Abstract
One of the rare examples of a single major gene underlying a naturally occurring behavioral polymorphism is the foraging locus of Drosophila melanogaster. Larvae with the rover allele, forR, have significantly longer foraging path lengths on a yeast paste than do those homozygous for the sitter allele, fors. These variants do not differ in general activity in the absence of food. The evolutionary significance of this polymorphism is not as yet understood. Here we examine the effect of high and low animal rearing densities on the larval foraging path-length phenotype and show that density-dependent natural selection produces changes in this trait. In three unrelated base populations the long path (rover) phenotype was selected for under high-density rearing conditions, whereas the short path (sitter) phenotype was selected for under low-density conditions. Genetic crosses suggested that these changes resulted from alterations in the frequency of the fors allele in the low-density-selected lines. Further experiments showed that density-dependent selection during the larval stage rather than the adult stage of development was sufficient to explain these results. Density-dependent mechanisms may be sufficient to maintain variation in rover and sitter behavior in laboratory populations.
Studies of population dynamics have for the most part mistakenly considered the behavior of organisms within a population to be homogeneous (1). However, individual differences in behavior are common and have consequences for the ecology and evolution of populations. A special case of individual differences is behavioral polymorphism, where individuals within a population can be categorized into morphs (phenotypes or strategists) according to their behavior(s). When the polymorphism has a heritable component, the relative selective advantages of the morphs under different environmental conditions can be measured (2). One condition, population density, varies over time and space and plays a significant role in the evolution of characteristics within populations (3). Population density affects a number of processes, for example, predator–prey (4) and parasite–host interactions (5), the spread of disease (6), competition (7), population regulation (8), and territoriality (9). There have been many theoretical studies of density-dependent selection, including those that model its effect on competitive ability (10–13). However, empirical studies that address whether behavioral polymorphisms affect fitness in a density-dependent manner are rare. The fruit fly Drosophila melanogaster is one of the few systems in which we are beginning to have a detailed empirical understanding of density-dependent selection (14–22). It is an ideal model system to study how a genetically characterized polymorphism in behavior responds to density-dependent selection.
The rover/sitter polymorphism in D. melanogaster is a model system that has been used to address both the mechanistic and evolutionary significance of behavioral phenotypes. The locomotion of rover larvae on yeast is significantly higher than that of sitter larvae, resulting in rovers having visibly longer foraging path lengths than sitters during a defined time interval (23). Adult rovers move farther from the food source after feeding than sitters (24). The behavioral differences found between rovers and sitters are found only in the presence of food. In the absence of food, the locomotory scores of rovers and sitters do not differ; that is, they both exhibit long path lengths (24, 25). Rovers move significantly more from food patch to food patch while feeding, whereas sitters tend to feed within a patch (26). The rover/sitter polymorphism is an example of a quantitative trait that varies in nature with approximate phenotypic frequencies of 70% rovers and 30% sitters in an orchard population in Toronto (23, 27). We have shown that differences in the locomotory component of foraging behavior in D. melanogaster larvae and adults result primarily from a single gene called foraging (for) (24, 28, 29). Despite advances on the mechanistic basis of rover/sitter differences (28, 30), the evolutionary significance of the polymorphism has remained refractory. In the present paper we examine whether differential selection occurs on the larval path-length phenotype after many generations of high- and low-density rearing conditions.
METHODS
Strains.
Strains were maintained in 6-oz (1 oz = 29.6 ml) plastic culture bottles on 45 ml of dead yeast, sucrose, and agar medium under conditions of 25 ± 1°C, 15 ± 1 mbar (100 Pa) vapor pressure deficit, and a 12:12 light:dark regime with lights on at 0800 hr (standard conditions). Rover and sitter field-derived strains were constructed from a population of 500 D. melanogaster adult flies collected from an orchard population in the Toronto area (24). After collection the population was bred in the laboratory for 1 year under standard conditions. More than 100 flies per bottle and more than 20 bottles were used to maintain the population. Flies from all bottles were mixed to produce progeny for larval behavioral tests. The foraging path lengths of 500 third-instar larvae were measured after 1 year using a procedure described previously (30). The resultant distribution of larval path lengths is shown in Fig. 1. Individual rover and sitter behaving male larvae were chosen to produce the homozygous forR/forR and fors/fors strains used in the experiments presented herein. Since for had been localized to chromosome 2 at cytological position 24A3–5 (28, 29), we crossed each male to a chromosome-2 balancer stock [In(2LR)SM1, al2 Cy cn2 sp2/In(2LR)bwV1, ds33k bwV1, described in ref. 31]. Previously, the balancer stock had been repeatedly backcrossed (10 times) to the orchard population to maintain variation in the genetic background. The chromosomes carrying the balancers were removed in the crossing procedure so that little to none of the genetic background from the balancer strain was left in the resultant lines that were homozygous for the forR or fors allele (24). The presence of homozygous forR or fors alleles was verified by measuring the larval foraging behavior of progeny from crosses to laboratory rover and sitter strains and to a strain Df(2L)edSz carrying a deficiency of for (28, 32–34). This ensured that each line had heterogeneous genetic backgrounds from the orchard population (≈60% of the genome) for all but chromosome 2.
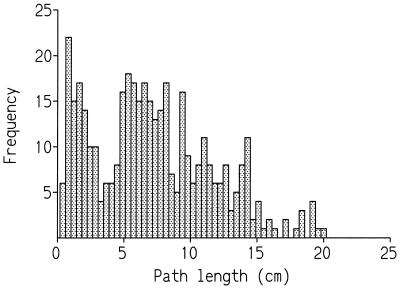
Figure 1.
Frequency distribution of larval foraging path lengths (n = 500). Larvae are the descendants of 500 adult flies collected from a natural population (see text).
Density-Selected Lines.
The r and K lines were kindly provided by L. Mueller (University of California, Irvine) and had undergone 286 fly generations of density-dependent selection at the time of our larval behavioral tests. Three r lines were maintained at low larval and adult densities and three K lines were maintained at high larval and adult densities in a serial transfer system described in ref. 14. Briefly, r lines were maintained by placing 50 3- to 6-day-old adults in half-pint (237-ml) glass culture bottles on 40 ml of dead yeast, sucrose, cornmeal flour, and agar medium under standard conditions for 24 hr, during which time adults laid eggs. Then all adults were discarded. Fourteen days later another 50 arbitrarily sampled 3- to 6-day-old adults were taken from 200–400 progeny and the whole process was repeated. K lines were maintained by a serial transfer system at about 800–1,200 adult flies (carrying capacity) per culture bottle. These conditions produced extreme crowding of larvae and adults in the K lines compared with little evidence of crowding of larvae and adults in the r lines.
The mr and mK Lines were established in our laboratory from a different base population than Mueller’s r and K lines. Flies were reared in 6-oz plastic culture bottles on 45 ml of dead yeast, sucrose, and agar medium and were maintained as described in ref. 14. Three mr lines and five mK lines were established, using equal numbers of a field-derived rover and a field-derived sitter strain. Each selection line underwent 74 fly generations of density-dependent selection prior to our behavioral tests.
CU and UU lines were established by Mueller et al. (35) from the B lines of Rose (36) that had been kept at low larval densities (60–100 larvae per vial) since 1980. Four CU lines were selected in crowded larval conditions (500 larvae per 8-dram (1 dram = 3.7 ml) vial for 12 generations followed by >1,000 larvae per 6-dram vial for 84 generations) and uncrowded adult conditions (≈50 adults per vial). Five UU lines were selected in uncrowded larval conditions (50–80 larvae per 8-dram vial) and uncrowded adult conditions (≈50 adults per vial) for 78 generations.
Genetic Crosses Between the mr and mK Lines.
A series of crosses between the mr and mK lines determined whether the pattern of dominance between the mr and mK lines fit that found in the forR and fors alleles. We also assayed the behavior of mr and mK larvae heterozygous for a deletion of for. Each of the independently generated mr (mr1, mr2, and mr3) and mK (mK1, mK2, mK3, mK4, and mK5) lines were crossed within and between each other and the larval foraging behavior of the progeny from each cross was measured. Crosses were grouped as follows: (i) the mr lines [mr1 × mr1, mr2 × mr2, and mr3 × mr3]; (ii) the mr line crosses [mr1 × mr2, mr1 × mr3, and mr2 × mr3]; (iii) the mK lines [mK1 × mK1, mK2 × mK2, mK3 × mK3, mK4 × mK4, and mK5 × mK5]; (iv) the mK line crosses (all 10 combinations of the mK lines crossed); and (v) the mr × mK crosses [all 15 combinations of the mr × mK lines crossed]. Several of the mr and mK lines were also crossed to Df(2L)edSz, and larval foraging behavior of progeny heterozygous for the mr and mK lines and this deficiency was assayed.
Larval Foraging Behavior: Path Length Assay.
The locomotory component of foraging behavior in third-instar larvae was quantified using a procedure described in ref. 37 that we briefly outline here. Foraging third-instar larvae (96 ± 2 hr posthatching) were collected and washed in distilled water. Larvae were individually placed in the center of one of six circular wells (0.5 mm deep with a 4.25 cm radius) on black rectangular Plexiglas plates (25 cm width × 37 cm length × 0.5 cm height) after each well had been filled with a thin homogeneous layer of aqueous yeast suspension (distilled water and Fleischmann’s bakers’ yeast in a 2:1 ratio by weight). Wells were then covered with Petri dish lids (8.5 cm diameter × 1.4 cm height). After 5 min the path lengths of foraging larvae were measured using an electronic digitizer. Behavioral testing occurred within an 8-hr interval beginning at 1100 hr at room temperature and under homogeneous overhead illumination. Selection was relaxed and lines were maintained under standard conditions (described above) for one to two generations prior to the performance of the behavioral assays.
Statistics.
Nested ANOVA was done to determine whether the effect of treatment (high or low density) was significant. One-way ANOVA followed by Student–Neuman–Keuls a posteriori tests (P = 0.05) were also performed. All data were normally distributed.
RESULTS
Significant variation was found between the high- and low-density lines (Fig. 2a). The low-density-selected r lines had significantly shorter larval path lengths than did the high-density K lines. There was a small, but nevertheless worrisome, possibility that these results arose by chance events (e.g., genetic drift). To determine whether we could reproduce the relative differences in path length found between the r and K lines we used a different base population to generated the mr and mK lines. The advantage of the mr and mK selected lines was that they were established with a known proportion (50:50) of field-derived rovers (forR/forR) and sitters (fors/fors). We found that the mr lines had significantly shorter larval path lengths than the mK lines after 74 generations of density-dependent natural selection (Fig. 2b). We did not expect the absolute path lengths found in the r and K lines to be found in the mr and mK lines, since the base populations used in these experiments likely differed in their initial frequencies of forR and fors and in minor modifier loci that act on for. In addition, environmental factors such as culture medium and rearing condition differences between the Mueller laboratory and ours may affect larval behavioral measures (38). The larval foraging paths of the UU lines (low larval density) were significantly lower than those of the CU lines (high larval density) (Fig. 2c). This showed that density-dependent selection at the larval stage of development was important for these differences in larval foraging behavior.
Figure 2.
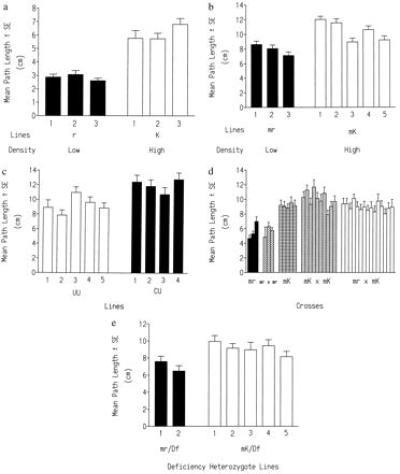
(a) Mean ± SE larval path lengths for the three low-density- (r) and high-density-selected (K) lines. The low-density lines had significantly shorter path lengths than the high-density lines (nested ANOVA, F(1, 4) = 74.5, P < 0.0001; Student–Neuman–Keuls, P = 0.05). Sample sizes ranged from 43 to 53 larvae per line. (b) Mean ± SE larval path lengths for the independently selected replicate low-density (mr) and high-density (mK) lines (n = 40 per line). The low-density mr lines had significantly shorter path lengths than the high-density mK lines (nested ANOVA, F(1, 6) = 8.3, P < 0.05). (c) Mean ± SE larval path lengths for the five UU (low larval and low adult density) and the four CU (high larval and low adult density) lines. UU lines had significantly lower path lengths than CU lines (nested ANOVA, F(1, 7) = 15.25, P < 0.01). (d) Mean ± SE larval path lengths of the mr, mr × mr, mK, mK × mK, and mK × mr crosses. Crosses made with mK lines had significantly higher path lengths than those with mr lines alone (see text). (e) Mean ± SE larval path lengths of the two mr/Df and the five mK/Df heterozygotes (17 < N < 39). Df stands for Df(2L)edSz. The recessive short path-length phenotype found in the mr lines is uncovered by this deficiency. The differences between the mr and mK lines were significant (nested ANOVA, F(1, 7) = 15.25, P < 0.01).
All possible combinations of crosses between the three mr and five mK lines resulted in 10 mK × mK, 15 mK × mr crosses, and 3 mr × mr crosses. We assayed the behavior of 25 larvae from each cross except in four cases where the sample size of larvae tested ranged between 14 and 24. The path lengths of progeny from the mK lines, the mK × mK crosses, and the mK × mr crosses did not differ from each other (one-way ANOVA, F(29, 693) = 1.27, not significant), and all were significantly longer than those from the mr lines (Student–Neuman–Keuls, P = 0.05). The mean ± SE path lengths for these data are shown in Fig. 2d. A nested ANOVA on mr, mK, mK × mK, and mK × mr showed a significant treatment effect [F(3, 29) = 12.55, P < 0.001]. The path lengths of the progeny from different combinations of mr parents (mr1 × mr2, mr2 × mr3, and mr1 × mr3) did not differ significantly from those of their parental mr lines (mr1, mr2, and mr3) (nested ANOVA F(1, 4) = 0.10, not significant). Thus the rover-like behavior found in the mK lines showed complete genetic dominance to the sitter-like one in the mr lines. This paralleled the relationship between the forR (rover) and fors (sitter) alleles. The long larval path-length rover phenotype was dominant to the sitter one in all comparisons despite the fact that the eight lines (three mr and five mk) were independently generated selected lines. Indeed, density-dependent selection had similar effects on genetic variation in foraging behavior in all mr and mK lines. Complementation analysis between the recessive path-length phenotype lines mr1, mr2, and mr3 showed a lack of complementation for short larval path length. This shows that the same allele was selected for in each of the three independently selected mr lines. The deficiency analysis supported the hypothesis that the sitter fors alleles were indeed selected, since larvae heterozygous for an mr line and the Df(2L)edSz had short path lengths (Fig. 2e). The third mr line over this deficiency was not tested, since larval size at 96 hr was too variable in these heterozygotes. Larvae heterozygous for the deficiency Df(2L)edSz and each of five mK chromosomes had significantly longer larval paths, supporting the hypothesis that the long path-length phenotype of the mK lines was dominant. It was not possible to determine whether the forR allele was selected for in each of the mK lines, since complementation and deficiency analysis cannot be applied to dominant phenotypes. An alternative approach, segregation analysis, was also not feasible for two reasons. First, the frequencies of forR alleles (if they were indeed selected for in the mK lines) were not fixed in these lines. Rather it was the relative frequencies of the forR and fors alleles that likely differed between the mr and mK lines. Second, even if the mK lines were homozygous for the forR allele the high degree of overlap between the long and short path-length phenotypes would make accurate classification of larvae as forR or fors extremely difficult. Therefore the results of the crossing analysis enables us to conclude that fors was selected for in the mr lines and that the pattern of inheritance between the mr and mK lines paralleled that found in for.
DISCUSSION
Morphological polymorphisms have been shown to be affected by density-dependent selection in, for example, aphids (39), damsel flies (40), and ungulates (41). However, the importance of genetically based behavioral polymorphisms for population regulation has rarely been investigated. Populations of wild house mice, Mus musculus domesticus, exhibit two heritable alternative strategies called “aggressive or active copers” and “nonaggressive or passive copers” (42). “Active copers” have a fitness advantage in established (called K type strategist) populations, whereas “passive copers” are better at establishing new populations (called r type strategist) (42). In our study, the rover phenotype was selected under high-density (K type) conditions, whereas the sitter was selected under low-density (r type) conditions. Both studies show a clear relationship between individual behavior and population dynamics (density-dependent selection). Our study is, to our knowledge, the first to show a genetic basis to this relationship.
One theoretical framework that these studies contribute to is the Chitty hypothesis (43) (also called the self-regulation or the genetic control hypothesis) for population regulation that is based on the idea that populations can be regulated by factors intrinsic to the organism. In this scenario, directional selection is thought to act on behavioral morphs in a density-dependent manner such that the gene pool changes with population size. Thus, as in the present study, behavioral morphs are differentially selected under high (K-selected) as compared with low (r-selected) population densities.
Although foraging behavior has been studied in detail by behavioral ecologists, little is known about the heritable basis of this trait and under what conditions individual differences in foraging behavior contribute to fitness. Two exceptions to this are the rover/sitter polymorphism investigated here and the foraging behavior of zebra finches (Taeniopygia guttato), which show heritable differences in the ability to discriminate between patches of food that differ in quality (44, 45).
In our study, density-dependent selection in the laboratory resulted in differences in larval foraging behavior in three independent experiments using three unrelated base populations. Low-density conditions selected for significantly shorter paths (sitter phenotype) in the m, mr, and UU lines and longer paths (rover phenotype) in the K, mK, and CU lines. The differences in behavior between the UU and CU lines suggested that density-dependent selection was important during the larval stages of development. Results of genetic crosses between the mr and mK lines and a strain carrying a deficiency of for demonstrated that some of the differences in behavior are attributable to variation at the for locus. The present study is of particular interest because it involves natural selection in the laboratory and not artificial selection. No artificial selection pressure on larval path length was placed on our high- and low-density treatment lines. The differences in path length resulted from high- compared with low-density rearing conditions, and these conditions constituted the selection pressure.
It is difficult to make a case for the direct action of selection on a trait (46). Indeed, a number of factors could be responsible for density-dependent selection on larval path length. High-density compared with low-density cultures would differ in, for example, the distribution and concentration of food, waste products, and abiotic factors such as the moisture content of the medium. Larval density in the medium is an important biotic factor that varies in time and space (47). It is low during the early stages of medium infestation but higher during later stages. Variation in larval density also occurs within and between fruits.
Drosophila larvae spend most of their lives foraging for food (23, 48). They move through the food by extending their anterior end and retracting their posterior ends. They feed by shovelling the food (yeast) with their mouth hooks. In the absence of food, both rover and sitter larvae have long paths that do not differ from each other. Within a patch of food rover larvae exhibit significantly longer foraging trails than sitters. When food has a patchy distribution, rover larvae forage for food by moving between patches, whereas sitters forage within a patch. At high densities, larvae are required to crawl around other larvae, drowned pupae, and adults to reach a nearby food patch. In contrast, under low-density conditions, increased locomotory behavior (rover behavior) is unnecessary, since food is continuously distributed and of relatively higher quantity and quality. Thus patch size, patch quality, and interpatch distance would differ in high-density compared with low-density cultures. Indeed, the energetic cost of locomotion in Drosophila larvae is extremely high (49, 50). In addition, larvae may be forced to look elsewhere for food under high-density conditions when food is limited. These factors should influence the success of rover compared with sitter larval behavior in high-density compared with low-density conditions.
Density-dependent selection has been shown to influence a number of traits in D. melanogaster populations. Mueller and Sweet (15) and Guo et al. (21) found that larval crowding increased pupation height (the distance larvae pupate from the food in vials) and larval feeding rate (see also refs. 15, 16, 21, and 35). The for gene does not have pleiotropic effects on these traits, although phenotypic correlations between larval path length and pupation behavior have been found in natural populations (51). Pupation height and larval feeding rate are polygenic characters influenced by many genes with additive effects on the major autosomes in D. melanogaster (23, 52–55). From the perspective of larval fitness, feeding and moving are two of the most important behaviors performed in the larval period. Both larval behaviors showed a significant response to density-dependent selection from larval crowding (feeding rate, ref. 35; locomotion while foraging, this study). Indeed, K lines evolved increased larval viability at high densities relative to the r lines (19).
Density-dependent selection on the frequency of the rover and sitter phenotypes is a mechanism that could act both in the field and in the laboratory. In the laboratory, density-dependent selection likely arises from fly rearing methods. In the laboratory, 50–100 flies are placed in a bottle with culture medium, where they mate and lay eggs. Eggs are laid on the surface of the medium for several weeks. The larval period lasts 4–5 days at 25°C. The first larvae to hatch will experience low larval density conditions in contrast to larvae that hatch later. Thus larval density is low in the initial stages but high in the later stages of culture growth. Surveys of laboratory and natural populations have shown that some are polymorphic for rover/sitter behavior (23, 56). Population numbers and larval density of D. melanogaster in the field also fluctuate both temporally (e.g., over the season) and spatially (between fruits). Indeed, population numbers may fluctuate dramatically as a consequence of density-dependent regulating mechanisms (57).
It should be possible to determine whether density-dependent selection affects the rover/sitter polymorphism in field populations because this polymorphism is found as a single gene (for). However, performing behavioral assays for rover/sitter phenotypic frequencies in the field is a difficult task. This is because the character being measured is a behavioral one and phenotypic frequencies will not always be representative of the true underlying genotypic frequencies due to, for example, incomplete penetrance. Proper aging of the larva and strict control of environmental conditions (i.e., food availability) are important for minimizing the probability of misclassifying a rover as a sitter or vice versa. These types of controls are impossible to implement in the field. Thus we are trying to develop DNA probes for rover and sitter alleles to assess their frequencies in the field. These probes can then be used to address the density-dependent selection hypothesis in a variety of natural populations whose density varies temporally and/or spatially. Experimental manipulations of population densities in the field along with careful monitoring of the polymorphism should enable us to further our understanding of how density-dependent selection contributes to the maintenance of the rover/sitter polymorphism.
Acknowledgments
Special thanks to Larry Mueller for sharing his selected fly lines with us and for valuable discussion. A. J. Hilliker and J. Shore commented on an earlier version of the manuscript. M. Licht, H. Rodd, D. Hyndman, P. De Franco, M. Blows, K. Williams, R. Swartz, and T. Hansell helped with the stock maintenance and behavioral tests. L. Crozier prepared the figures. H.S.P. and K.H. were supported by Natural Sciences and Engineering Council of Canada fellowships. Research was supported by grants from the Natural Sciences and Engineering Council of Canada, the Medical Research Council of Canada, and the Human Frontier Science Program to M.B.S.
References
- 1.Sutherland W J. From Individual Behavior to Population Ecology. New York: Oxford Univ. Press; 1996. [Google Scholar]
- 2.Ford E B. Ecological Genetics. New York: Wiley; 1964. [Google Scholar]
- 3.MacArthur R M, Wilson E O. The Theory of Island Biogeography. Princeton, NJ: Princeton Univ. Press; 1967. [Google Scholar]
- 4.Berg A, Lindberg T, Gunnar K. J Anim Ecol. 1992;61:469–476. [Google Scholar]
- 5.Pacala S W, Hassell M P. Am Nat. 1991;138:584–605. [Google Scholar]
- 6.Côté I M, Gross M R. Behav Ecol Soc. 1993;33:269–274. [Google Scholar]
- 7.Arcese P, Smith J N M. J Anim Ecol. 1988;57:119–136. [Google Scholar]
- 8.Sinclair A R E. In: Ecological Concepts. Cherrett J, editor. Oxford: Blackwell; 1989. pp. 197–241. [Google Scholar]
- 9.Elliott J M. Quantitative Ecology and the Brown Trout. Oxford: Oxford Univ. Press; 1994. [Google Scholar]
- 10.Anderson W W. Am Nat. 1971;105:489–498. [Google Scholar]
- 11.Charlesworth B. Ecology. 1971;52:469–474. [Google Scholar]
- 12.Anderson W W, Arnold J. Am Nat. 1983;121:649–655. [Google Scholar]
- 13.Asmussen M A. Genetics. 1983;103:335–350. doi: 10.1093/genetics/103.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mueller L D, Ayala F J. Proc Natl Acad Sci USA. 1981;78:1303–1305. doi: 10.1073/pnas.78.2.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mueller L D, Sweet V F. Evolution (Lawrence, Kans) 1986;40:1354–1356. doi: 10.1111/j.1558-5646.1986.tb05761.x. [DOI] [PubMed] [Google Scholar]
- 16.Joshi A, Mueller L D. Evolution (Lawrence, Kans) 1988;42:1090–1093. doi: 10.1111/j.1558-5646.1988.tb02527.x. [DOI] [PubMed] [Google Scholar]
- 17.Mueller L D. Am Nat. 1988;132:786–809. [Google Scholar]
- 18.Mueller L D. Proc Natl Acad Sci USA. 1988;85:4383–4386. doi: 10.1073/pnas.85.12.4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bierbaum T J, Mueller L D, Ayala F J. Evolution (Lawrence, Kans) 1989;43:382–392. doi: 10.1111/j.1558-5646.1989.tb04234.x. [DOI] [PubMed] [Google Scholar]
- 20.Mueller L D. Evol Ecol. 1990;4:290–297. [Google Scholar]
- 21.Guo P Z, Mueller L D, Ayala F J. Proc Natl Acad Sci USA. 1991;88:10905–10906. doi: 10.1073/pnas.88.23.10905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mueller L D, Guo P Z, Ayala F J. Science. 1991;253:433–435. doi: 10.1126/science.1907401. [DOI] [PubMed] [Google Scholar]
- 23.Sokolowski M B. Behav Genet. 1980;10:291–302. doi: 10.1007/BF01067774. [DOI] [PubMed] [Google Scholar]
- 24.Pereira H S, Sokolowski M B. Proc Natl Acad Sci USA. 1993;90:5044–5046. doi: 10.1073/pnas.90.11.5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sokolowski M B, Hansell K P. Genetica (The Hague) 1992;85:205–209. doi: 10.1007/BF00132272. [DOI] [PubMed] [Google Scholar]
- 26.Sokolowski M B, Hansell R I C, Rotin D. Behav Genet. 1983;13:169–177. doi: 10.1007/BF01065665. [DOI] [PubMed] [Google Scholar]
- 27.Sokolowski M B. Anim Behav. 1982;30:1252–1253. [Google Scholar]
- 28.de Belle J S, Hilliker A J, Sokolowski M B. Genetics. 1989;123:157–164. doi: 10.1093/genetics/123.1.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Belle J S, Sokolowski M B, Hilliker A J. Genome. 1993;36:94–101. doi: 10.1139/g93-013. [DOI] [PubMed] [Google Scholar]
- 30.de Belle J S, Sokolowski M B. Heredity. 1987;59:73–83. [Google Scholar]
- 31.Lindsely D L, Zimm G G. The Genome of Drosophila melanogaster. New York: Academic; 1992. [Google Scholar]
- 32.Reuter G, Szidonya J. Chromosoma. 1983;88:277–285. doi: 10.1007/BF00292904. [DOI] [PubMed] [Google Scholar]
- 33.Szidonya J, Reuter G. Genet Res. 1988;51:197–208. [Google Scholar]
- 34.Szidonya J, Reuter G. Drosophila Inf Serv. 1988;67:77–79. [Google Scholar]
- 35.Mueller L D, Graves L, Rose M R. Funct Ecol. 1993;7:469–479. [Google Scholar]
- 36.Rose M R. Evolution (Lawrence, Kans) 1984;38:1004–1010. doi: 10.1111/j.1558-5646.1984.tb00370.x. [DOI] [PubMed] [Google Scholar]
- 37.Pereira H S, MacDonald D E, Hilliker A J, Sokolowski M B. Genetics. 1995;141:263–270. doi: 10.1093/genetics/141.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sokolowski M B. In: Techniques for the Genetic Analysis of Brain and Behavior. Goldowitz D, Wahlsten D, Winer R E, editors. Amsterdam: Elsevier; 1992. pp. 479–512. [Google Scholar]
- 39.Harrison R G. Annu Rev Ecol Syst. 1980;11:95–118. [Google Scholar]
- 40.Cordero A. J Anim Ecol. 1992;61:769–780. [Google Scholar]
- 41.Moorcroft P R, Albon S D, Permberon J M, Stevenson I R, Clutton-Brock T H. Proc R Soc London B. 1996;263:31–38. doi: 10.1098/rspb.1996.0006. [DOI] [PubMed] [Google Scholar]
- 42.Benus R F, Bohus J M, Koolhaas J M, van Oortmerssen G A. Experientia. 1991;47:1008–1019. doi: 10.1007/BF01923336. [DOI] [PubMed] [Google Scholar]
- 43.Chitty D. Can J Zool. 1960;38:99–113. [Google Scholar]
- 44.Lemon W C. Nature (London) 1991;352:153–155. [Google Scholar]
- 45.Lemon W C. Evol Ecol. 1993;7:421–428. [Google Scholar]
- 46.Clarke B. Genetics. 1975;79:101–113. [PubMed] [Google Scholar]
- 47.Ruiz-Dubreuil G, Burnet B, Connolly K, Furness P. Heredity. 1996;76:55–64. doi: 10.1038/hdy.1996.7. [DOI] [PubMed] [Google Scholar]
- 48.Green C H, Burnet B, Connolly K J. Anim Behav. 1983;31:282–291. [Google Scholar]
- 49.Berrigan D, Lighton J R B. J Exp Biol. 1993;179:245–259. doi: 10.1242/jeb.179.1.245. [DOI] [PubMed] [Google Scholar]
- 50.Berrigan D, Pepin D J. J Insect Physiol. 1995;41:329–337. [Google Scholar]
- 51.Sokolowski M B. J Insect Physiol. 1985;31:857–864. [Google Scholar]
- 52.Sokolowski M B, Bauer S J. Heredity. 1989;62:177–183. doi: 10.1038/hdy.1989.26. [DOI] [PubMed] [Google Scholar]
- 53.Sokolowski M B, Hansell R I C. Behav Genet. 1983;13:267–280. doi: 10.1007/BF01071872. [DOI] [PubMed] [Google Scholar]
- 54.Bauer S J, Sokolowski M B. J Hered. 1984;75:131–134. [Google Scholar]
- 55.Bauer S J, Sokolowski M B. Behav Genet. 1988;18:81–97. doi: 10.1007/BF01067077. [DOI] [PubMed] [Google Scholar]
- 56.Carton Y, Sokolowski M B. J Insect Behav. 1992;5:161–175. [Google Scholar]
- 57.Mueller L D, Huynh P T. Ecology. 1994;75:430–437. [Google Scholar]