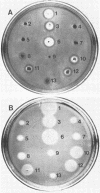
Abstract
The RP4::mini-Mu plasmid pULB113, transferred from Escherichia coli strain MXR, was stable and transfer proficient in Erwinia amylovora strain EA303, E. carotovora subsp. atroseptica strain ECA12, E. carotovora subsp. carotovora strain ECC193, and E. chrysanthemi strain EC183. The plasmid mobilized an array of Erwinia sp. chromosomal markers (E. amylovora: his+,ilv+,rbs+,ser+,thr+;E. chrysanthemi:arg+,his+,ilv+,leu+; E. carotovora subsp. atroseptica: arg+,gua+,leu+,lys+,pur+,trp+; E. carotovora subsp. carotovora: arg+,gua+,leu+,lys+,out+[export of enzymes],pur+,trp+), suggesting random interactions of the plasmid with the chromosomes. In E. carotovora subsp. carotovora, pULB113-mediated two-factor crosses revealed linkage between three auxotrophic markers and the out loci. The export of pectate lyase, polygalacturonase, and cellulase and the maceration of potato tuber tissue occurred with Out+, but not Out-, strains of E. carotovora subsp. carotovora, indicating the importance of enzyme export in plant tissue maceration. Erwinia sp. donors harboring pULB113 complemented mutations in various biosynthetic and catabolic genes (arg, gal, his, leu, met, pro, pur, thy) in Escherichia coli recA strains. Escherichia coli transconjugants harbored pULB113 primes as indicated by the cotransfer of Erwinia genes and pULB113 markers and a change in plasmid mass. Moreover, the PstI and SmaI cleavage patterns of selected pULB113 primes were different from those of pULB113. pULB113 primes carried DNA insertions ranging from 3 to about 160 kilobases. These findings indicate that pULB113 is useful for in vivo gene cloning and genetic analysis of various enterobacterial phytopathogens.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andro T., Chambost J. P., Kotoujansky A., Cattaneo J., Bertheau Y., Barras F., Van Gijsegem F., Coleno A. Mutants of Erwinia chrysanthemi defective in secretion of pectinase and cellulase. J Bacteriol. 1984 Dec;160(3):1199–1203. doi: 10.1128/jb.160.3.1199-1203.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann B. J. Linkage map of Escherichia coli K-12, edition 7. Microbiol Rev. 1983 Jun;47(2):180–230. doi: 10.1128/mr.47.2.180-230.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A. K. Acceptance by Erwinia spp. of R plasmid R68.45 and its ability to mobilize the chromosome of Erwinia chrysanthemi. J Bacteriol. 1980 Apr;142(1):111–119. doi: 10.1128/jb.142.1.111-119.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A. K., Brown M. A. Generalized transduction in the enterobacterial phytopathogen Erwinia chrysanthemi. J Bacteriol. 1980 Sep;143(3):1444–1449. doi: 10.1128/jb.143.3.1444-1449.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A. K., Starr M. P. Donor strains of the soft-rot bacterium Erwinia chrysanthemi and conjugational transfer of the pectolytic capacity. J Bacteriol. 1977 Dec;132(3):862–869. doi: 10.1128/jb.132.3.862-869.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A. K., Starr M. P. Gene transmission among strains of Erwinia amylovora. J Bacteriol. 1973 Dec;116(3):1100–1106. doi: 10.1128/jb.116.3.1100-1106.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee A. K., Thurn K. K., Feese D. A. Tn5-Induced Mutations in the Enterobacterial Phytopathogen Erwinia chrysanthemi. Appl Environ Microbiol. 1983 Feb;45(2):644–650. doi: 10.1128/aem.45.2.644-650.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collmer A., Whalen C. H., Beer S. V., Bateman D. F. An exo-poly-alpha-D-galacturonosidase implicated in the regulation of extracellular pectate lyase production in Erwinia chrysanthemi. J Bacteriol. 1982 Feb;149(2):626–634. doi: 10.1128/jb.149.2.626-634.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currier T. C., Morgan M. K. Direct DNA repeat in plasmid R68.45 is associated with deletion formation and concomitant loss of chromosome mobilization ability. J Bacteriol. 1982 Apr;150(1):251–259. doi: 10.1128/jb.150.1.251-259.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faelen M., Resibois A., Toussaint A. Mini-mu: an insertion element derived from temperate phage mu-1. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):1169–1177. doi: 10.1101/sqb.1979.043.01.132. [DOI] [PubMed] [Google Scholar]
- Hirsch P. R., Beringer J. E. A physical map of pPH1JI and pJB4JI. Plasmid. 1984 Sep;12(2):139–141. doi: 10.1016/0147-619x(84)90059-3. [DOI] [PubMed] [Google Scholar]
- Hohn B., Collins J. A small cosmid for efficient cloning of large DNA fragments. Gene. 1980 Nov;11(3-4):291–298. doi: 10.1016/0378-1119(80)90069-4. [DOI] [PubMed] [Google Scholar]
- Holloway B. W. Plasmids that mobilize bacterial chromosome. Plasmid. 1979 Jan;2(1):1–19. doi: 10.1016/0147-619x(79)90002-7. [DOI] [PubMed] [Google Scholar]
- Kado C. I., Liu S. T. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981 Mar;145(3):1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotoujansky A., Lemattre M., Boistard P. Utilization of a thermosensitive episome bearing transposon TN10 to isolate Hfr donor strains of Erwinia carotovora subsp. chrysanthemi. J Bacteriol. 1982 Apr;150(1):122–131. doi: 10.1128/jb.150.1.122-131.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Lejeune P., Mergeay M., Van Gijsegem F., Faelen M., Gerits J., Toussaint A. Chromosome transfer and R-prime plasmid formation mediated by plasmid pULB113 (RP4::mini-Mu) in Alcaligenes eutrophus CH34 and Pseudomonas fluorescens 6.2. J Bacteriol. 1983 Sep;155(3):1015–1026. doi: 10.1128/jb.155.3.1015-1026.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandels M., Andreotti R., Roche C. Measurement of saccharifying cellulase. Biotechnol Bioeng Symp. 1976;(6):21–33. [PubMed] [Google Scholar]
- Pugashetti B. K., Chatterjee A. K., Starr M. P. Isolation and characterization of Hfr strains of Erwinia amylovora. Can J Microbiol. 1978 Apr;24(4):448–454. doi: 10.1139/m78-074. [DOI] [PubMed] [Google Scholar]
- Pugashetti B. K., Starr M. P. Conjugational transfer of genes determining plant virulence in Erwinia amylovora. J Bacteriol. 1975 May;122(2):485–491. doi: 10.1128/jb.122.2.485-491.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riess G., Masepohl B., Puehler A. Analysis of IS21-mediated mobilization of plasmid pACYC184 by R68.45 in Escherichia coli. Plasmid. 1983 Sep;10(2):111–118. doi: 10.1016/0147-619x(83)90063-x. [DOI] [PubMed] [Google Scholar]
- Riley M., O'Reilly C., McConnell D. Physical map of Salmonella typhimurium LT2 DNA in the vicinity of the proA gene. J Bacteriol. 1984 Feb;157(2):655–657. doi: 10.1128/jb.157.2.655-657.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoonejans E., Toussaint A. Utilization of plasmid pULB113 (RP4::mini-Mu) to construct a linkage map of Erwinia carotovora subsp. chrysanthemi. J Bacteriol. 1983 Jun;154(3):1489–1492. doi: 10.1128/jb.154.3.1489-1492.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeberg A. H., Wiedemann B. Transfer of the chromosomal bla gene from Enterobacter cloacae to Escherichia coli by RP4::mini-Mu. J Bacteriol. 1984 Jan;157(1):89–94. doi: 10.1128/jb.157.1.89-94.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprenger G. A., Lengeler J. W. L-Sorbose metabolism in Klebsiella pneumoniae and Sor+ derivatives of Escherichia coli K-12 and chemotaxis toward sorbose. J Bacteriol. 1984 Jan;157(1):39–45. doi: 10.1128/jb.157.1.39-45.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr M. P., Chatterjee A. K., Starr P. B., Buchanan G. E. Enzymatic degradation of polygalacturonic acid by Yersinia and Klebsiella species in relation to clinical laboratory procedures. J Clin Microbiol. 1977 Oct;6(4):379–386. doi: 10.1128/jcm.6.4.379-386.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teather R. M., Wood P. J. Use of Congo red-polysaccharide interactions in enumeration and characterization of cellulolytic bacteria from the bovine rumen. Appl Environ Microbiol. 1982 Apr;43(4):777–780. doi: 10.1128/aem.43.4.777-780.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C. M. Molecular genetics of broad host range plasmid RK2. Plasmid. 1981 Jan;5(1):10–19. doi: 10.1016/0147-619x(81)90074-3. [DOI] [PubMed] [Google Scholar]
- Van Gijsegem F., Toussaint A. Chromosome transfer and R-prime formation by an RP4::mini-Mu derivative in Escherichia coli, Salmonella typhimurium, Klebsiella pneumoniae, and Proteus mirabilis. Plasmid. 1982 Jan;7(1):30–44. doi: 10.1016/0147-619x(82)90024-5. [DOI] [PubMed] [Google Scholar]
- Van Gijsegem F., Toussaint A. In vivo cloning of Erwinia carotovora genes involved in the catabolism of hexuronates. J Bacteriol. 1983 Jun;154(3):1227–1235. doi: 10.1128/jb.154.3.1227-1235.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willetts N. S., Crowther C., Holloway B. W. The insertion sequence IS21 of R68.45 and the molecular basis for mobilization of the bacterial chromosome. Plasmid. 1981 Jul;6(1):30–52. doi: 10.1016/0147-619x(81)90052-4. [DOI] [PubMed] [Google Scholar]
- Zink R. T., Kemble R. J., Chatterjee A. K. Transposon Tn5 mutagenesis in Erwinia carotovora subsp. carotovora and E. carotovora subsp. atroseptica. J Bacteriol. 1984 Mar;157(3):809–814. doi: 10.1128/jb.157.3.809-814.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]