Abstract
The evolutionary origin of centriole/kinetosomes, centrosomes, and other microtubule organizing centers (MTOCs), whether by direct filiation or symbiogenesis, has been controversial for >50 years. Centrioles, like mitochondria and chloroplasts, duplicate independently of the nucleus and constitute a heritable system independent of chromosomal DNA. Nucleic acids endogenous to the MTOC would support evolutionary origin by symbiogenesis. To date, most reports of centrosome-associated nucleic acids have used generalized reagents such as RNases and nucleic acid dyes. Here, from a library of RNAs extracted from isolated surf clam (Spisula solidissima) centrosomes, we describe a group of centrosome-associated transcripts representing a structurally unique intron-poor collection of nuclear genes skewed toward nucleic acid metabolism. Thus, we resolve the debate over the existence of centrosome-associated RNA (cnRNA). A subset of cnRNAs contain functional domains that are highly conserved across distant taxa, such as nucleotide polymerase motifs. In situ localization of cnRNA65, a molecule with an RNA polymerase domain, showed it is present in the intact oocyte nucleus (germinal vesicle). Its expression, therefore, precedes the appearance of γ-tubulin-containing centrosomes. At this stage, the in situ signal resembles the nucleolinus, a poorly understood organelle proposed to play a role in spindle formation. After oocyte activation and germinal vesicle breakdown, cnRNA65 persists as a cytoplasmic patch within which γ-tubulin-stained centrosomes can be seen. These observations provoke the question of whether cnRNAs and the nucleolinus serve as cytological progenitors of the centrosome and may support a symbiogenetic model for its evolution.
Keywords: centrosome, evolution, nucleolinus
The centrosome is the major microtubule organizing center (MTOC) of animal cells and many protists. It is composed of paired centrioles embedded within a pericentriolar matrix, the composition of which is poorly understood. The evolutionary origin of the centrosome–centriole system is unknown. If integrated into eukaryotes by symbiogenesis, comparable to mitochondria and chloroplasts, it is expected to have retained remnant nucleic acids of a once-independent genome (1, 2). Other investigators posit the centrosome evolved by direct filiation (3). The discovery of specific centrosomal nucleic acids, although not resolving questions of evolutionary origin, would be an indispensable step toward resolving the organelle's structure and function. The ability to isolate centrosomes from parthenogenetically activated surf clam (Spisula solidissima) oocytes in biochemical quantities (4) permitted a direct approach and led to this description of a unique family of centrosome-associated RNAs (cnRNAs).
During the course of this study, we observed that the in situ staining pattern in unactivated oocytes for one of these cnRNAs precisely resembled the shape, size, and position of the nucleolinus. The nucleolinus was described >100 years ago (5), but almost nothing is known of its function or composition. It may have at least limited capacity for independent division (6) and, like the centrosome, is dysregulated in aneuploid cells (7). It has been suggested that, after the breakdown of the oocyte nucleus [germinal vesicle (GV)] in meiosis I, the nucleolinus remains in the vicinity of the forming maturation spindle and may play a role in spindle formation (8, 9). The nucleolinus, like the GV and nucleolus, eventually disappears after oocyte activation. Any relationship to the centrosome at the molecular level is therefore noteworthy and may help us to establish a developmental link between the MTOC during cell division and intranuclear structures present before oocyte activation.
Results
Characterization of the cnRNA Library.
Preparation of the cnRNA library was described previously (10). To control for the amplification and subsequent cloning of genomic DNA contaminants, equal quantities of nonreverse-transcribed cnRNA were used in parallel experiments. No detectable PCR product or cloned inserts were obtained from these controls. For the present report, whole ooplasmic RNA was also cloned for comparison by precisely the same methods. Fifteen of these clones were selected randomly for sequencing and, as expected, 14 were identified as ribosomal RNAs in BLAST analysis (average probability value of <e−130). The remaining clone was identified as similar to “fetal Alzheimer protein” (e−72).
A total of 156 clones were generated from centrosome-associated RNA, average size 599 nt ± 20.2 (SEM). GenBank accession nos. and additional data are given in supporting information (SI) Dataset S1. Thirty-six of these overlapped with other sequences in the set, leaving a total of 120 distinct clones. This represents a maximum, because extended sequencing will likely reveal more overlaps. BLAST analysis of the cnRNA fraction revealed that only one-fourth (39) of the sequences had significant database matches. The lack of significant matches was determined to not be a general feature of Spisula sequences because of either divergence or underrepresentation of molluscan species in the databases, because BLAST analysis returned interspecific matches in 18 of the 20 previously reported Spisula sequences used as controls. Thus, the paucity of database matches for Spisula cnRNAs is particular to this set of transcripts.
This subset of 39 sequences that exhibited database identities could be divided into three categories (Table S1). The largest group (19) matched uncharacterized BAC clones, hypothetical proteins, or other uncharacterized chromosomal sequences from a wide variety of species. Sixteen of the 39 identities were related to nucleic acid or genome structure and metabolism. Examples include nucleotide polymerases (cnRNAs 65, 118), an Entamoeba snRNP (cnRNA48), clam (Venus) and pig microsatellite sequences (cnRNAs 123, 154, 226), and retroelements from a variety of species (cnRNAs 15, 93, 142, 171). The third category containing only four sequences had no clear relationship to centrosomes, the cytoskeleton, or nucleic acid metabolism.
PCR Screen for Enrichment in Centrosomes.
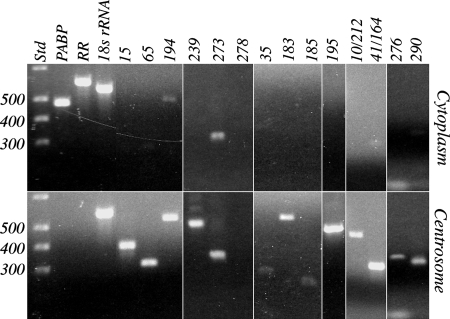
Thirty-six cnRNAs were examined by PCR for enrichment in isolated centrosomes vs. whole oocyte cytoplasm (which included centrosomes). Selection of these 36 for the screen was made arbitrarily, by using default settings in MacVector DNA analysis software to identify target sequences with compatible PCR primer pairs. PCR product sizes ranged from 582 to 139 bp. We would expect that, if cnRNAs are enriched in centrosomes, their PCR products should be generated from centrosome- but not cytoplasmic-derived templates. Primers for S. solidissima PABP and RR were used as controls to represent generalized cytoplasmic RNAs. PCR product for these two should be generated from cytoplasmic template but only minimally from centrosomal template (depending on the purity of the centrosome preparation). We also probed for Spisula 18S rRNA as an additional control for template level in the starting reaction mixture. Agarose gel images exemplifying the range of results are shown in Fig. 1. Full results are given in Table S2, along with other details of the experiment. In 7 of the 36 cases, we were unable to detect PCR product in either centrosomal or whole-cytoplasmic template samples. Almost all of these were in the low range of product sizes (220 to 155 bp), so it is possible they were undetected, at least in part, because of stoichiometrically low ethidium bromide incorporation. In every one of the remaining 29 cases where cnRNA PCR product was detected, the amount was greater in centrosomal template samples than in cytoplasmic. The difference was profound in approximately one-third of these. The results of our PCR screen indicate that cnRNAs are substantially enriched in the centrosome fraction of Spisula oocytes.
Fig. 1.
Differential expression of cnRNAs in centrosomes vs. whole oocyte cytoplasm. Equal quantities of RNA extracted from either whole oocyte cytoplasm or isolated centrosomes were reverse transcribed and used as template in PCR with primers targeting known cytoplasmic RNAs or putative cnRNAs. Conditions such as number of cycles, template input, and product size were determined empirically in preliminary experiments to yield product in the linear range of amplification. The examples shown here are outcomes from several separate experiments and were chosen to represent the full range of results observed. Full results, which can be cross-referenced to these images, are summarized in Table S2. Gel lanes in the upper half of the figure show PCR products obtained from cytoplasmic template for both controls (PABP, RR, 18S rRNA) and the cnRNAs (clones indicated across top). Corresponding lanes (Lower) show PCR products obtained from centrosome-derived template. A 100-bp DNA reference ladder is shown in the lanes marked Std of both Upper and Lower; 300- to 500-bp bands are marked on the left. Note that these gel images are taken from separate experiments, so a direct comparison horizontally across lanes is not a precise representation of product size.
Structural Analysis of Full-Length cnRNA15 and cnRNA194.
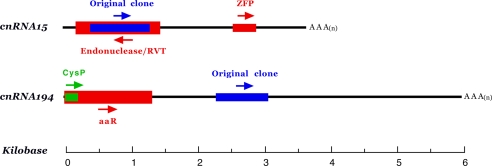
We have reported on cnRNA11, the first fully sequenced RNA extracted from isolated centrosomes (10). Although divergent across almost all of its 3.5-kb length, cnRNA11 contains a highly conserved reverse transcriptase (RVT) domain, most closely related to those found in plants and sea urchins. We are now able to describe the structural characteristics of two additional full-length cnRNA sequences; cnRNA15 and cnRNA194 (Fig. 2). As is cnRNA11, both are relatively large transcripts of 3.5 and 6.0 kb, respectively. BLAST analysis of full-length cnRNA15 revealed similarities to two recent database entries, a sea urchin (Strongylocentrotus purpuratus) endonuclease/RVT (2e−115) and a newly described Spisula zinc finger protein (5e−98). The conserved RVT domain includes a putative enzymatic active site and nucleic acid- and NTP-binding sites. Analysis of the translated cnRNA15 antisense sequence indicated similarity to presenilin genes from a variety of species. This observation is of interest, because presenilin was recently shown to localize with centrosomes and is thought to play a role in chromosome segregation (11, 12). cnRNA194 exhibits similarity to a cathepsin-like cysteine protease from the scutociliate endosymbiont of echinoids, Uronema marinum (4e−28) and the flowering plant Anthurium andraenum (2e−20). In addition, a ligand-binding domain present in a wide range of amino acid receptors was revealed, including bacterial amino acid-binding proteins, mammalian atrial natriuretic factor receptor, and glutamate receptors from sea urchins and primates. We generated monoclonal antibodies to peptides in the putative protein coding domains of both molecules, but the results of Western blot analysis were negative. We were therefore unable to determine whether cnRNAs 15 and 194 are translated in the cell.
Fig. 2.
Structural analysis of cnRNAs 15 and 194. Sense and antisense strands are defined by the orientation of the 5′ caps and 3′ polyadenylation sites. Blue bars represent the 692- and 671-nt sequences originally cloned from isolated centrosomes; arrows indicate 5′ to 3′ orientation. cnRNA15 includes a 314-nt putative protein coding domain (red) with 85% identity at the amino acid level to a newly described Spisula zinc finger protein (ZFP; GenBank accession no. AB231865). The homology domain in cnRNA15 does not correspond to the C2H2 zinc-binding motif region. In addition, a 1,128-nt domain is present in the antisense strand that exhibits 40% identity (at the amino acid level) to S. purpuratus endonuclease/RVT (GenBank accession no. XP792610). An amino acid-binding domain similar to that found in glutamate receptors and other amino acid-binding proteins from a variety of species is present in the 5′ end of cnRNA194, overlapping with a cathepsin-like cysteine protease (CysP) protease domain (green) [GenBank accession nos. EU069824 (cnRNA15) and EU069825 (cnRNA194).]
In Situ Localization of cnRNAs: a Link Between the Centrosome and Nucleolinus?
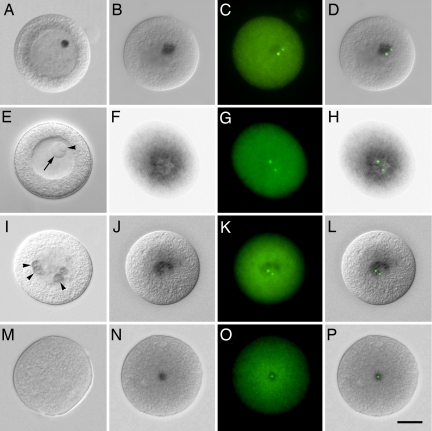
In situ hybridization of cnRNAs 15, 65, and 239 all revealed a discrete patch of signal in the cytoplasm of 7-to 10-min activated oocytes (Fig. 3B, J, and N), similar to that observed for cnRNA11 (10). When hybridized cells were colabeled with antibodies to the centrosomal marker protein, γ-tubulin, the centrosomes were seen to be embedded within or on the edge of the cnRNA hybridization patch (Fig. 3 D, L, and P). In cases where two centrosomes were visible and widely separated, one of the pair may not be associated with the hybridization patch. There was a key difference between the expression of cnRNAs 15 and 65 and that reported for cnRNA11. The latter was not observed in unactivated oocytes, whereas cnRNAs 15 and 65 clearly were. Their expression, therefore, precedes the appearance of γ-tubulin-containing centrosomes in Spisula oocytes.
Fig. 3.
In situ localization of cnRNAs. (A) Differential interference contract microscopy (DIC) image of an unactivated oocyte labeled for cnRNA65. A dense circular patch within the GV is seen, similar in size and position to the nucleolinus. Note that both the nucleolinus and nucleolus are morphologically indistinct after cells were subjected to the hybridization regimen. A DIC image of a different unfixed oocyte is therefore shown in E for comparison with A. An arrow points to the nucleolus, and the nucleolinus is indicated by an arrowhead. B shows the distribution of cnRNA65 at 7 min postactivation. A distinct patch, although slightly more diffuse than seen in GV oocytes, is visible in the cytoplasm. Centrosomes (labeled with anti-γ-tubulin antibody in C) appear within or “attached to” the cnRNA hybridization patch. (D) Overlay of B and C. F (DIC) and G (immunofluorescence) show cnRNA65 and γ-tubulin, respectively, at 24 min postactivation (overlay in H). A series is shown for cnRNA15 in I–L: I, unactivated oocyte, arrowheads highlight ring-like hybridization patterns; J and K, cnRNA15 and γ-tubulin, respectively, at 7 min postactivation; L, overlay of J and K. (M) Hybridization control using cnRNA239 sense probe. (N) cnRNA239 (using antisense probe) at 7 min postactivation; O, γ-tubulin staining in the same cell; P, overlay. (Scale bar in P: 20 μm.]
cnRNA65 in unactivated oocytes was a well defined spherical patch within the GV (Fig. 3A) and may be localized to the nucleolar subdomain known as the nucleolinus. Although the patch of label was of the precise size, shape, and placement as the nucleolinus seen in living or fixed cells (compare Fig. 3 A and E), the nucleolus and nucleolinus are not morphologically distinct in specimens after they undergo the hybridization regimen, and there are no known markers of the nucleolinus to demonstrate a colocalization. A direct connection between cnRNA65 and the nucleolinus, therefore, cannot be made at this time. However, we were able to draw an indirect link between the nucleolinus and centrosomes (and by inference, cnRNA65), as follows: After oocyte activation, the nucleolinus persists as a distinct morphological entity in the cytoplasm after dissolution of the nuclear envelope and nucleolus. The nucleolinus dissolves as the meiosis I spindle begins to develop. Time-lapse video sequences show that the position of the nucleolinus in the cytoplasm at the time of its dissolution consistently coincides with the internal meiosis I spindle pole (Movie S1). Thus, the nucleolinus coincides with the spindle pole and thereby with the centrosome. cnRNA65 staining coincides with centrosomes in the activated oocyte cytoplasm but first appears as a spherical patch in the unactivated oocyte nucleus of the precise size and shape of the nucleolinus. Although the development of specific markers will be required before a direct connection can be demonstrated, this series of overlapping links leading backward from the first meiotic spindle to the nucleolinus in unactivated oocytes is highly suggestive.
By 24-min postactivation, cnRNA65 hybridization signal takes on a donut, or ring pattern, with the meiotic spindle poles on opposite sides (Fig. 3 F–H). We cannot be certain at this time, but the transition from solid patch (as in Fig. 3D) to ring appears to occur coincidentally with formation of the first meiotic spindle, rather than preceding or being predictive of the latter.
cnRNA15 in the unactivated oocyte GV is distributed in several patches among the chromosomes. These patches sometimes appear as discrete rings (arrowheads in Fig. 3I). After activation, cnRNA15 coalesces into a more dense ring or crescent shape with the centrosomes embedded or “attached” (Fig. 3 J–L). Later, at the time when cnRNA65 had formed a ring in the cytoplasm (24 min postactivation), we no longer detected cnRNA15. The localization of cnRNAs 11, 15, 65, and 239 with γ-tubulin-stained foci confirms their association with centrosomes observed in biochemical preparations. However, we can already see from just these four molecules that cnRNA expression is complex and dynamic rather than static in nature.
Identification of Genomic DNA Corresponding to the cnRNAs.
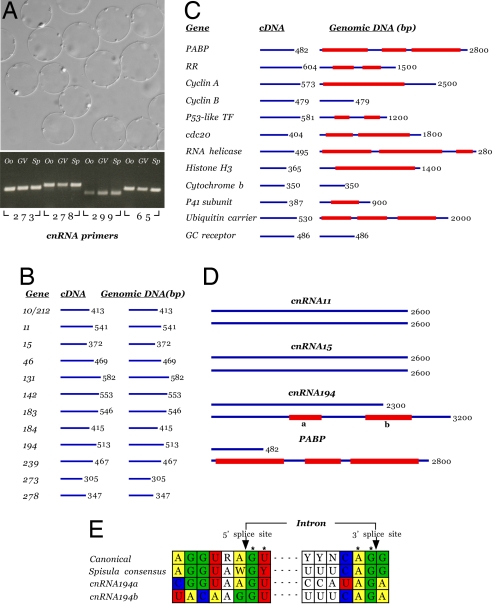
We addressed the question of whether there is DNA in the cell corresponding to cnRNAs and, if so, whether it is present in the nucleus or cytoplasm [given a history of reports on DNA associated with centrioles and basal bodies (13)]. A PCR approach was taken by using as template genomic DNA extracted from sperm, whole oocytes, or isolated GVs. Our results were unclear in only 4 of 30 cases, the PCR product appearing as a smear or series of small faint bands on agarose gels. In the remaining 26 cases, we found that cnRNA sequences were present in genomic DNA extracted from oocytes and sperm and isolated GVs in virtually equal quantities (Fig. 4A). Unexpectedly, the genomic PCR products were in every case identical in size to their counterparts generated from cDNA. That is, they encompassed no introns. We examined the possibility that the targeted sequences were by chance too small to include introns by repeating the experiment with primers for a panel of characterized cytoplasmic gene products. The predicted product sizes for our controls were similar to those of the cnRNA PCR products (460 vs. 482 bp, respectively). Our results indicate that the lack of introns was not due to target sequence size (Fig. 4 B and C). The amplified control products encompassed an average of two introns in genomic DNA in 9 of 12 cases. Sequencing confirmed the identity of the PCR products and revealed conserved eukaryotic splice sites at the putative intron/exon borders (Fig. 4E). Thus, a PCR protocol revealing multiple introns in 75% of control sequences failed to discover a single intron in similarly sized regions of all 26 cnRNA genes examined.
Fig. 4.
Screen of genomic DNA for cnRNA sequences. (A Lower) Results of a PCR screen for cnRNAs 273, 278, 299, and 65 in genomic DNA extracted from oocytes (Oo), sperm (Sp), and isolated GV. (Upper) Isolated GVs from which genomic DNA was extracted are shown in the DIC micrograph. In addition to the four examples shown, DNA corresponding to 22 other cnRNAs was demonstrated to be present in all three genomic preparations. However, introns were not detected in any of the 26 cases examined. These results are diagrammed for 12 cnRNAs with amplification products of 300 bp or more (average = 460 bp) in B. The results using 12 control genes with similar PCR product sizes in cDNA (average = 482 bp) are illustrated in C. Multiple introns are revealed with canonical eukaryotic splice sequences (“Spisula consensus” in E). In subsequent analysis, cnRNAs 11 and 15 were found to be intronless >2.6-kb regions (D). Two small sequences (359 and 562 bp) were discovered within a 3.2-kb region of cnRNA194 genomic DNA (GenBank accession no. EU078900) that were not present in cDNA. (E) Sequence analysis indicates these are bona fide introns with conserved splice sites, including the invariant 5′-GU and 3′ AG residues (R = A or G; Y = C or U; W = A or T; N = any nucleotide). Results for PABP are redrawn to scale in D to highlight the relative paucity of intronic sequence in the cnRNAs vs. control Spisula sequences.
To be sure we did not overlook cnRNA introns in our initial analysis, the full-length sequences now available for cnRNAs 11, 15, and 194 were used to target extended regions in genomic DNA. For both cnRNA11 and -15, introns were still not revealed, even in a 2.6-kb product (Fig. 4D). We were able to discover two small introns of 359 and 562 bp within a 3.2-kb region of cnRNA194. Both contained likely splice sites, including the invariant GU and AG at the 5′ and 3′ borders of the putative intron, but they otherwise varied from both the eukaryotic canonical sequence and the Spisula consensus generated from our analysis of the control cytoplasmic sequences. Together, these results demonstrate a quantitative difference between the cnRNAs and other Spisula genes, which can be summarized as follows: A PCR scan encompassing 5,736 nt of control Spisula genes resulted in the capture of 13,337 intronic nucleotides, for a ratio of 1 to 2.32. In contrast, a search through 10,667 cnRNA nucleotides yielded only 920 intronic nucleotides for a ratio of 1 to 0.09. Nuclear DNA corresponding to the cnRNAs has an exceedingly low intronic content, making it likely they evolved under a different set of selection pressures.
Discussion
More than 50 years after “pentosenucleic acid” was reported to be in the polar regions of the isolated sea urchin mitotic apparatus (14), it is now clear that centrosomes (and mitotic spindles) do contain RNA. The early evidence for centrosomal RNA was largely indirect, but these studies (reviewed in refs. 2 and 13) have now been bolstered by this and other recent reports identifying specific sequences, using in situ localization and involving controlled functional analyses (10, 15–17). Some of the RNAs described in these recent studies are known cytoplasmic transcripts targeted to centrosomes for distribution to other parts of the cell, and their corresponding genomic sequences all exhibit the intron/exon structure typical of animal genomes. Others appear more specifically associated with the centrosome and mitotic apparatus. The cnRNAs described in this article and an earlier report (10) coisolate with centrosomes, are biochemically enriched in the centrosome fraction of cells, localize with the centrosome in situ, and exhibit structural features at variance with other previously characterized Spisula genes. Moreover, one of these molecules may colocalize with the nucleolinus, an enigmatic organelle that appears to herald the position of the first meiotic spindle pole. On this matter and broad questions of cnRNA function, we consider our observations to be only a starting point but one from which we may now rationally target individual molecules with sequence-specific reagents.
We can envision only four possible sources for these cnRNAs. The first three are: (i) artifact from cytoplasmic transcripts adhering to centrosomes during cell fractionation, (ii) artifact from contamination of isolated centrosome preparations, and (iii) artifact derived from a contemporary viral infection of oocytes (with viral particles aggregated at the centrosome after microtubular transport). The first two possibilities are eliminated by the combined results of in situ localizations, BLAST analysis, and the presence of intron-poor nuclear genes in both oocytes and sperm. These same observations make it unlikely that cnRNAs are derived from a contemporary viral infection of oocytes, especially because they resemble no known viruses any more than they do bacterial, plant, or mammalian genes. That cnRNA194 present in nuclear DNA contains introns, albeit meager ones, also warrants against a contemporary viral source (although viral genomes do contain introns, they are extremely rare). The data support the fourth possibility as being most likely: (iv) cnRNAs represent a class or family of molecules truly associated with the centrosome.
The question of whether cnRNAs represent an organellar genome or are the original source of elements integrated into the nuclear genome must remain the subject of debate for now. Our observations and other recent discoveries suggest that a broader perspective may be useful in considering this riddle. The preconceived notion of a centrosomal genome seems to be that it should exist as a distinct locus in the middle of the organelle, perhaps contained within the centrioles, and that it should be tightly tethered and stationary. We suggest this is a poor model on which to base a hypothesis, a model almost without precedent in living systems. Even the eukaryotic genome in its membrane-bounded compartment fails to fit this description. It exists interchangeably as dispersed euchromatin and condensed heterochromatin, depending on transcriptional activity, differentiation state, phase of the cell cycle, and moment-to-moment physiological conditions. The eukaryotic genome is sometimes anchored and sometimes translocated within the intact nucleus. At other times, it is transported around the cytoplasm in highly regulated patterns with the mitotic apparatus. cnRNAs are likewise dynamic, sometimes existing as a discrete patch associated with (or perhaps antecedent to) the centrosome, at other times transported around the cytoplasm in patterns associated with the mitotic apparatus. One point of certainty, as attested to by 50 years of debate is that, if cnRNAs are derived from an organellar genome, we should expect a conclusion to be reached not in a concise hypothesis testing but by the accumulation of functional data and knowledge of what the sequences encode and how they are related to intra- and interspecific genes. This was no less true for nucleic acids found in mitochondria, first discovered in 1924 (18) but not accorded “genome status” until almost 50 years later.
Materials and Methods
Source and Handling of Oocytes.
Gravid S. solidissima were obtained from the Marine Resources Center at the Marine Biological Laboratory. Gametes were collected by dissection. Oocytes were rinsed several times in filtered sea water and activated with 0.14 vol of 0.5 M KCl. Centrosomes were isolated from activated oocytes as described (4, 19, 20). These centrosomes are functionally competent to form asters, undergo centriole duplication, and have been extensively characterized. Unactivated oocyte GVs used in PCR screens of genomic DNA were isolated according to methods reported by Maul and Avdalovic (21). Oocytes for in situ hybridization were fixed for 2 hours in 4% paraformaldehyde in 3-(N-morpholino) propanesulfonic acid (MOPS) buffer (40 mM MOPS, 0.5 M NaCl, 10 mM sodium acetate, 1 mM EDTA, pH 7.4) at 4°C. They were then washed by settling and resuspension twice in MOPS buffer and once in 0.5 M NaCl, dehydrated in a graded series of ethanols to 70% and stored at −20°C in 70% ethanol until use. Methods for in situ hybridization and immunofluorescence have been detailed (10). Briefly, samples were hybridized with digoxigenin-labeled RNA probes for 3 days at temperatures and probe concentrations determined to be optimal for each probe and target. The alkaline phosphatase reaction product was visualized after development for 3 days at 4°C with gentle rocking.
General Molecular.
RNA was extracted from the peak sucrose density gradient fraction of isolated centrosome preparations by using Qiagen RNeasy reagents, reverse-transcribed (Retroscript, Ambion), and amplified by random PCR using primers and methods described by Froussard (22) and Von Eggeling and Spielvogel (23). PCR products were blunt-cloned into PCR-Script plasmid (Stratagene). Transformation resulted in the growth of 315 separate bacterial colonies. All 315 were numbered in the order they were sampled and assayed for insert-containing plasmids. Those containing inserts were sequenced and subcloned.
Equal quantities of total RNA isolated from whole oocyte lysate and RNA isolated from peak centrosome fractions were reverse-transcribed and used as PCR template to test for enrichment of putative cnRNAs. Internal controls, primer pairs for known Spisula cytoplasmic RNAs, included poly(A)-binding protein, ribonucleotide reductase, and 18S rRNA. PCR conditions were such as determined in preliminary experiments to generate reaction product in the linear range of yield for putative cnRNAs: 20-ng template; small amplification products (≈150–600 bp); 30 cycles of 94°C × 30 sec, 55°C × 30 sec, 72°C × 1 min. PCR primers used in this study are listed in Table S2.
Full-length sequence data for cnRNAs 15 and 194 were obtained by RACE by using Invitrogen Gene Racer reagents. Accelrys MacVector software was used for ORF and other structural analyses. Databases accessed for similarity searches included nonredundant eukaryotic, prokaryotic, and viral nucleotide (via BLASTn, BLASTx, and tBLASTx programs); dbEST (via tBLASTx), human genome (BLASTn); human RefSeq protein (BLASTx); mouse genome (BLASTn); mouse RefSeq protein (BLASTx); Zebrafish mRNA and reference proteins (BLASTn, BLASTx); nematode mRNA and protein (BLASTn, BLASTx); plant (Triticum and Arabidopsis) DNA and protein, respectively (BLASTn, BLASTx); and insect (Drosophila), fungal (all), malarial (Plasmodium falciparum and Plasmodium yoelii), and microbial (all) DNA and protein databases (via BLASTn and BLASTx).
Supplementary Material
Acknowledgments.
We thank those who supported our efforts in the wake of Hurricane Katrina. Our laboratory was resurrected in part from the dining room table of Bruce and Sharon Waddell (Slidell, LA). We thank our colleagues in the Department of Cell and Molecular Biology at Tulane University, Drs. Carol Burdsal, David Mullin, Peter Cserjesi, and Ken Muneoka, for lending us bench space and facilities from October, 2005 through May, 2006. Dr. Robert Palazzo (Reusselear Polytechnic Institute, Troy, NY) supplied the isolated centrosomes from which RNA was originally extracted and many insightful thoughts and comments during the course of our studies. Dr. Gloria Giarratano assisted in the recloning of our cnRNA library. This work was supported by National Institutes of Health Grant GM075163 and post-Katrina emergency recovery funds from the Society for Developmental Biology.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. are listed in Dataset S1).
This article contains supporting information online at www.pnas.org/cgi/content/full/0802293105/DCSupplemental.
References
- 1.Chapman MJ, Dolan MF, Margulis L. Centrioles and kinetosomes: Form, function, and evolution. Q Rev Biol. 2000;75:409–429. doi: 10.1086/393621. [DOI] [PubMed] [Google Scholar]
- 2.Chapman MJ, Alliegro MC. A symbiogenetic basis for the centrosome? Symbiosis. 2007;44:23–32. [Google Scholar]
- 3.Bornens M, Azimzadeh J. Origins and Evolution of Eukaryotic Endomembranes and Cytoskeleton. In: Jekely G, editor. Austin, TX: Landes Bioscience; 2007. [Google Scholar]
- 4.Palazzo RE, Vogel JM. Isolation of centrosomes from Spisula solidissima oocytes. Methods Cell Biol. 1999;61:35–56. doi: 10.1016/s0091-679x(08)61974-3. [DOI] [PubMed] [Google Scholar]
- 5.Montgomery TH. Comparative cytological studies, with especial regard to the morphology of the nucleolus. J Morph. 1899;15:265. [Google Scholar]
- 6.Carleton HM. Observations on an intra-nucleolar body in columnar epithelium cells of the intestine. Q J Microsc Sci. 1920;64:329–341. [Google Scholar]
- 7.Love R, Walsh JR. Nucleolinar morphology in normal diploid, neoplastic, and aneuploid cells in vitro. Cancer Res. 1970;30:990–997. [PubMed] [Google Scholar]
- 8.Allen RD. Fertilization and artificial activation in the egg of the surf calm, Spisula solidissima. Biol Bull. 1953;105:213–239. [Google Scholar]
- 9.Allen RD. The role of the nucleolus in spindle formation. Biol Bull. 1951;101:214. [Google Scholar]
- 10.Alliegro MC, Alliegro MA, Palazzo RE. Centrosome-associated RNA in surf clam oocytes. Proc Natl Acad Sci USA. 2006;103:9034–9038. doi: 10.1073/pnas.0602859103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li J, Xu M, Zhou H, Ma J, Potter H. Alzheimer presenilins in the nuclear membrane, interphase kinetochores, and centrosomes suggest a role in chromosome segregation. Cell. 1997;90:917–927. doi: 10.1016/s0092-8674(00)80356-6. [DOI] [PubMed] [Google Scholar]
- 12.Nizzari M, et al. Amyloid precursor protein and presenilin1 interact with the adaptor GRB2 and modulate ERK 1,2 signaling. J Biol Chem. 2007;282:13833–13844. doi: 10.1074/jbc.M610146200. [DOI] [PubMed] [Google Scholar]
- 13.Marshall WF, Rosenbaum JL. Are there nucleic acids in the centrosome? Curr Top Dev Biol. 2000;49:187–205. doi: 10.1016/s0070-2153(99)49009-x. [DOI] [PubMed] [Google Scholar]
- 14.Mazia D. The organization of the mitotic apparatus. Symp Soc Exp Biol. 1955;9:335–357. [Google Scholar]
- 15.Groisman I, et al. CPEB, maskin, and cyclin B1 mRNA at the mitotic apparatus: implications for local translational control of cell division. Cell. 2000;103:435–447. doi: 10.1016/s0092-8674(00)00135-5. [DOI] [PubMed] [Google Scholar]
- 16.Lambert JD, Nagy LM. Asymmetric inheritance of centrosomally-localized mRNAs during embryonic cleavages. Nature. 2002;420:682–686. doi: 10.1038/nature01241. [DOI] [PubMed] [Google Scholar]
- 17.Blower MD, Nachury M, Heald R, Weis K. A Rae1-containing ribonucleoprotein complex is required for mitotic spindle assembly. Cell. 2005;121:223–234. doi: 10.1016/j.cell.2005.02.016. [DOI] [PubMed] [Google Scholar]
- 18.Bresslau E, Scremin L. Die kerne der trypanosomen und ihr verhalten zur nuclealreaktion. Archiv Protistenkunde. 1924;48:509–515. [Google Scholar]
- 19.Schnackenberg BJ, Palazzo RE. Identification and function of the centrosome centromatrix. Biol Cell. 1999;91:429–438. [PubMed] [Google Scholar]
- 20.Schnackenberg BJ, Hull DR, Balczon RD, Palazzo RE. Reconstruction of microtubule potential in centrosomes isolated from Spisula solidissima oocytes. J Cell Sci. 2000;113:943–953. doi: 10.1242/jcs.113.6.943. [DOI] [PubMed] [Google Scholar]
- 21.Maul GG, Avdalovic N. Nuclear envelope proteins from Spisula solidissima germinal vesicles. Exp Cell Res. 1980;130:229–240. doi: 10.1016/0014-4827(80)90059-2. [DOI] [PubMed] [Google Scholar]
- 22.Froussard P. A random-PCR method (rPCR) to construct whole cDNA library from low amounts of RNA. Nucleic Acids Res. 1992;20:2900. doi: 10.1093/nar/20.11.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Von Eggeling F, Spielvogel H. Applications of random PCR. Cell Mol Biol. 1995;41:653–670. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.