Abstract
All of the defective interfering (DI) RNAs of mouse hepatitis virus (MHV) contain both the 5' and 3' ends of the viral genomic RNA, which presumably include the cis sequences required for RNA replication. To define the replication signal of MHV RNA, we have used a vaccinia virus-T7 polymerase-transcribed MHV DI RNA to study the effects of sequence deletion on DI RNA replication. Following infection of susceptible cells with a recombinant vaccinia virus expressing T7 RNA polymerase, various cDNA clones derived from a DI RNA (DIssF) of the JHM strain of MHV, which is a 3.5-kb naturally occurring DI RNA, behind a T7 promoter were transfected. On superinfection with a helper MHV, the ability of various DI RNAs to replicate was determined. Serial deletions from the middle of the RNA toward both the 5' and 3' ends demonstrated that 859 nucleotides from the 5' end and 436 nucleotides from the 3' end of the MHV RNA genome were necessary for RNA replication. Surprisingly, an additional stretch of 135 nucleotides located at 3.1 to 3.3 kb from the 5' end of the genome was also required. This stretch is discontiguous from the 5'-end cis replication signal and is present in all of the naturally occurring DI RNAs studied so far. The requirement for a long stretch of 5'- and 3'-end sequences predicts that the subgenomic MHV mRNAs cannot replicate. The efficiency of RNA replication varied with different cDNA constructs, suggesting possible interaction between different regions of DI RNA. The identification of MHV RNA replication signals allowed the construction of an MHV DI-based expression vector, which can express foreign genes, such as the chloramphenicol acetyltransferase gene.
Full text
PDF




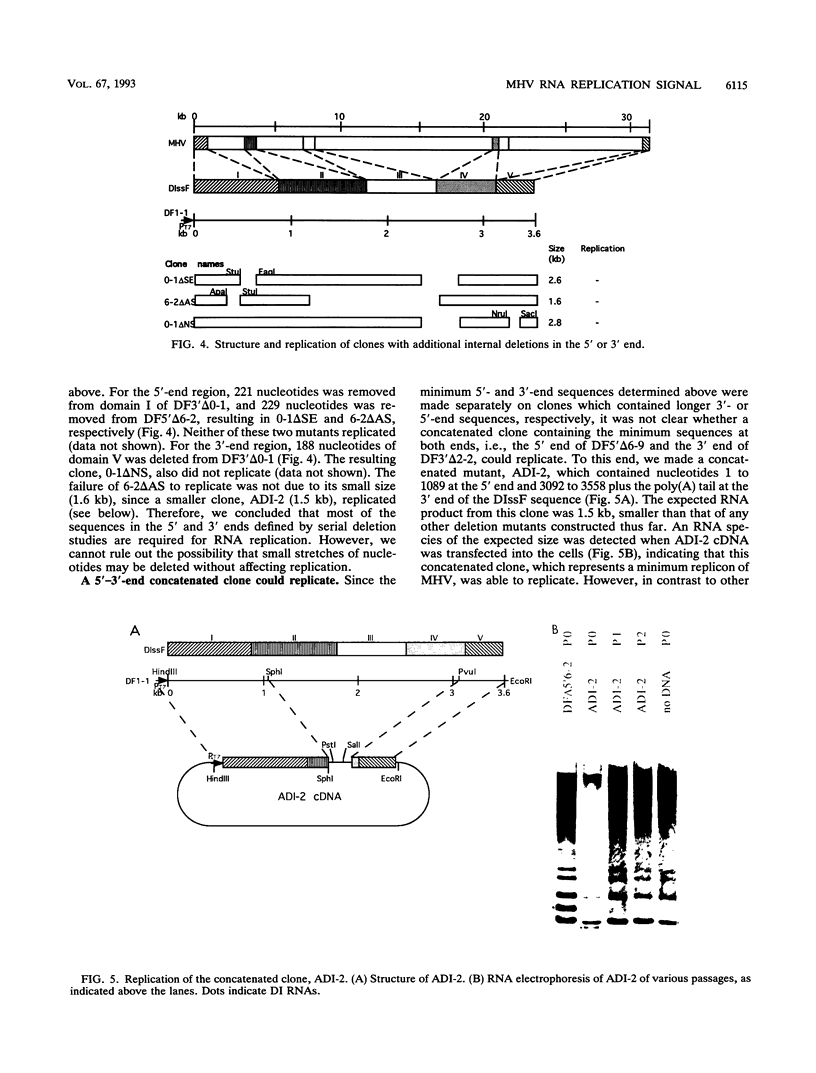


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Fosmire J. A., Hwang K., Makino S. Identification and characterization of a coronavirus packaging signal. J Virol. 1992 Jun;66(6):3522–3530. doi: 10.1128/jvi.66.6.3522-3530.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuerst T. R., Niles E. G., Studier F. W., Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci U S A. 1986 Nov;83(21):8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuya T., Macnaughton T. B., La Monica N., Lai M. M. Natural evolution of coronavirus defective-interfering RNA involves RNA recombination. Virology. 1993 May;194(1):408–413. doi: 10.1006/viro.1993.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano N., Fujiwara K., Hino S., Matumoto M. Replication and plaque formation of mouse hepatitis virus (MHV-2) in mouse cell line DBT culture. Arch Gesamte Virusforsch. 1974;44(3):298–302. doi: 10.1007/BF01240618. [DOI] [PubMed] [Google Scholar]
- Jang S. K., Davies M. V., Kaufman R. J., Wimmer E. Initiation of protein synthesis by internal entry of ribosomes into the 5' nontranslated region of encephalomyocarditis virus RNA in vivo. J Virol. 1989 Apr;63(4):1651–1660. doi: 10.1128/jvi.63.4.1651-1660.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong Y. S., Makino S. Mechanism of coronavirus transcription: duration of primary transcription initiation activity and effects of subgenomic RNA transcription on RNA replication. J Virol. 1992 Jun;66(6):3339–3346. doi: 10.1128/jvi.66.6.3339-3346.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. N., Lai M. M., Makino S. Generation and selection of coronavirus defective interfering RNA with large open reading frame by RNA recombination and possible editing. Virology. 1993 May;194(1):244–253. doi: 10.1006/viro.1993.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koetzner C. A., Parker M. M., Ricard C. S., Sturman L. S., Masters P. S. Repair and mutagenesis of the genome of a deletion mutant of the coronavirus mouse hepatitis virus by targeted RNA recombination. J Virol. 1992 Apr;66(4):1841–1848. doi: 10.1128/jvi.66.4.1841-1848.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M. M. Coronavirus: organization, replication and expression of genome. Annu Rev Microbiol. 1990;44:303–333. doi: 10.1146/annurev.mi.44.100190.001511. [DOI] [PubMed] [Google Scholar]
- Lai M. M., Stohlman S. A. RNA of mouse hepatitis virus. J Virol. 1978 May;26(2):236–242. doi: 10.1128/jvi.26.2.236-242.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. J., Shieh C. K., Gorbalenya A. E., Koonin E. V., La Monica N., Tuler J., Bagdzhadzhyan A., Lai M. M. The complete sequence (22 kilobases) of murine coronavirus gene 1 encoding the putative proteases and RNA polymerase. Virology. 1991 Feb;180(2):567–582. doi: 10.1016/0042-6822(91)90071-I. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibowitz J. L., Weiss S. R., Paavola E., Bond C. W. Cell-free translation of murine coronavirus RNA. J Virol. 1982 Sep;43(3):905–913. doi: 10.1128/jvi.43.3.905-913.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luytjes W., Krystal M., Enami M., Parvin J. D., Palese P. Amplification, expression, and packaging of foreign gene by influenza virus. Cell. 1989 Dec 22;59(6):1107–1113. doi: 10.1016/0092-8674(89)90766-6. [DOI] [PubMed] [Google Scholar]
- MANAKER R. A., PICZAK C. V., MILLER A. A., STANTON M. F. A hepatitis virus complicating studies with mouse leukemia. J Natl Cancer Inst. 1961 Jul;27:29–51. [PubMed] [Google Scholar]
- Makino S., Fujioka N., Fujiwara K. Structure of the intracellular defective viral RNAs of defective interfering particles of mouse hepatitis virus. J Virol. 1985 May;54(2):329–336. doi: 10.1128/jvi.54.2.329-336.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Joo M., Makino J. K. A system for study of coronavirus mRNA synthesis: a regulated, expressed subgenomic defective interfering RNA results from intergenic site insertion. J Virol. 1991 Nov;65(11):6031–6041. doi: 10.1128/jvi.65.11.6031-6041.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Shieh C. K., Keck J. G., Lai M. M. Defective-interfering particles of murine coronavirus: mechanism of synthesis of defective viral RNAs. Virology. 1988 Mar;163(1):104–111. doi: 10.1016/0042-6822(88)90237-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Shieh C. K., Soe L. H., Baker S. C., Lai M. M. Primary structure and translation of a defective interfering RNA of murine coronavirus. Virology. 1988 Oct;166(2):550–560. doi: 10.1016/0042-6822(88)90526-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Taguchi F., Fujiwara K. Defective interfering particles of mouse hepatitis virus. Virology. 1984 Feb;133(1):9–17. doi: 10.1016/0042-6822(84)90420-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S., Yokomori K., Lai M. M. Analysis of efficiently packaged defective interfering RNAs of murine coronavirus: localization of a possible RNA-packaging signal. J Virol. 1990 Dec;64(12):6045–6053. doi: 10.1128/jvi.64.12.6045-6053.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMaster G. K., Carmichael G. G. Analysis of single- and double-stranded nucleic acids on polyacrylamide and agarose gels by using glyoxal and acridine orange. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4835–4838. doi: 10.1073/pnas.74.11.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer F., Weber H., Weissmann C. Interactions of Q beta replicase with Q beta RNA. J Mol Biol. 1981 Dec 15;153(3):631–660. doi: 10.1016/0022-2836(81)90411-3. [DOI] [PubMed] [Google Scholar]
- Moss B., Elroy-Stein O., Mizukami T., Alexander W. A., Fuerst T. R. Product review. New mammalian expression vectors. Nature. 1990 Nov 1;348(6296):91–92. doi: 10.1038/348091a0. [DOI] [PubMed] [Google Scholar]
- Pachuk C. J., Bredenbeek P. J., Zoltick P. W., Spaan W. J., Weiss S. R. Molecular cloning of the gene encoding the putative polymerase of mouse hepatitis coronavirus, strain A59. Virology. 1989 Jul;171(1):141–148. doi: 10.1016/0042-6822(89)90520-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park K. H., Huang T., Correia F. F., Krystal M. Rescue of a foreign gene by Sendai virus. Proc Natl Acad Sci U S A. 1991 Jul 1;88(13):5537–5541. doi: 10.1073/pnas.88.13.5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattnaik A. K., Ball L. A., LeGrone A. W., Wertz G. W. Infectious defective interfering particles of VSV from transcripts of a cDNA clone. Cell. 1992 Jun 12;69(6):1011–1020. doi: 10.1016/0092-8674(92)90619-n. [DOI] [PubMed] [Google Scholar]
- Percy N., Barclay W. S., Sullivan M., Almond J. W. A poliovirus replicon containing the chloramphenicol acetyltransferase gene can be used to study the replication and encapsidation of poliovirus RNA. J Virol. 1992 Aug;66(8):5040–5046. doi: 10.1128/jvi.66.8.5040-5046.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddell S. Coronavirus JHM: coding assignments of subgenomic mRNAs. J Gen Virol. 1983 Jan;64(Pt 1):113–125. doi: 10.1099/0022-1317-64-1-113. [DOI] [PubMed] [Google Scholar]
- Stohlman S. A., Baric R. S., Nelson G. N., Soe L. H., Welter L. M., Deans R. J. Specific interaction between coronavirus leader RNA and nucleocapsid protein. J Virol. 1988 Nov;62(11):4288–4295. doi: 10.1128/jvi.62.11.4288-4295.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokomori K., La Monica N., Makino S., Shieh C. K., Lai M. M. Biosynthesis, structure, and biological activities of envelope protein gp65 of murine coronavirus. Virology. 1989 Dec;173(2):683–691. doi: 10.1016/0042-6822(89)90581-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot R. J., van der Most R. G., Spaan W. J. The fitness of defective interfering murine coronavirus DI-a and its derivatives is decreased by nonsense and frameshift mutations. J Virol. 1992 Oct;66(10):5898–5905. doi: 10.1128/jvi.66.10.5898-5905.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Most R. G., Bredenbeek P. J., Spaan W. J. A domain at the 3' end of the polymerase gene is essential for encapsidation of coronavirus defective interfering RNAs. J Virol. 1991 Jun;65(6):3219–3226. doi: 10.1128/jvi.65.6.3219-3226.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Most R. G., Heijnen L., Spaan W. J., de Groot R. J. Homologous RNA recombination allows efficient introduction of site-specific mutations into the genome of coronavirus MHV-A59 via synthetic co-replicating RNAs. Nucleic Acids Res. 1992 Jul 11;20(13):3375–3381. doi: 10.1093/nar/20.13.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]