Kuraoka I, Kobayashi T, Nakatsu Y, Nagai A, Enjoji T, Masutani C, Sugasawa K, Hanaoka F, Yasui A, Tanaka K. J Biol Chem. 1995;270:4152–4157. doi: 10.1074/jbc.270.8.4152. [DOI] [PubMed] [Google Scholar]
- 14.Lee S-H, Kim D-K, Driss R. J Biol Chem. 1995;270:21800–21805. doi: 10.1074/jbc.270.37.21800. [DOI] [PubMed] [Google Scholar]
- 15.Carty M P, Zernik-Kobak M, MaGrath S, Dixon K. EMBO J. 1994;13:2114–2123. doi: 10.1002/j.1460-2075.1994.tb06487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu V F, Weaver D T. Mol Cell Biol. 1993;13:7222–7231. doi: 10.1128/mcb.13.12.7222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brush G S, Morrow D M, Hieter P, Kelly T J. Proc Natl Acad Sci USA. 1996;93:15075–15080. doi: 10.1073/pnas.93.26.15075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li R, Botchan M R. Cell. 1993;73:1207–1221. doi: 10.1016/0092-8674(93)90649-b. [DOI] [PubMed] [Google Scholar]
- 19.He Z, Brinton B T, Greenblatt J, Hassell J A, Ingles C J. Cell. 1993;73:1223–1232. doi: 10.1016/0092-8674(93)90650-f. [DOI] [PubMed] [Google Scholar]
- 20.Dutta A, Ruppert J M, Aster J C, Winchester E. Nature (London) 1993;365:79–82. doi: 10.1038/365079a0. [DOI] [PubMed] [Google Scholar]
- 21.Lin J, Chen J, Elenbaas B, Levine A J. Genes Dev. 1994;8:1235–1246. doi: 10.1101/gad.8.10.1235. [DOI] [PubMed] [Google Scholar]
- 22.Henricksen L A, Umbricht C B, Wold M S. J Biol Chem. 1994;269:11121–11132. [PubMed] [Google Scholar]
- 23.Din S, Brill S, Fairman M P, Stillman B. Genes Dev. 1990;4:968–977. doi: 10.1101/gad.4.6.968. [DOI] [PubMed] [Google Scholar]
- 24.Ko L J, Prives C. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 25.Friedberg E C, Walker G C, Siede W. DNA Repair and Mutagenesis. Washington, DC: Am. Soc. Microbiol.; 1995. [Google Scholar]
- 26.Lu X, Lane D P. Cell. 1993;75:765–778. doi: 10.1016/0092-8674(93)90496-d. [DOI] [PubMed] [Google Scholar]
- 27.Maltzman W, Czyzyk L. Mol Cell Biol. 1984;4:1689–1694. doi: 10.1128/mcb.4.9.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sancar A. Annu Rev Genet. 1995;29:69–105. doi: 10.1146/annurev.ge.29.120195.000441. [DOI] [PubMed] [Google Scholar]
- 29.Dunn W C, Regan J D. Mol Pharmacol. 1979;15:367–374. [PubMed] [Google Scholar]
- 30.Fornace A J, Jr, Seres D S. Radiat Res. 1983;93:107–111. [PubMed] [Google Scholar]
- 31.Leiter L M, Chen J, Marathe T, Tanaka M, Dutta A. Oncogene. 1996;12:2661–2668. [PubMed] [Google Scholar]
- 32.Lu H, Levine A J. Proc Natl Acad Sci USA. 1995;92:5154–5158. doi: 10.1073/pnas.92.11.5154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thut C J, Chen J L, Klemin R, Tjian R. Science. 1995;267:100–104. doi: 10.1126/science.7809597. [DOI] [PubMed] [Google Scholar]
- 34.Miller S D, Moses K, Jayaraman L, Prives C. Mol Cell Biol. 1997;17:2194–2201. doi: 10.1128/mcb.17.4.2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang X W, Yeh H, Schaeffer L, Roy R V M, Egly J-M, Wang Z, Friedberg E C, Evans M K, Taffe B G, Bohr V A, Weeda G, Joeijmakers J H J, Forrester K, Harris C C. Nat Genet. 1995;10:188–195. doi: 10.1038/ng0695-188. [DOI] [PubMed] [Google Scholar]
- 36.Ford J M, Hanawalt P C. Proc Natl Acad Sci USA. 1995;92:8876–8880. doi: 10.1073/pnas.92.19.8876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin Y-L, Chen C, Keshav K F, Winchester E, Dutta A. J Biol Chem. 1996;271:17190–17198. doi: 10.1074/jbc.271.29.17190. [DOI] [PubMed] [Google Scholar]