Abstract
The involvement of the antioxidant enzymes superoxide dismutase (SOD), catalase (CAT), and glutathione peroxidase in radiobiological processes has been described at the enzyme activity level. We irradiated radiation-resistant (RR) and radiation-sensitive (RS) mice and studied antioxidant enzymes at the transcriptional and activity level. In addition, aromatic hydroxylation and lipid peroxidation parameters were determined to study radiation resistance at the oxidation level. RS BALB/c/J Him mice and RR C3H He/Him mice were whole-body-irradiated with x-rays at 2, 4, and 6 Gy and killed 5, 15, and 30 min after irradiation. mRNA was isolated from liver and hybridized with probes for antioxidant enzymes and β-actin as a housekeeping gene control. Antioxidant enzyme activities were determined by standard assays. Parameters for aromatic hydroxylation (o-tyrosine) and lipid peroxidation (malondialdehyde) were determined by HPLC methods. Antioxidant transcription was unchanged in contrast to antioxidant activities; SOD and CAT activities were elevated within 15 min in RR animals but not in RS mice, at all doses studied. Glutathione peroxidase activity was not different between RR and RS mice and was only moderately elevated after irradiation. No significant differences were found between RR and RS animals at the oxidation level, although a radiation dose-dependent increase of oxidation products was detected in both groups. We found that ionizing irradiation led to increased antioxidant activity only minutes after irradiation in the absence of increased transcription of these antioxidant enzymes. RR animals show higher antioxidant enzyme activities than do RS mice, but oxidation products are comparable in RS and RR mice. As unchanged transcription of antioxidant enzymes could not have been responsible for the increased antioxidant enzyme activities, preformed antioxidant enzymes should have been released by the irradiation process. This would be in agreement with previous studies of preformed, stored SOD. The finding of higher SOD and CAT activities in RR than in RS animals could point to a role for these antioxidant enzymes for the process of radiation sensitivity.
Keywords: superoxide dismutase, catalase, glutathione peroxidase, malondialdehyde, lipid peroxidation
The transcription, expression, and increased activity of antioxidant enzymes (AOEs) after ionizing irradiation are well known. However, the biological meaning, i.e., the role of the AOEs superoxide dismutase (SOD), catalase (CAT), and glutathione peroxidase (GSH-PX) for radioresistance in biological systems, still remains unclear and equivocal (1–9). According to current opinion, the AOE response to ionizing irradiation is part of an SOS reaction in the bacterial system that has been described in detail (10, 11). In this reaction, active oxygen species generated by the radiation process lead to the induction of global regulatory genes as, for example, oxyR regulon responding to hydrogen peroxide (12) or the soxRS and soxQ regulons responding to superoxide (13). These regulons are rapidly and readily induced and control a series of genes, including those encoding for AOEs (14). This biological principle was also found to be relevant in the mammalian system (15–22). Most studies of higher organisms examined the AOE response hours and days after irradiation and are, therefore, confounded by influences, such as inflammation, secondary to radiation per se. No study on early events of AOE transcription has been reported to date, to our knowledge, although it is known that as soon as 30 min after ionizing irradiation SOD activity can be significantly elevated (18). When they approached this problem, de Toledo and coworkers (7) showed by in vitro studies that mRNA levels for Cu-Zn SOD and CAT in fibroblasts were not increased 1–6 hr after ionizing irradiation at low (3.6 Gy) or high (20 Gy) doses. In contrast to other “early response genes” (15, 17), no early transcriptional data on AOEs are available in literature.
We were interested in whether early (within 5–30 min) transcription and activity of liver SOD, CAT, and GSH-PX were increased after in vivo x-ray whole-body irradiation. Furthermore, we wanted to investigate whether transcription was correlated with the activity of AOEs, and we also tried to find differences in the transcription and activity between a radiation-sensitive (RS; BALB/c) and a radiation-resistant (RR; C3H) mouse strain after ionizing irradiation (23). The determination of hydroxyl radical attack and lipid peroxidation complemented our studies to reveal differences at the lipid and protein oxidation level, particularly as they could have been modified by different AOE levels.
MATERIALS AND METHODS
Animals and Irradiation Protocol.
We used 60 female BALB/c/J Him (RS) and 60 female C3H/He Him (RR) mice (mean age of 90 ± 3 days) in the experiments. The differences in their responses to ionizing irradiation is well known and documented (23). This different radiation sensitivity between the two strains consists of different survival rates after whole-body ionizing irradiation and was confirmed in our laboratory by a pilot study. All experiments were carried out with the permission of the German Committee on Animal Experiments (Bonn). Animals were divided into an RR and an RS control group, and the other six (three RS and three RR) groups were irradiated with 2, 4, and 6 Gy. Mice were killed by neck dislocation after 5, 15, and 30 min. Each group consisted of six animals. Whole-body irradiation was performed with an x-ray tube 200 RT (C.H.F. Mueller, Bonn). We applied 200 kV, 20 mA, 0.1 mm Cu filters, and a dose rate of 2 Gy/min.
mRNA Isolation from Mouse Liver: Northern Blot and Slot Blot Analyses.
The organs were obtained at autopsy and immediately frozen in liquid nitrogen. Frozen liver samples were ground, and mRNA extraction was performed with use of the QuickPrep Micro mRNA Purification kit (Pharmacia). Subsequently, mRNA was applied to a 1.4% agarose gel after denaturation with glyoxal and dimethyl sulfoxide according to the method of McMaster and Carmichael (24) and electrophoresed at 3–4 V/cm for 2.5 hr in circulating 0.01 M phosphate buffer (pH 7.0). RNA was then transferred to a positively charged nylon membrane (Hybond N+; DuPont, catalogue no. NEF-986) by capillary blotting (25), fixed with 0.05 M sodium hydroxide for 5 min at room temperature, and equilibrated at pH 7.0 with three washes in 2× standard saline citrate.
Probes for human β-actin (ATCC 9800), SOD (ATCC 39786), CAT (ATCC 57439), and GSH-PX (ATCC 104830) were used for Northern blot and slot blot analyses.
For transfection, an aliquot of frozen competent cells (Escherichia coli HB 101) was thawed to O°C, and 5 μl of each plasmid (5 μg/100 μl) was incubated with the competent cells on ice, followed by an incubation step at 90 sec at 42°C; then the tubes were returned to the ice bath for 2 min. Subsequently, 1 ml of prewarmed (37°C) SOC medium was added and incubated for 60 min on a shaker. Then tubes were centrifuged for 10 min at 4,500 × g, and the pellet was resuspended in Luria–Bertani (LB) medium at three different dilutions and plated in LB agar medium plus ampicillin (100 μg/ml), in which they were allowed to grow overnight at 37°C.
A single bacterial colony was transferred to 4 ml of LB medium containing ampicillin, and the culture was incubated overnight at 37°C with vigorous shaking. One milliliter of the tube was inoculated in 500 ml of LB medium containing ampicillin prewarmed to 37°C in a 2-liter flask; this culture was incubated under vigorous shaking until the OD at 600 nm was 0.4.
For the isolation from the plasmids (pBRM for SOD, psP64 for CAT, and pBluescript for GSH-PX), the Plasmid Maxi Kit (Qiagen, Hilden, Germany), based upon a modified alkaline lysis procedure, was used, and then the plasmid DNA was bound to an anion-exchange resin under appropriate low salt and pH conditions. Plasmid DNA is eluted by this principle by a high salt buffer, concentrated, and delated by isopropanol precipitation.
For digestion with restriction enzymes, 3 μg of plasmid; 2 μl of SuRE/Cut buffers for restriction endonucleases (Boehringer Mannheim); 2 μl each of BamHI for SOD, EcoRI and PstI for CAT, and EcoRI and XhoI for GSH-PX (Boehringer Mannheim); and 9 μl of water were incubated for 3 hr in a thermoblock at 37°C. After the reaction was stopped, the solution was electrophoresed on a 1% agarose gel in 0.15 M Tris-borate buffer (pH 8.0) to show the length of fragments.
Using QIAquick gel extraction (Qiagen, 28704), a method with selective DNA binding properties of a silica gel membrane, the DNA probes were isolated and quantified at 260 nm.
The probes were denatured before labeling by boiling for 5 min and subsequent cooling on ice, and were labeled with fluorescein-12-dUTP using the Renaissance Random Primer Fluorescein-12 dUTP Labeling Kit (DuPont, catalogue no. NEL-203).
After fixation of bound RNA, the nylon membrane was incubated in prehybridization solution [0.25 M phosphate buffer, pH 7.2, containing 5% (wt/vol) SDS, 1 mM EDTA, and 0.5% blocking reagent (from DuPont, catalogue no. NEL-203)] for 12 hr at 65°C in a hybridization oven. The blots were hybridized overnight at 65°C with each of the labeled probes (50 ng/ml of the prehybridization buffer).
After hybridization, nonspecifically bound material was removed by posthybridization washes with 0.5× and 0.1× prehybridization buffer (10 min each at 65°C). The 0.5× and 0.1× prehybridization buffers were brought to 65°C before use, and the second wash was performed at room temperature.
Hybridized blots were blocked with 0.5% blocking reagent in 0.1 M Tris⋅HCl (pH 7.2) and 0.15 M NaCl for 1 hr at room temperature. Membranes were then incubated with anti-fluorescein horseradish peroxidase antibody (DuPont, catalogue no. NEL-203) at a 1:1,000 dilution in the solution given above for 1 hr under constant shaking.
Membranes were washed four times for 5 min each in the solution given above.
The Nucleic Acid Chemiluminescence Reagent (DuPont, catalogue no. NEL-201) was added to the membranes and incubated for 1 min. Excess detection reagent was removed by the use of filter paper, and the membrane was placed in Sarawrap paper and exposed to autoradiography Reflection films (DuPont, catalogue no. NEF-496) for 15 min at room temperature.
Slot blot analysis was performed according to the method of White and Bancroft (26). This procedure consisted of placing 2 μg of total RNA (dissolved) in 10 μl of double-distilled water mixed with 500 μl of 100% formamide, 162 μl of 37% formaldehyde, and 100 μl of 10× Mops buffer. This mixture was incubated for 10 min at 65°C and then cooled on ice. Samples were placed onto the membrane by the Manifolds filtration equipment (slot blot apparatus Bio slot, Bio-Rad), and the hybridization was performed as described above.
Probes showed the expected insert lengths of 0.88 for SOD, 0.80 for CAT, and 0.15 for GSH-PX (Fig. 1).
Figure 1.
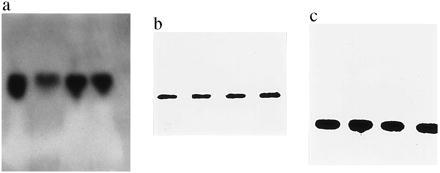
The Northern blot pattern for SOD (a) reveals a single band each in an RR unirradiated mouse (lane 1) or an RR mouse irradiated with 6 Gy (lane 2), in an RS unirradiated mouse (lane 3), and in an RS mouse irradiated with 6 Gy (lane 4). (b) Lanes 5–8 show the pattern for CAT, and lanes 9–12 represent the Northern blot pattern of GSH-PX transcription. (c) Lanes are the same as those given for SOD.
Denistometry of films was performed using the Hirschmann elscript 400 densitometer (Bonn).
Determination of Activities of Antioxidant Enzymes.
Sample preparation for the determination of liver AOEs was given in a previous paper (27).
SOD was determined by a spectrophotometrical, commercially available assay (SOD-525, Bioxytech S.A., Bonneuil/Marne, France).
CAT was determined spectrophotometrically following a standard method (28), and GSH-PX was evaluated by using a commercially available, spectrophotometrical kit (Bioxytech S.A., Bonneuil/Marne, France).
Determination of Malondialdehyde.
Malondialdehyde was determined in liver by an HPLC method using the thiobarbituric acid principle as described (29, 30). Briefly, tissue was added to 1 ml of 10% trichloroacetic acid and 50 μl of butylhydroxytoluol (2 mg/ml methanol), heated for 30 min under constant shaking to 95°C, and spun down at 5,000 × g. The trichloroacetic acid extract (150 μl) and 150 μl of thiobarbituric acid reagent (60 mg/10 ml of H2O) were heated for 30 min at 95°C. After cooling, the mixture was extracted with 0.6 ml of n-butanol, and the extract was centrifuged. For HPLC determination of malondialdehyde, we used a Lichrosphere RP18 column (Hewlett–Packard) (125 × 4 mm); the eluent was 50 mM phosphate buffer, pH 6.8/MeOH (6:4), the flow was 1 ml/min, and detection was carried out with a fluorescence detector (Jasco, Tokyo, model ModFP) 920 at the excitation of 525 nm and emission of 550 nm.
Determination of o-Tyrosine.
The determination of o-tyrosine was carried out after acid hydrolysis of liver samples by an HPLC method given in a previous paper (31, 32). Briefly, The hydrolysates were evaporated to dryness on a Pierce Reactitherme at 60°C and redissolved in distilled water. This solution was run on HPLC on a HP 1050 high performance liquid chromatograph with electrochemical detector HP 1049A (Hewlett–Packard). The column used was a Spherisorb ODS 2 (250 × 0.5 mm). For the elution, water/acetonitrile (99:1) was used in a linear gradient. The potential applied was 0.9 V.
Statistical Methods.
The ANOVA with subsequent Kruskal–Wallis test and Student’s t test was applied, and a level of P < 0.05 was considered significant.
RESULTS
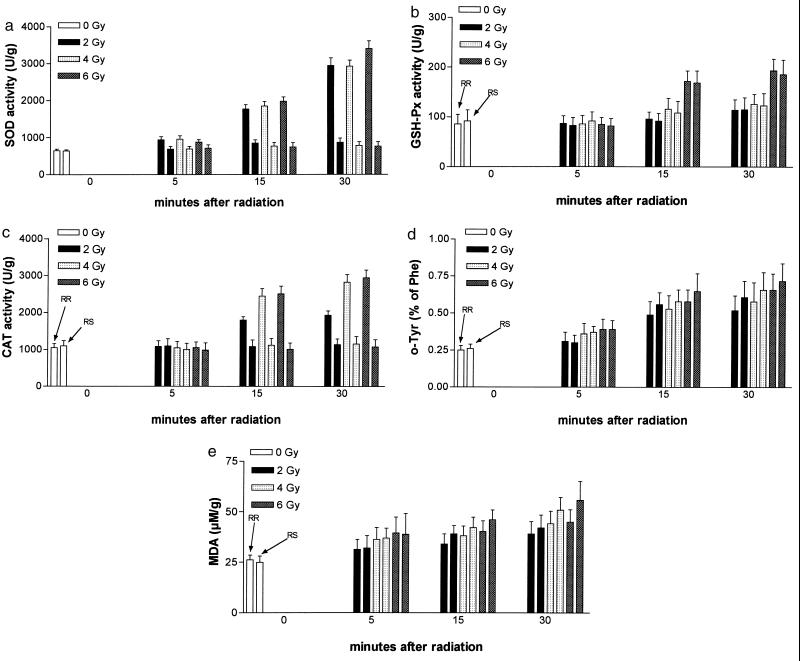
The results for SOD transcription and activity are shown in Table 1 and Fig. 2. No significant differences at the transcriptional level, normalized versus the housekeeping gene β-actin, were found between the groups studied. SOD activities, however, were significantly elevated as early as 5 min after irradiation and increased with time in both RR and RS mice. Activities in RR mice, however, were significantly higher at all time points.
Table 1.
Mean (±SD) of SOD activity and mRNA-SOD/mRNA-β-actin activity
| Dose, Gy | Mice | SOD activity, units/g
|
mRNA-SOD/mRNA-β-actin activity
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Minutes after radiation
|
Minutes after radiation
|
||||||||
| 0 | 5 | 15 | 30 | 0 | 5 | 15 | 30 | ||
| 0 | RR | 642 ± 45 | 2.26 ± 0.17 | ||||||
| RS | 636 ± 35 | 2.17 ± 0.21 | |||||||
| 2 | RR | 941 ± 86 | 1,780 ± 118 | 2,960 ± 212 | 2.12 ± 0.21 | 2.09 ± 0.32 | 2.06 ± 0.31 | ||
| RS | 685 ± 76 | 846 ± 96 | 870 ± 117 | 2.24 ± 0.31 | 2.16 ± 0.39 | 2.04 ± 0.29 | |||
| 4 | RR | 960 ± 94 | 1,860 ± 119 | 2,940 ± 167 | 2.36 ± 0.32 | 1.86 ± 0.50 | 2.16 ± 0.31 | ||
| RS | 695 ± 68 | 766 ± 102 | 780 ± 103 | 2.22 ± 0.34 | 1.96 ± 0.48 | 2.31 ± 0.40 | |||
| 6 | RR | 880 ± 67 | 1,990 ± 114 | 3,420 ± 211 | 2.16 ± 0.35 | 2.01 ± 0.43 | 2.14 ± 0.36 | ||
| RS | 712 ± 92 | 746 ± 117 | 762 ± 125 | 2.08 ± 0.39 | 2.16 ± 0.31 | 2.16 ± 0.35 | |||
Figure 2.
Results of AOE activities (a, SOD; b, GSH-PX; c, CAT) and product levels (d, o-tyrosine; e, malondialdehyde). 0, Before irradiation. The first of the corresponding pairs reflects the RR group, and the second reflects the RS group of mice.
As shown in Table 2, CAT transcription, as expressed by mRNA-CAT/mRNA-β-actin, was not different between any groups and time points studied.
Table 2.
Mean (±SD) of CAT activity and mRNA-CAT/mRNA-β-actin activity
| Dose, Gy | Mice | CAT activity, units/g
|
mRNA-CAT/mRNA-β-actin activity
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Minutes after radiation
|
Minutes after radiation
|
||||||||
| 0 | 5 | 15 | 30 | 0 | 5 | 15 | 30 | ||
| 0 | RR | 1,046 ± 112 | 1.21 ± 0.09 | ||||||
| RS | 1,100 ± 140 | 1.26 ± 0.14 | |||||||
| 2 | RR | 1,086 ± 152 | 1,807 ± 89 | 1,942 ± 117 | 1.06 ± 0.21 | 1.27 ± 0.25 | 1.24 ± 0.21 | ||
| RS | 1,094 ± 202 | 1,086 ± 176 | 1,140 ± 152 | 1.21 ± 0.31 | 1.31 ± 0.24 | 1.22 ± 0.12 | |||
| 4 | RR | 1,047 ± 172 | 2,460 ± 202 | 2,840 ± 211 | 1.09 ± 0.30 | 1.30 ± 0.41 | 1.26 ± 0.31 | ||
| RS | 1,004 ± 166 | 1,120 ± 184 | 1,160 ± 202 | 1.08 ± 0.31 | 1.33 ± 0.29 | 1.27 ± 0.22 | |||
| 6 | RR | 1,057 ± 151 | 2,520 ± 209 | 2,960 ± 213 | 0.99 ± 0.26 | 1.21 ± 0.20 | 1.37 ± 0.31 | ||
| RS | 986 ± 201 | 1,010 ± 176 | 1,076 ± 204 | 0.97 ± 0.31 | 1.09 ± 0.37 | 1.38 ± 0.28 | |||
Again, significant differences were found at the activity level (Fig. 2). No difference for CAT activity was found at 5 min after irradiation, but in RR mice, a significant rise was observed from 15 min, and this pattern was observed at all irradiation doses.
Table 3, listing the results of GSH-PX transcription (mRNA-GSH-PX/mRNA-β-actin) and activity, shows that no differences between groups at the transcriptional level were found. Unirradiated mice showed comparable activities. at no irradiation dose was a significant difference between RR and RS mice detectable, and a moderate rise in activity over time was observed at all radiation doses.
Table 3.
Mean (±SD) of GSH-PX activity and mRNA-GSH-PX/mRNA-β-actin activity
| Dose, Gy | Mice | GSH-PX activity, units/g
|
mRNA GSH-PX/mRNA-β-actin activity
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Minutes after radiation
|
Minutes after radiation
|
||||||||
| 0 | 5 | 15 | 30 | 0 | 5 | 15 | 30 | ||
| 0 | RR | 86 ± 19 | 0.41 ± 0.16 | ||||||
| RS | 92 ± 22 | 0.42 ± 0.12 | |||||||
| 2 | RR | 87 ± 15 | 96 ± 14 | 114 ± 21 | 0.42 ± 0.08 | 0.49 ± 0.02 | 0.42 ± 0.07 | ||
| RS | 83 ± 16 | 92 ± 15 | 115 ± 24 | 0.43 ± 0.12 | 0.48 ± 0.09 | 0.41 ± 0.08 | |||
| 4 | RR | 86 ± 17 | 116 ± 22 | 126 ± 20 | 0.51 ± 0.14 | 0.47 ± 0.09 | 0.51 ± 0.12 | ||
| RS | 92 ± 18 | 108 ± 24 | 123 ± 25 | 0.50 ± 0.12 | 0.51 ± 0.12 | 0.56 ± 0.14 | |||
| 6 | RR | 85 ± 14 | 172 ± 21 | 194 ± 23 | 0.46 ± 0.12 | 0.43 ± 0.08 | 0.48 ± 0.04 | ||
| RS | 82 ± 15 | 169 ± 24 | 186 ± 29 | 0.48 ± 0.13 | 0.42 ± 0.12 | 0.46 ± 0.12 | |||
The results of aromatic hydroxylation (o-tyrosine levels) and lipid peroxidation (malondialdehyde levels) are given in Table 4 and Fig. 2.
Table 4.
Mean (±SD) of o-tyrosine representing aromatic hydroxylation and malondialdehyde reflecting lipid peroxidation
| Dose, Gy | Mice |
o-tyrosine, % of phenylalanin
|
Malondialdehyde, μM/g
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Minutes after radiation
|
Minutes after radiation
|
||||||||
| 0 | 5 | 15 | 30 | 0 | 5 | 15 | 30 | ||
| 0 | RR | 0.25 ± 0.03 | 26.2 ± 2.4 | ||||||
| RS | 0.26 ± 0.03 | 25.0 ± 3.1 | |||||||
| 2 | RR | 0.31 ± 0.06 | 0.49 ± 0.09 | 0.52 ± 0.10 | 31.4 ± 4.9 | 34.1 ± 5.1 | 39.2 ± 6.1 | ||
| RS | 0.30 ± 0.05 | 0.56 ± 0.08 | 0.61 ± 0.11 | 32.1 ± 6.1 | 39.2 ± 4.1 | 42.2 ± 6.4 | |||
| 4 | RR | 0.36 ± 0.07 | 0.53 ± 0.09 | 0.58 ± 0.13 | 36.4 ± 5.9 | 38.2 ± 4.9 | 44.3 ± 6.2 | ||
| RS | 0.37 ± 0.04 | 0.58 ± 0.08 | 0.66 ± 0.12 | 37.0 ± 4.9 | 42.3 ± 5.1 | 51.0 ± 6.4 | |||
| 6 | RR | 0.39 ± 0.07 | 0.58 ± 0.08 | 0.66 ± 0.11 | 39.6 ± 7.9 | 40.4 ± 5.2 | 45.1 ± 6.2 | ||
| RS | 0.39 ± 0.06 | 0.65 ± 0.12 | 0.72 ± 0.12 | 38.9 ± 10.4 | 46.2 ± 4.8 | 56.0 ± 9.3 | |||
o-Tyrosine and malondialdehyde were not significantly different when unirradiated or irradiated RR and RS mice were compared. Irradiation, however, increased significantly and in a dose-dependent manner aromatic hydroxylation and lipid peroxidation.
DISCUSSION
No AOE tested showed increased transcription in the early phase up to 30 min after irradiation. This finding is compatible with the only relevant work on transcription of AOEs by de Toledo and coworkers (7), who did not find transcription of AOEs 1 hr after ionizing irradiation. This finding cannot be assigned to a general suppression of transcription in the early postirradiation phase as proposed by Woloschak and coworkers (18) as accumulation of specific mRNAs takes place as soon as 15 min after irradiation. Radiation is known to generate active oxygen species immediately, and in turn, one would expect that AOE as a part of the antioxidant defense would be transcribed immediately, either directly by the presence of active oxygen species or indirectly mediated by the induction of the SOS early response genes to irradiation. These early response genes are indeed expressed immediately (within minutes) after irradiation (10, 15, 17), but one wonders why AOEs, if they do play a role in the defense against ionizing radiation, are not transcribed at the early phase. We show (Table 4) that, minutes after irradiation, hydroxyl radical attack as well as lipid peroxidation has already taken place, but no increased AOE transcription was detectable.
Determining the activity of AOE, however, we found that activities of SOD were increased at a very early postirradiation period, followed by increased CAT and GSH-PX activities. This early AOE response, i.e., increased activity (transcription not tested), was published before supporting our findings (1, 16).
The posttranscriptional effects, the presence of preformed AOEs or conformational changes by irradiation or its products, generated could have led to increased AOE activity (33). In eukaryotic cells, MnSOD, an SOD isoenzyme, is preformed and stored as a precursor (34). Radiation may well have led to the activation of liver AOE in our model. On the other hand, extracellular AOEs such as extracellular SOD (35) exist, and imbibition of the liver by extracellular fluid after irradiation could have been responsible for increased enzyme activities detected. This extracellular influence is, however, highly unlikely as we could show in a parallel study that plasma levels of AOE were not increased after irradiation (unpublished results and data not shown). We were most probably detecting this activity as we used by purpose an assay for nonspecific SOD activity and not an assay for isoenzymes.
Another main goal of the study was to investigate the differences of AOE transcription in RS and RR mouse strains. Neither in the unirradiated nor in the irradiated state transcriptional could differences be found. However, when activities were considered in this context, SOD activities were significantly higher in RR animals as early as 5 min after irradiation (Table 1). CAT was significantly higher after 15 min in the RR animals, but no difference was observed for GSH-PX activities. All three AOEs were comparable in the unirradiated state. These findings suggest a role for SOD and CAT in radioresistance mechanisms, as has already been proposed.
The fact that GSH-PX did not differ between the RR and RS mouse strains in our study supports previous reports demonstrating that GSH-PX may not be important for radiation resistance (4).
Lipid peroxidation and hydroxyl radical attack were found to be in a radiation dose-dependent manner (Table 4), but no significant differences (RS had insignificantly higher oxidation products) between RR and RS groups were detectable. One might expect lower levels of oxidation products in RR animals because of the increased AOE activities; on the other hand, the influence of other oxidoreductases, such as thioredoxin (36) or metallothioneins (37), has to be taken into account and may well modulate generation of oxidation products. Again, baseline levels (unirradiated mice) did not show different levels of oxidation.
We have shown in a simple, nonsophisticated model that no increase of AOE at the transcriptional level occurred (most probably as all transcriptional activity is needed for the “emergency/SOS gene transcripts” given above) but that increased AOE response in terms of activity was detected. This phenomenon may be caused by either activation of preformed AOEs or imbibition of the irradiated liver with extracellular fluid. AOE activities were higher in the RR animals, with the mechanism for the RR/RS difference remaining unclear (but increased AOE activities were not associated with lower levels of lipid peroxidation and hydroxyl radical attack). These findings, however, support the hypothesis that SOD and CAT may be involved in the radiation resistance mechanism.
Acknowledgments
We are highly indebted to Dr. H. Rink (Department of Radiobiology, University of Bonn, Bonn) for his kind advice and support of the irradiation procedures. We are also indebted to the Red Bull Company, Salzburg, Austria, for generous support of the study.
ABBREVIATIONS
- SOD
superoxide dismutase
- CAT
catalase
- GSH-PX
glutathione peroxidase
- RR
radiation resistant
- RS
radiation sensitive
- AOE
antioxidant enzyme
References
- 1.Oberley L, St. Clair D K, Autor A P, Oberley T D. Arch Biochem Biophys. 1987;254:69–80. doi: 10.1016/0003-9861(87)90082-8. [DOI] [PubMed] [Google Scholar]
- 2.Benderitter M, Maingon P, Abadie C, Assem M, Maupoil V, Briot F, Horiot J C, Rochette L. Radiat Res. 1995;144:64–72. [PubMed] [Google Scholar]
- 3.Yamaoka K, Edamatsu R, Mori A. Free Radical Biol Med. 1991;11:299–306. doi: 10.1016/0891-5849(91)90127-o. [DOI] [PubMed] [Google Scholar]
- 4.Liebmann J, Fisher J, Lipschultz C, Kuno R, Kaufman D C. Cancer Res. 1995;55:4465–4470. [PubMed] [Google Scholar]
- 5.Jaworska A, Rosiek O. Int J Radiat Biol. 1991;60:899–906. [PubMed] [Google Scholar]
- 6.Richardson M E, Siemann M S, Siemann D W. Int J Radiat Oncol Biol Phys. 1994;29:387–392. doi: 10.1016/0360-3016(94)90295-x. [DOI] [PubMed] [Google Scholar]
- 7.de Toledo S M, Azzam E I, Gassmann M K, Mitchel R E J. Int J Radiat Biol. 1995;67:135–143. doi: 10.1080/09553009514550171. [DOI] [PubMed] [Google Scholar]
- 8.Biaglow J E, Mitchell J B, Held K. Int J Radiat Oncol Biol Phys. 1992;22:665–669. doi: 10.1016/0360-3016(92)90499-8. [DOI] [PubMed] [Google Scholar]
- 9.Riley P A. Int J Radiat Biol. 1994;65:27–33. doi: 10.1080/09553009414550041. [DOI] [PubMed] [Google Scholar]
- 10.Little J W, Mount D W. Cell. 1982;29:11–22. doi: 10.1016/0092-8674(82)90085-x. [DOI] [PubMed] [Google Scholar]
- 11.Davies K J A, Lin S W. In: Bacterial Gene Expression During Oxidative Stress. Nohl H, Esterbauer H, Rice Evans C, editors. London: Richelieu; 1994. pp. 563–579. [Google Scholar]
- 12.Christman M F, Morgan R W, Jacobson F S, Ames B N. Cell. 1985;41:753–761. doi: 10.1016/s0092-8674(85)80056-8. [DOI] [PubMed] [Google Scholar]
- 13.Tsaneva I R, Weiss B. J Bacteriol. 1990;172:4197–4205. doi: 10.1128/jb.172.8.4197-4205.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greenberg J T, Monach P A, Chou J H, Josephy P D, Demple B. Proc Natl Acad Sci USA. 1990;87:6181–6185. doi: 10.1073/pnas.87.16.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fornace A J., Jr Annu Rev Genet. 1992;26:507–526. doi: 10.1146/annurev.ge.26.120192.002451. [DOI] [PubMed] [Google Scholar]
- 16.Colburn N C. In: Biological Consequences of Oxidative Stress. Spatz L, Bloom A D, editors. New York: Oxford Univ. Press; 1992. pp. 121–137. [Google Scholar]
- 17.Vrdoljak E, Borchardt P E, Bill C A, Stephens L C, Tofilon P J. Int J Radiat Biol. 1994;66:739–746. [PubMed] [Google Scholar]
- 18.Woloschak G E, Chang-Liu C M, Shearin Jones P, Jones C A. Cancer Res. 1990;50:339–344. [PubMed] [Google Scholar]
- 19.Weichselbaum R R, Hallahan D E, Sukhatme V, Dritschillo A, Sherman M L, Kufe D W. J Natl Cancer Inst. 1991;83:480–484. doi: 10.1093/jnci/83.7.480. [DOI] [PubMed] [Google Scholar]
- 20.Holbrook N J, Fornace A J. New Biol. 1991;3:825–833. [PubMed] [Google Scholar]
- 21.Papathanasiou M A, Kerr N C K, Robbins J H, McBride O W, Alamo J R I, Barret S F, Hickson I D, Fornace A J. Mol Cell Biol. 1991;11:1009–1016. doi: 10.1128/mcb.11.2.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boothman D A, Meyers M, Fukunaka N, Lee S W. Proc Natl Acad Sci USA. 1993;90:7200–7204. doi: 10.1073/pnas.90.15.7200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roderick T H. Radiat Res. 1963;20:631–639. [PubMed] [Google Scholar]
- 24.McMaster G K, Carmichael G G. Proc Natl Acad Sci USA. 1977;74:4835–4841. doi: 10.1073/pnas.74.11.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Southern E M. J Mol Biol. 1975;98:503–511. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 26.White B A, Bancroft F C. J Biol Chem. 1982;257:8569–8575. [PubMed] [Google Scholar]
- 27.Hayn M, Kremser K, Singewald N, Nemethova M, Lubec G. Life Sci. 1996;59:537–544. doi: 10.1016/0024-3205(96)00334-7. [DOI] [PubMed] [Google Scholar]
- 28.Batist G, Mersereau W, Malashenko B A, Chiuc R C. Circulation. 1989;80:10–13. [PubMed] [Google Scholar]
- 29.Wong H Y, Knight J A, Hopfer S M, Zaharia O, Leach C N, Jr, Sunderman F W., Jr Clin Chem. 1987;33:214–220. [PubMed] [Google Scholar]
- 30.Lubec B, Hayn M, Denk W, Bauer G. Free Radical Biol Med. 1996;21:219–223. doi: 10.1016/0891-5849(96)00018-4. [DOI] [PubMed] [Google Scholar]
- 31.Kelly F, Lubec G. Pediatr Res. 1995;38:286–291. doi: 10.1203/00006450-199509000-00003. [DOI] [PubMed] [Google Scholar]
- 32.Lubec G, Weninger M, Anderson S. FASEB J. 1994;8:1166–1169. doi: 10.1096/fasebj.8.14.7958623. [DOI] [PubMed] [Google Scholar]
- 33.Harris E D. FASEB J. 1992;6:2675–2683. doi: 10.1096/fasebj.6.9.1612291. [DOI] [PubMed] [Google Scholar]
- 34.Wisp J R, Clark J C, Burhans M S, Kropp K E, Korfhagen T R, Whitsett J A. Biochim Biophys Acta. 1989;994:30–36. doi: 10.1016/0167-4838(89)90058-7. [DOI] [PubMed] [Google Scholar]
- 35.Marklund S L. Proc Natl Acad Sci USA. 1982;79:7634–7638. doi: 10.1073/pnas.79.24.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lunn C A, Pigiet V P. Int J Radiat Biol. 1987;51:29–38. doi: 10.1080/09553008714550461. [DOI] [PubMed] [Google Scholar]
- 37.Hilburger C L, Gager S R, Rider B J, White L E, George W J, Agrawal K C. Int J Radiat Oncol Biol Phys. 1994;29:397–402. doi: 10.1016/0360-3016(94)90297-6. [DOI] [PubMed] [Google Scholar]