Abstract
A reverse transcriptase (RT) cDNA, designated HERV-K-T47D-RT, was isolated from a hormonally treated human breast cancer cell line. The protein product putative sequence is 97% identical to the human endogenous HERV-K retroviral sequences. Recombinant T47D-RT protein was used to generate polyclonal antibodies. The expression of HERV-K-T47D-RT protein increased in T47D cells after treatment with estrogen and progesterone. The RT-associated DNA polymerase activity was substantially increased after over-expressing a chimeric YFP-HERV-K-T47D-RT protein in cells. This RT-associated polymerase activity was significantly reduced by mutating the active site sequence YIDD to SIAA. Moreover, the endogenous RT activity observed in T47D cells was decreased by HERV-K-T47D-RT-specific siRNA, confirming the dependence of the endogenous enzymatic activity. To assess HERV-K-T47D-RT expression in human breast tumors, 110 paraffin sections of breast carcinoma biopsies were stained and subjected to confocal analysis. Twenty-six percent (28/110) of the tumor tissues and 18% (15/85) of the adjacent normal tissue, from the same patients, expressed the RT. HERV-K-T47D-RT expression significantly correlates with poor prognosis for disease-free patients and their overall survival. These results imply that HERV-K-T47D-RT might be expressed in early malignancy and might serve as a novel prognostic marker for breast cancer. Furthermore, these results provide evidence for the possible involvement of endogenous retrovirus in human breast carcinoma.
Introduction
Breast cancer is the most common cancer among women in Western society, and the second most deadly malignancy among women [1]. Approximately 10% of breast cancers are due to known genetic pre-disposition [2,3], and approximately one third of the familial cases are probably due to mutations in the BRCA1 or BRCA2 genes [4]. Since the discovery of the mouse mammary tumor virus (MMTV) as an etiologic agent of murine mammary tumors [5], there has been an interest in the possible existence of similar infectious agents in humans. The human genome contains a large variety of endogenous retroviral sequences (HERVs; approximately 8% of the human genome) [6], of which the majority are highly defective [7,8]. In contrast to many defective HERV sequences, the HERV-K family shows a conservation of seeming intact retroviral genes [9]. These transcripts possess open reading frames (ORFs) that can potentially encode for the complete Gag, Pol, and Env proteins. Therefore, out of all known endogenous retrovirus families, the HERV-K is the most likely to encode for complete and possibly infectious retroviruses [10]. Retroviral-like particles were shown to be present in different types of human tissues and cell lines. Human teratoma-derived particles (HTDV/HERV-K) were first observed in human teratocarcinoma cell lines. These particles contain high-molecular weight RNA and a reverse transcriptase (RT) [11]. It has been previously shown by others and by us that the human breast carcinoma-derived cell line T47D release retroviral-like particles that resemble type B virions [12,13]. These particles possess low RT activity and cross-react with antibodies against the MMTV envelope protein, gp52 [14,15]. RT-encoding sequences with identity to MMTV and HERV-K10 were detected using polymerase chain reaction (PCR) amplification of peripheral mononuclear cells cDNA (prepared from cellular mRNA) and genomic DNA, with primers for conserved RT regions. These sequences were divided into six groups, designated human endogenous MMTV-like (HML) 1 through 6 [16]. Three different retroviral sequences were isolated from purified T47D particles [17]. One of the proviral pol sequences showed an uninterrupted ORF that encodes for 241 amino acids with 65% identity to HERV-K10 [17]. Expression of an mRNA that encodes for a HERV-K RT ORF was shown in particles released from hormonally treated T47D cells [18,19]. On the basis of the HERV-K sequences, an RT with low activity was expressed from human bone marrow cells [20]. HERV-K-env transcripts were detected in several breast cancer cell lines and breast tumor tissues but not in nonmalignant breast tissues [21]. The expression of HERV-K-env transcripts was 5- to 10-fold higher in breast cancer cell lines that were treated with estradiol and progesterone, relative to untreated cells. HERV-K-env expression was significantly higher in most breast cancer tissues than in normal breast tissues [22]. Despite a lot of circumstantial evidence [17,18, 21,23], there is still no conclusive evidence for retroviral involvement in human breast neoplasia. Because RT is a crucial enzyme in the retroviral reproductive cycle, there is high importance to isolate an RT-encoding gene from human breast carcinoma cell lines and to confirm the existence of an active RT enzyme in these cells. In this work, an endogenous RT enzyme was cloned from the breast carcinoma cell line T47D, and its intracellular induction by steroid hormones and its activity were characterized. We have also determined the level of HERV-K-T47D-RT protein expression in 110 breast cancer human tissue biopsies and showed a significant positive correlation with the patient's disease-free interval and overall survival in breast cancer.
Materials and Methods
Cell Culture
The mammary carcinoma cell lines: T47D [24], MDA-MB-231, BT549 (obtained from American Type Culture Collection, Manassas, VA), the 293T cells (a human embryonic kidney cell line stably transfected with SV40 large T-antigen), and the mouse mammary tumor cell line (Mm5MT) [25] were all maintained in DMEM supplemented with 10% heat-inactivated fetal calf serum (FCS; Invitrogen, Carlsbad, CA), 1% sodium pyruvate, and 1% penicillin-streptomycin. The human mammary epithelial cell line (HB2), which is a clonal derivative of a nontumorigenic mammary epithelial cells line, MTSV1-7 [26], was maintained in DMEM supplemented with heat-inactivated 10% FCS, 10 µg/ml insulin, and 0.5 µg/ml hydrocortisone. For hormone stimulation studies, cells were grown in phenol red-free DMEM (Invitrogen) and were treated with 10-8 M β-estradiol (Sigma-Aldrich, St. Louis, MO) supplemented with 1% dialyzed FCS for 48 hours followed by treatment with 10-8 M progesterone (Sigma-Aldrich) for 48 hours. Mm5MT cells were treated with 10-6 M dexamethasone (Difco, Detroit, MI).
Cloning and Purification of Recombinant RT
Two primer sets were used to amplify HERV-K-RT genes by reverse transcription-polymerase chain reaction (RT-PCR). The short (1.4 kb) RT segment was amplified by primer set based on the HERV-K-pol published sequence [18] HERV-K-short-sense 5′-GGGAATTCCATATGCCACTAACTTGGAAAACAGAAAAAC-3′ and HERV-K-short-antisense 5′-GGCGCAAGCTTGTTCTCTCGGCCCTGTGTAA-3′. The sense and antisense primers contain either an NdeI or HindIII restriction enzyme site, respectively (underlined in the sequences). The full-length RT (1.8 kb) was amplified with a primer set based on the HERV-K (clone 10.1) published sequence [20]. The sense HERV-K-RT primer was 5′-GGAATTCGCATGCAATAAATCAAGA AAGAGAAGGAATAG-3′ and the antisense one was 5′-CCGTCGAAGCTTTA AGCATGAAGTTCTTGTGC-3′. These primers contain either an SphI or an HindIII restriction enzyme site, respectively (underlined in the sequences). The antisense primer provides a UAA stop codon (marked in bold in the sequence). This primer set was also used to amplify HERV-K-RT/HTDV from the pcGPK31ΔLTR vector containing the HERV-K pol, gag, and env genes [27], generously given by Dr. Ralf Tonjes (Paul-Ehrlich-Institut, Langen, Germany). The PCR products were digested with the proper restriction enzymes (NdeI or SphI and HindIII) to allow the in-frame fusion of the RT ORFs into a pT5M-based plasmid, which was engineered to express a six-His tag fused to the amino-terminus of the recombinant protein. All PCR products were sequenced.
The two versions of the recombinant His-tagged proteins were expressed in BL21(DE3) pLysS strain of Escherichia coli (Stratagene, La Jolla, CA) by induction with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). The His-Tag HERV-K-T47D-RT-short recombinant protein (42 kDa) was purified under denaturing conditions. The bacteria pellet was lysed with 100 mM sodium phosphate, 10 mM Tris, 8 M urea, pH 8.0, and was purified with Ni-NTA superflow (Qiagen, Valencia, CA) following the manufacturer's instructions (the equilibration and washing steps were performed with the same buffer). The protein was eluted with stepwise decreasing pH solutions of pH 6.3, 5.9, and 4.5 containing 100 mM sodium phosphate, 10 mM Tris, and 8 M urea. The denatured protein was refolded by a gradual removal of urea employing a stepwise dialysis from 8 M urea to no urea in PBS.
The full-length HERV-K/HTDV recombinant protein (66 kDa) and a recombinant MMTV-RT protein were expressed in bacteria and purified in native conditions as described in detail previously [28].
Generation of Polyclonal Antibodies
To generate HERV-K-T47D-RT antiserum, the purified His-tagged short HERV-K-T47D-RT was injected into two rabbits, using standard immunization procedures [29]. For purification, the sera were absorbed with an acetone powder of BL21(DE3) pLysS bacteria prepared as described in detail [30].
Reverse Transcription-Polymerase Chain Reaction
Total RNA was extracted from 1 x 107 hormonally treated T47D cells, using TRI Reagent (Sigma-Aldrich). The first-strand cDNA was synthesized by incubating 5 µg of total RNA with the random primers (GTGAGCTCC, CCAGTACAG) and RT (Superscript II, Invitrogen) according to the manufacturer's instructions (in a volume of 20 µl). All PCR reactions were performed using 5 µl of the RT products with DynaZyme DNA polymerase (Finnzymes, Helsinki, Finland) with the appropriate primers according to the manufacturer's instructions. All PCR reactions were conducted for 35 cycles, each of 1 minute at 94°C, 1 minute at the primer-adjusted melting temperature (Tm), and 2 minutes at 72°C. Negative controls were performed with identical aliquots of cDNA synthesis reaction that was performed with no RT present. The PCR products were analyzed by electrophoresis on Tris-acetate-EDTA 1% agarose gels for fragments with the appropriate lengths.
Plasmid Construction and Sequencing
To generate the yellow fluorescence protein (YFP) tag T47D-HERV-K-RT, the full-length RT-encoding gene was amplified by RT-PCR (as described previously) with the following set of primers: 5′-CCGCGCCCCTCGAGCT AATAAATCAAGAAAGAGAAGGAATAG-3′ and 5′-CCGTCGAAGCTTTA AGCATGAAGTTCTTGTGC-3′. The sense and antisense primers contain a XhoI and HindIII restriction enzyme sites, respectively (underlined in the sequences). The antisense primers provide a UAA stop codon (marked in bold in the antisense sequences). The PCR product, predigested with XhoI and HindIII, was ligated in-frame into the XhoI and HindIII-cleaved pYFP-C1 expression vector (Clontech Laboratories Inc., Palo Alto, CA). The complete sequence of the HERV-K-T47D-RT gene and the short HERV-K-T47D-RT gene were confirmed by DNA sequencing of the pYFP-HERV-K-T47D-RT plasmid and the pT5-HERV-K-T47D-RT-short plasmid, respectively. For full-length sequencing, we used several internal sequencing primers. New primers include the sense DNA oligonucleotides pEYFP5′-F (5′-CATGGTCCTGCTGGAGTTCGTG-3′), T47D-RT-642-F (5′-GACTGGC AATAGCATCTGATAAG-3′), T47D-RT-1521-F (5′-GTACAGGCTACAAGGGA TGTTG-3′) and the antisense primer T47D-RT-1350-R (5′-GTAGTCAATTTTAAGAACTG GAAGATC-3′). The nucleotide sequence of the full-length HERV-K-T47D-RT gene in this study has been deposited in the GenBank database under accession number DQ821442.
The mutated YFP-HERV-K-T47D-RT was produced by site-directed mutagenesis using the Stratagene QuickChange kit (Stratagene), following the manufacturer's protocol. Using the pYFP-HERV-K-T47D-RT plasmid as a template, three point mutations were introduced with the following oligos: sense - 5′-GAGAGAAAAGTTTTCAGACTGTTATATTATTC ATTCTATTGCTGCTATTTTATGTGCTGCAGAAACGAAAG-3′; and antisense - 5′-CTTTCGTTTCTGCAGCACATAAAATAGCAGCAATAGAATGAATAATA TAACAGTCTGAAAACTTTTCTCTC-3′ (all mutated nucleotides were A to C transitions, marked in bold and underlined). The mutations were verified by sequencing.
Construction of siRNA Expression Vectors
The mammalian expression vector pSUPER.retro (pSR) (Oligoengine, Seattle, WA) was used for the expression of siRNA [31]. To produce an intact target recognition sequence to suppress HERV-K-T47D-RT expression by pSUPER. HERV-K-T47D-RT vector, the siRNA sequences were designed to correspond to nucleotide positions 202–223 of the HERV-K-T47D-RT-short gene (marked in bold in the following sequences): sense strand - 5′-GATCCCCATCAGTGGCCGCTACCAAATTCAAGAGATTTGGTAGCGGCCACTGATTTTTTGGAAA-3′ and antisense strand - 5′-TCGATTTCCAAAAAATCAG TGGCCGCTACCAAATCTCTTGAATTTGGTAGCGGCCACTGATGGG-3′. Selection of the 202–223 nucleotide region was based on its location within the HERV-K-T47D-RT-encoding gene and its conservation in the HERV-K/HTDV gene sequence. siRNA was annealed as previously described [32]. The oligo-annealing buffer was made with 100 mM potassium acetate, 30 mM HEPES-KOH (pH 7.4), and 2 mM Mg-acetate. pSUPER vector was digested with XhoI and BglII restriction endonucleases. Subsequently, the oligos of HERV-K-T47D-RT siRNA were inserted. The sequence of the plasmids of the pSUPER. HERV-K-RT siRNA was confirmed by sequencing. The control pSUPER.retro-GFP vector was kindly given by Dr. Shilo's laboratory (Tel Aviv University, Tel-Aviv, Israel). All transient transfections were performed using 293T cells. Cells were cotransfected with 4 µg of the pYFP-HERV-K-T47D-RT plasmid and with 15 µg of the pSUPER-siRNA or pSUPER empty vector as negative control.
Cell Transfection
293T cells were transiently transfected at 60% to 70% confluence by calcium phosphate precipitation transfection [33]. T47D cells (8 x 106) were transfected with 20 µg of DNA (recombinant or empty vector) by electroporation (0.25 V, 960 µF) and immediately plated in tissue culture dishes containing growth medium (supplemented DMEM). G418 (2 mg/ml) was added 24 hours after transfection, and stable transfectants were selected within 14 days or after. The same procedure was used to generate all stable transfectants.
Northern Blot Analysis
The DNA probes for hybridization were prepared as follows: HERV-K-RT probe was generated by digestion of the pT5-HERV-K-T47D-RT-short vector with NdeI and HindIII. MMTV-RT probe was generated by digestion of the pUC112N-MMTV-RT-(3A) vector [28] with NotI and HindIII. HERV-K. gag and HERV-K- env probes were generated by PCR with the pcGPK31ΔLTR vector [27], using the following primers, respectively:
gag:
sense - 5′-GGGAATTCCATATGGAAGCTAGGGT GATAATGGGGC-3′;
antisense - 5′-CCGTCGA AGCTTACTCCACTATGACCTTACCGGC-3′.
env:
sense - 5′-GGGAATTCCATATGGAAGCTAGGGTGATAATGGGGC-3′;
antisense - 5′-CCGTCGAAGCTTAGAGCTG TTGGGTACACCTG-3′.
18S cDNA was purchased from Clontech Laboratories Inc.
Total RNA was prepared from cultured cells using TRI Reagent (Sigma-Aldrich). Samples containing 20 µg of RNA were separated on 1.2% agarose/2% formaldehyde gels and were blotted to nylon membrane Hybond-N (Amersham Pharmacia, Buckinghamshire, England). The membrane was prehybridized for 4 hours in 5 x SSC, 5 x Denhardt's solution, 50% formamide, 0.1% SDS, and 200 µg/ml denatured salmon sperm DNA at 42°C. The DNA probes were labeled with [α-32P]dCTP using NEBlot Kit and were added to the hybridization buffer for overnight incubation at 42°C. Then, the membrane was washed three times at 42°C in 2 x SSC, 0.1% SDS for 20 minutes, followed by 1 to 0.1 x SSC, 0.1% SDS at 42°C (salt concentration and number of washing were adjusted for each probe). For reprobing, the membrane was stripped in 0.1% SDS at 100°C for 30 minutes.
Preparation of Cell Lysates
Confluent cells (5 x 106) were washed in PBS and resuspended in 150 µl of lysis buffer [20 mM Tris-HCl, pH 7.8, 100 mM NaCl, 50 mM NaF, 1% NP40, 0.1% SDS, 2 mM EDTA, 10% glycerol and protease inhibitors cocktail EDTA-free (Boehringer, Mannheim, Germany)]. Cell lysates were left on ice for 10 minutes and then centrifuged at 14,000g for 5 minutes at 4°C, and protein concentration was quantified by BCA kit (Pierce, Rockford, IL).
Western Blot Analysis
SDS-PAGE was performed in reducing conditions in 8% or 9% acrylamide gels (acrylamide/bisacrylamide ratio, 40:1 w/w). Approximately 100 to 200 µg of whole-cell protein extract were applied to each lane. For Western blot analysis, proteins were electrophoretically transferred to 0.2-µm-thick nitrocellulose membranes. Then, the membranes were blocked for 1 hour at room temperature in 5% milk/PBS. Primary antibodies were used as follows: anti-T47D-RT at a 1:250 dilution, anti-GFP (Santa Cruz Biotechnology, Santa Cruz, CA) 1:200, and anti-β-actin 1:1500 (Chemicon, Temecula, CA), all diluted in milk/PBS for 16 hours at 4°C. The membranes were washed in 0.2% Tween-PBS (PBST) and incubated with HRP-conjugated antirabbit IgG diluted 1:40,000 or antimouse IgG diluted 1:20,000 (Jackson ImmunoResearch Laboratories, West Grove, PA). Membranes were washed in 0.2% PBST, and enhanced chemiluminescence (Pierce) was performed. For reincubation, the membrane was subjected to stripping with a fresh solution of Ponceau S stain (0.2% Ponceau in a solution of 3% trichloroacetic acid in H2O).
Immunofluorescence Staining and Confocal Analysis
Cells. Hormonally treated or untreated T47D cells (1 x 104) were grown in Lab-Tek chamber slides (Nalgene Nunc International, Naperville, IL). After 2 days, the cells were washed twice with PBS and fixed for 10 minutes at room temperature in 4% paraformaldehyde/PBS, washed three times with PBS, and permeabilized for 10 minutes in 0.5% Triton X-100 in PBS. Then, blocking was done in 1% BSA/10% normal donkey serum/PBS for 30 minutes at room temperature. The cells were subsequently incubated with the primary antibodies. The antibodies anti-T47D-RT or preimmune rabbit serum [normal rabbit serum (NRS)] were diluted 1:10 in primary antibody dilution buffer (green buffer; purchased from Biomeda Corporation, Foster City, CA) and incubated for 16 hours at 4°C. The cells were then washed three times in PBS and incubated for 1 hour in the dark with the secondary antibody (rhodamine- or FITC-conjugated donkey antirabbit IgG diluted 1:200 in PBS). Slides were then washed in PBS and mounted with coverslips using Gel-Mount (Biomeda).
Tissues. Tissues from malignant breast lesions were formalin-fixed and embedded in paraffin. All the tissues were obtained from the Oncology Department, Tel Aviv Sourasky Medical Center (Tel Aviv, Israel) with the approval of the institutional Helsinki Committee. Serial sections (5-µm thick) were prepared from the blocks and processed as described. The slides were deparaffinized, washed in PBS, and incubated with AutoZyme (Biomeda) for 10 minutes at 37°C. The tissue sections were then blocked and stained as described previously for cells. For live cells YFP visualization, cells were grown on 12-mm round glass coverslips, compatible for confocal microscopy. Samples were analyzed using a 410 Zeiss (Obercocen, Germany) confocal laser scan microscope (CLSM) with the following configuration: 25mW krypton/argon (488 and 568 nm) and HeNe (633 nm) laser lines. When comparing fluorescence intensities, identical CLSM parameters (e.g., scanning line, laser light, contrast, and brightness) were used nearly identical to methods used previously for breast cancer analysis [34,35]. To compare the relative levels of protein expression, we have used the average area intensity (AAI) image analysis procedure for cell immunostains, and we also used mean square deviation of the intensity value, entropy, and log2 entropy calculations for the tissue stains. The image analysis calculations were done on several microscopic fields (3–10), using the MICA software (Cytoview, Petach Tikva, Israel). Variance was analyzed by 2-tailed analysis of variance.
RT Assay
Colorimetric reverse transcriptase activity was assayed by using the colorimetric HS sensitivity RT kit (Cavidi, Upsalla, Sweden) in accordance with the manufacturer's directions. Radioactive RNA-dependent DNA polymerase (RDDP) activity was assayed by measuring the incorporation of [3H]dTTP to the poly(rA)·oligo(dT)12–18 template primer as in the study of Shaharabany et al. [36].
Statistics
The results are expressed as the mean ± SEM. Student's t test was used to analyze statistical differences between the control and experimental groups. Differences were considered statistically significant at P < .05. Analyses of overall survival were performed with Kaplan-Meier analysis [37]. Computations were performed using SAS (SAS Institute, Cary, NC). Cutoff point was selected as was previously described in Altstock et al. [38].
Results
Isolation of the Coding Sequence for the HERV-K-T47D-RT—Its Cloning, Expression, Purification, and Generation of Polyclonal Antibodies
To clone the specific mRNA and express its cognate active RT, total RNA was isolated from T47D cells that were treated with the steroid hormones estrogen and progesterone. The transcripts were amplified by RT-PCR, using two primers sets: primers for shorter ORF transcript [18], and primers for full-length transcript [20]. The PCR products were cloned and sequenced.
The putative amino acid sequence of HERV-K-T47D-RT-encoding gene shows a 97% identity with the published sequence encoded by the HERV-K-pol gene from T47D particles [18] and by the human HERV-K endogenous retroviral sequence from teratocarcinoma GH cell line (HTDV-human teratocarcinoma-derived virus) [39]. HERV-K-T47D-RT possesses low identity with MMTV-RT (41%). HERV-K-Phylogenetic comparison shows that the HERV-K-T47D-RT sequence belongs to an HERV-K subgroup called HML-2 (human MMTV like-2; Figure W1). Bioinformatic analysis demonstrated that HERV-K-RT sequence is present in many chromosomes in the human genome. The HERV-K-RT sequence has more than 98% similarity to chromosomes: 1, 3, 5, 6, 7, 8, 10, 11, 19, 21, and 22. The HERV-K-T47D-RT amino acid sequence is almost identical to the HERV-K-pol gene that was isolated from particles released by T47D cells [18]. However, our newly cloned sequence is much different (59%) from the HERV-K-T47D-1 that was also isolated from T47D particles by Seifarth et al. [17]. T47D-RT exhibits the catalytically central DNA polymerase YIDD motif, a derivate of the YXDD motif, which is conserved in all HERV-K RT sequences [40]. The short transcript of HERV-K-T47D-RT was cloned into the same E. coli expression vector pT5, for producing 6His-RT fusion protein. The 6His-HERV-K-T47D-RT fragment was used to generate specific rabbit antibodies (designated anti-T47D-RT). The recombinant full-length HERV-K-RT protein (66 kDa) cross-reacted with the anti-T47D antibody (Figure W2). However, the anti-T47D-RT antibody was shown to be specific to HERV-K-RT and did not cross-react with the recombinant MMTV-RT (Figure W2).
Hormonally Induced Expression of the HERV-K-T47D-RT
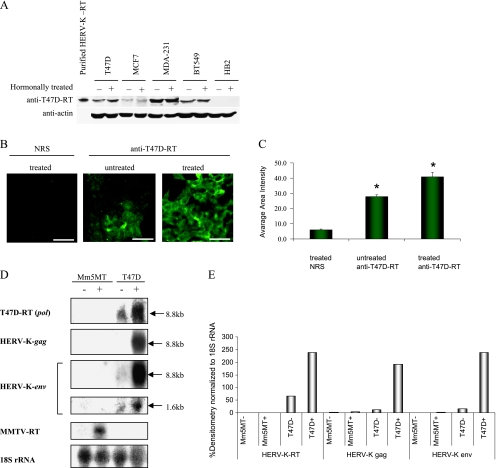
The HERV-K viral expression is increased when T47D cells are treated with the steroid hormones estrogen and progesterone, as described by our group and others [12,13,18]. Therefore, the effect steroid hormones treatment on RT protein expression was assessed. The HERV-K-T47D-RT protein expression level in response to hormonal treatment was examined in several breast carcinoma cell lines (T47D,MCF7, MDA-MB-231, and BT549) and in the normal human breast cell line HB2 (Figure 1A). The cells were treated with β-estradiol for 48 hours, followed by progesterone treatment for an additional 48 hours. A twofold increase in the level of the HERV-K-T47D-RT protein was observed in T47D cells after hormonal treatment (as was measured by densitometry; data not shown). The normal human breast cell line HB2 did not express any detectable levels of the HERV-K-T47D-RT with or without hormone treatment. The highest basal level of HERV-K-T47D-RT protein expression was observed in the metastatic breast cancer cell lines MDA-MB-231 and BT549. The level of protein did not increase significantly after hormonal treatment in these cell lines (Figure 1A). Because a significant hormonal induction was seen only in T47D cells, we have further characterized this induction by a single-cell resolution, using anti-T47D-RT antibodies by immunofluorescence technique. Ten different fields were taken from each staining, and the average fluorescent area intensity was calculated. HERV-K-T47D-RTexpression increased by 30% in response to hormone treatment (P = .006; Figure 1, B and C). To our knowledge, this is the first time that a hormonal induction of a viral-related RT protein is observed in human breast carcinoma cells. We have also tested the hormone-induced expression of the HERV-K viral gag, pol (RT), and env mRNA in T47D cells. T47D cells were hormonally treated as described previously. Mm5MT cells (mouse mammary carcinoma cell line expressing MMTV), treated with dexamethasone, served as a positive control for the MMTV RT probe and a negative one for the HERV-K-T47D-RT. Northern blot analyses demonstrated that, in response to hormonal treatment, expression levels of HERV-K-T47D pol (RT), HERV-K-gag and -env transcripts increased significantly, albeit to different levels indicating different mRNA stabilities (Figure 1, D and E). The MMTV-RT probe served as a negative control, and 18S rRNA probe served as a loading control. As could be expected from other retroviruses, the HERV-K-env probe reacted with the full mRNA (8.8 kb) and with spliced mRNA of approximately 1.6 kb. The spliced env mRNA was expressed 20-fold lower relative to the unspliced mRNA. As expected, no cross-hybridization was detected between the HERV-K-T47D-RT and the MMTV-RT transcripts (Figure 1D).
Figure 1.
Hormonal-induced expression of the HERV-K-T47D-RT protein. (A) Western blot analysis of a blot containing purified HERV-K-RT and extracts from different human breast cell lines: T47D, MCF7, MDA-MB-231, BT549, and HB2. The cells were either untreated (-) or treated (+) with steroid hormones, as described in the Materials and Methods section. The blot was incubated with the anti-T47D-RT antibody. Afterwards, the blot was striped and incubated with anti-β-actin antibodies (serving as a loading control). (B) Immunofluorescence staining: T47D were untreated (-) or treated (+) with β-estradiol and progesterone as described in the Materials and Methods section. The cells were fixed and stained with anti-T47D or with preimmune serum (NRS) as control. The stained cells were analyzed using CLSM. Scale bars, 50 µm. (C) Comparison between anti-HERV-K-T47D-RT and NRS staining (*P = .00001, Student's t test) and untreated versus treated cells' HERV-KT47D-RT staining levels (P = .0008, Student's t test) using average fluorescence intensity per area by MICA software on 10 fields of each staining. (D) The level of HERV-K-T47D, HERV-K-T47D-gag, and HERV-K-T47D-env mRNA expression was examined by Northern blot analysis of total cellular RNA isolated from either Mm5MT cells untreated (-) or hormonally treated (+) with dexamethasone, or from T47D cells untreated or treated with β-estradiol and progesterone as described in the Materials and Methods section. The same blot was separately probed with randomly primed 32P-labeled HERV-K-T47D-pol(RT), HERV-K-T47D-gag, HERV-K-T47D-env, MMTV-pol DNA, or with 18S rRNA as control. To avoid remnant signal, the blot was stripped between each probe and exposed overnight to a film to confirm that there is no remnant signal from the other probes. The RT, gag, and env were hybridized to the same-sized 8.8-kb band, representing the full transcript. The env probe was hybridized with an additional 1.6-kb transcript representing the alternative splice variant, demonstrating a 20-fold less-intensified signal. To better demonstrate the 1.6-kb faint band, the image was intensified. (E) The mRNA expression was quantified by densitometric scanning. The signals were normalized to the intensity of signal obtained with the 18S rRNA probe.
Over-Expression of HERV-K-T47D-RT Shows an Induction of the RT-Associated RNA-Dependent DNA Polymerase Activity That Is Abolished By Active Site Mutations
Because of the high sequence identity, a full HERV-K-RT/HTDV sequence [27,39] that was isolated from teratocarcinoma cells was recombinantly expressed. The purified HERV-K-RT enzyme was tested by in vitro assay system. It possesses an RNA-dependent DNA polymerase (RDDP) activity that is the hallmark activity of all known RTs [20,28]. Similar to the data available to all the recombinant HERV-RTs examined so far that were shown to have low catalytic activities [20,41], this level of the HERV-K-RTactivity is significant, albeit low, in comparison to MMTV-RT (data not shown).
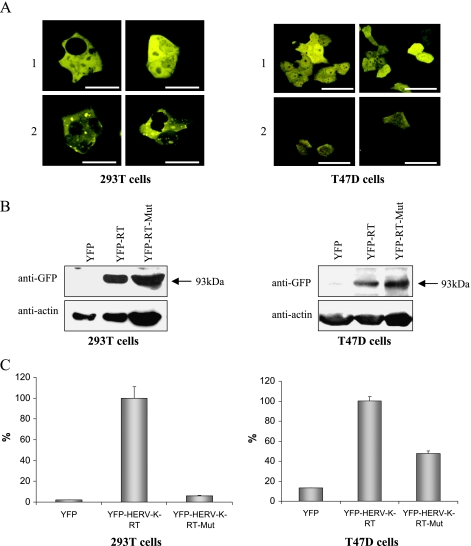
To further demonstrate that the HERV-K-T47D-RT exhibits also an in vivo enzymatic activity in mammalian cells, we constructed a plasmid encoding for the chimeric protein YFP-HERV-K-T47D-RT. Because the N-terminal extension is not expected to change the activity level of the RT [20] and may even stabilize it [28], we have chosen to use a YFP chimeric protein for easier identification of the positive clones. The YFP-HERV-K-T47D-RT plasmid was used to transiently transfect 293T cells and stably transfect T47D cells. An empty pYFP-C1 plasmid served as control. The chimeric protein expressing the wild type (WT) YFP-HERV-K-T47D-RT was expressed in both cells, as seen by CLSM analysis (Figure 2A). The chimeric YFP-HERV-K-T47D-RT protein was localized to the cytoplasm and showed lower intensity compared with cells transfected with the YFP-expressing vector alone (as expected for a chimeric protein). The YXDD motif, where X is an isoleucine in the specific case of HERV-K, is highly conserved among various retroviral RTs [40,42]. Several studies have shown that Y → S substitutions in the YXDD motif can abolish the polymerase activity of HIV-RT [43] and MuLV-RT [44]. To prove the HERV-K-T47D-RT-specific activity, we have mutated the YIDD motif, starting at position 184 of the chimeric YFP-HERV-K-T47D-RT, to SIAA using the direct point mutagenesis methodology. The mutated YFP-HERV-K-T47D-RT-expressing plasmid was also used to transiently transfect 293T cells and stably transfect T47D cells. Protein extracts were purified from all transfected cells and were subjected to Western blot analysis using anti-GFP-Ab (which cross-reacts with YFP; Figure 2B). In the cells transfected with the control plasmid, a lower band of 27-kDa YFP was observed (not shown). The amounts of total protein subjected to the Western blot analysis were normalized to achieve equal expression levels of the different RTs. The same extracts were also subjected to highly sensitive RT RNA-dependent DNA polymerase (RDDP) colorimetric activity assay (Figure 2C). The results represent a typical experiment that was repeated three times, with triplicates or duplicates each time. A significant polymerase activity was observed in both extracts of 293T cells and T47D cells transfected with the WT YFP-HERV-K-T47D-RT protein. Extracts of cells, transfected with the mutated YFP-HERV-K-T47D-RT, exhibited very low activity levels similar to the control. The basal activity in T47D cells (Figure 2C) is most probably due to the activity of the endogenous RT. These results indicate that the HERV-K-T47D-RT is enzymatically active in mammalian cells and that the observed activity is specific to this RT enzyme.
Figure 2.
Over-expression of the chimeric YFP-HERV-K-T47D-RT enzyme. (A) CLSM analysis of 293T cells (left panel) and T47D cells (right panel) transiently or stably transfected, respectively, either with pYFP-C1 (row 1) or with pYFP-HERV-K-T47D-RT (row 2). Scale bars, 50 µm. (B) Western blot analysis of protein extracts obtained from 293T (left panel) and T47D cells (right panel) transfected with pYFP-C1, with wild type (WT) pYFP-HERV-K-T47D-RT, or with the mutated pYFP-HERV-K-T47D-RT. The blots were incubated with anti-GFP antibody. As a loading control, the same blots were incubated with anti-β-actin antibody. (C) RT-associated DNA polymerase colorimetric activity assay was performed on the same protein extracts as in panel (B), from either the 293T cells (left) or the T47D cells (right). The activity was normalized to the HERV-KT47D-RT-transfected cells' activity that was set to 100%. The RT assay was repeated three times, and this is a representative experiment.
HERV-K-T47D-RT Expression Knockdown Suppresses the Cellular RT Expression Levels and Its Enzymatic Activity
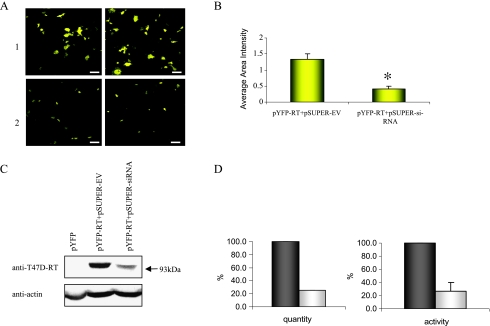
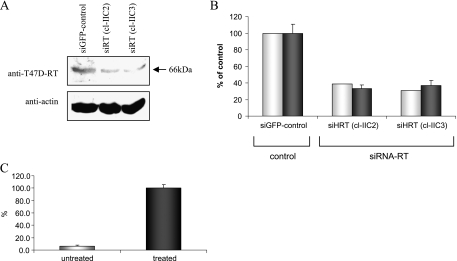
To prove that the endogenous RT-associated activity, seen in the transfected T47D cells, is derived specifically from the HERV-K-T47D-RT activity, we have used RNA interference to knockdown its expression. The efficiency of the siRNA was confirmed by transient cotransfection of 293T cells with the pYFP-HERV-K-T47D-RT plasmid and either the pSUPER-siRNA plasmid or an empty pSUPER plasmid as control. The data show that the exogenous YFP-HERV-K-T47D-RT expression levels and the RT-associated polymerase activity were significantly reduced by 70% to 75% with DNA of the pSUPER-siRNA compared to the pSUPER empty vector (Figure 3, A–D). Because the siRNA was efficient in reducing the exogenous YFP-HERV-K-T47D-RT levels, the hairpin siRNA was recloned to pRetro-super expression system to reduce the endogenous HERV-K-T47D-RT levels. T47D cells were stably transfected with the specific pRetro-siRNA-RT or with irrelevant pRetro-siRNA-GFP. The T47D pRetro-siRNA-RT stable clones, which showed a significant reduction in HERV-K-T47D-RT protein expression levels (compared with the control siRNA-GFP; Figure 4A), were also subjected to the RT-associated polymerase activity assay (Figure 4B). The results demonstrate that the RT polymerase activity decreased significantly by 60% to 80% compared with the control siRNA, with a high correlation to the protein expression levels (Figure 4B). This suggests that most of the endogenous enzymatic activity obtained in T47D cells is specific to the HERV-K-T47D-RT protein. To further examine the endogenous enzymatic activity, retroviral-like particles were purified from high-spin pellet of hormonally treated T47D cells supernatant. Representative results of three separate experiments showed a 20-fold induction in the specific RT polymerase activity in the pellet of hormonally treated T47D cells compared with the pellet of untreated T47D cells (Figure 4C). These results indicate that the endogenous HERV-K-RT is also active in particles released from hormonally treated T47D cells.
Figure 3.
Knockdown of the chimeric YFP-HERV-K-T47D-RT enzyme. (A) Confocal analysis of the 293T cells cotransfected with: 1) exogenous RT-expressing gene and pSUPER empty vector (EV) or 2) exogenous RT-expressing gene and pSUPER-siRNA. Two representative fields are shown. Scale bars, 50 µm. (B) Comparison of the average fluorescence intensity per area, of 10 different fields (MICA software), between the two transfections shown previously (A). *P = .0001 (analysis of variance). (C) Western blot analysis of an extract of 293T cells transfected with: 1) pYFP-C1, 2) cotransfected with YFP-HERV-K-T47D-RT and with pSUPER-EV, and 3) cotransfected with YFP-HERV-K-T47D-RT and pSUPER-siRNA. The blots were incubated with anti-T47D-RT antiserum (upper panel) and anti-β-actin (lower panel). (D, left panel) The protein expression levels were quantified by densitometric scanning. The signals were normalized to the intensity of signal obtained with the anti-β-actin. Gray bars represent percentage protein levels in cells transfected with pSUPER-EV (100%), and the white bars represent percentage protein levels in cells transfected with pSUPER-siRNA. (D, right panel) Colorimetric RT activity assay was performed with protein extracts form the same transfected cells as in panel (C) (a representative experiment out of three). The activity of the control cells (pSUPER-EV transfectant) were set to 100% (gray bars).
Figure 4.
The effects of RNA interference on the endogenous expression of HERV-K-T47D-RT. (A) Western blot analysis of protein extracts purified from different clones of T47D cells stably transfected with the control pRetro-GFP-siRNA or with pRetro-RT-siRNA (two different clones). The blots were incubated with the anti-T47D-RT antiserum. As a loading control, the same blots were incubated with anti-β-actin antibody. (B) Densitometric quantitation of the Western blot data shown in panel (A), normalized with β-actin (white bars). RT colorimetric activity assay was performed on the same protein extracts as in panel (A) (gray bars; a representative experiment out of three). (C) Colorimetric RT activity assay was performed on the solubilized T47D cell-derived viral-like pellets (a representative experiment out of three). Pellets were obtained by centrifuging (100,000g) T47D cell condition medium that was either untreated (white) or treated with β-estradiol and progesterone (gray).
Detection of the HERV-K-T47D-RT Protein in Breast Cancer Tissues and Its Implications as a Prognostic Marker
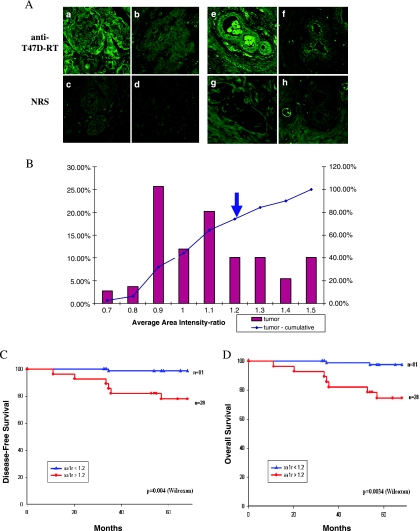
To investigate the expression of the HERV-K-T47D-RT in human breast carcinoma, 110 breast cancer biopsies from patients with 5 to 10 years of follow-up were immunostained. Serial sections of breast carcinoma were immunostained with anti-T47D-RT antibody or with NRS as a control. Subsequently, Nomarski and fluorescence CLSM images of both staining were acquired (three to five microscopic fields for each case). Representative image pairs, showing the varying levels of the HERV-K-T47D-RT protein expression versus the NRS control staining, are shown in Figure 5A. HERV-K-T47D-RT staining in the tumor tissue is shown in Figure 5A (a–d). The first two pairs of images demonstrate positive HERV-K-T47D-RT expression, where the RT staining is higher than the NRS control (Figure 5A, a vs c and b vs d, respectively). HERV-K-T47D-RTstaining in normal tissues adjacent to the tumor tissue is shown Figure 5A (e–h). The fourth pair of images demonstrates a typical section with no detectable HERV-K-T47D-RT expression (Figure 5A, f and h). We initially used an image analysis program (MICA) to calculate the simplest pixel-based image numerical descriptors, such as the AAI value, mean square deviation of the intensity value, entropy, and log2 entropy. A histogram of the AAI ratio (anti-T47D-RT/NRS) demonstrates the distribution of staining levels in the tumor tissue (Figure 5B). The cutoff level between positive and negative HERV-K-T47D-RT stainings (>1.2) was determined based on the 25% of the patients with the highest expression level, as we have previously done for another marker [38]. Of 110 tumors, 28 (26%) express the HERV-K-T47D-RT. In the adjacent normal tissue of the same sections, 15 (18%) of 85 tissues express the RT protein. Interestingly, there was a high correlation between the positive staining for the HERV-K-RT in the adjacent normal tissue and the positive staining in the tumor tissue.
Figure 5.
HERV-K T47D-RT expression in human breast carcinoma tissues. (A) Paraffin-embedded sections from breast carcinoma biopsies were subjected to indirect immunoflourescence staining using anti-T47D-RT antibody (a, b, e, and f) or NRS (normal rabbit serum) (c, d, g, and h) and analyzed by CLSM. Representative image pairs of anti-T47D-RT and NRS are presented: RT-positive (a, c) or RT-negative (b, d) tumor tissue; RT-positive (e, g) or RT-negative (f, h) normal adjacent tissue serial sections. (B) The distribution of the tumors' HERV-K T47D-RT staining level (AAI ratio; bars) and their cumulative percentages (lines). The cutoff point between positive and negative staining is marked by arrow (AAI = 1.2). (C) Kaplan-Meier analysis of disease-free survival for two groups of patients: HERV-K T47D-RT-positive (as measured by AAI ratios), at levels above the cutoff of 1.2 (red line); and HERV-K T47D-RT-negative, at levels below the cutoff (blue line). (D) Kaplan-Meier analysis of overall survival for the same two groups of patients as in panel (C): HERV-K T47D-RT-positive (red line) and HERV-K T47D-RT-negative (blue line). P values were calculated using log-rank and Wilcoxon tests.
A Kaplan-Meier analysis showed that patients, whose tumors overexpressed HERV-K-T47D-RT, had a significantly shorter metastasis-free survival (P = .004; Figure 5C) and shorter overall survival (P = .0034; Figure 5D) in comparison with other patients whose tumors did not over-express HERV-K-T47D-RT. The cutoff values of AAI ratios = 1.2 (see Materials and Methods section) produced statistically significant results (Figure 5, C and D). The other image analysis calculations produced very similar results (data not shown). These results clearly indicate that HERV-K-T47D-RT expression correlates with breast cancer patients' poor prognosis. Furthermore, we have also analyzed the potential correlations between HERV-K-T47D-RT overexpression and other clinical parameters, including age, clinical stage, menopausal, estrogen receptor status, and progesterone receptor status (patients with incomplete clinical parameters were included in the unknown group). HERV-K-T47D-RT over-expression correlates with age (P = .04) and distant metastasis (P = .05; Table 1). These results suggest that the HERV-K-T47D-RT can be reliably used as a novel independent prognostic marker for both metastasis and death risk in human breast carcinoma.
Table 1.
Correlation of HERV-K-T47D-RT Expression to Clinicopathologic Characteristics.
| Variable | HERV-K-T47D-RT* | P† | |
| - | + | ||
| Stage | |||
| 1 | 30 (39%) | 8 (30%) | .36 |
| 2 | 46 (61%) | 19 (70%) | |
| Unknown | 5 | 2 | |
| Total | 81 | 29 | |
| Age (yr) | |||
| <55 | 34 (45%) | 6 (22%) | .04 |
| 56–80 | 42 (55%) | 21 (78%) | |
| Unknown | 5 | 2 | |
| Total | 81 | 29 | |
| Death | |||
| Yes | 20 (25%) | 12 (41%) | .09 |
| No | 61 (75%) | 17 (59%) | |
| Total | 81 | 29 | |
| LR | |||
| Yes | 44 (54%) | 15 (52%) | .8 |
| No | 37 (46%) | 14 (48%) | |
| Total | 81 | 29 | |
| DM | |||
| Yes | 16 (20%) | 11 (38%) | .05 |
| No | 65 (80%) | 18 (62%) | |
| Total | 81 | 29 | |
| ER | |||
| Yes | 51 (63%) | 17 (59%) | .68 |
| No | 30 (37%) | 12 (41%) | |
| Total | 81 | 29 | |
| PGR | |||
| Yes | 44 (54%) | 15 (52%) | .81 |
| No | 37 (46%) | 14 (48%) | |
| Total | 81 | 29 | |
| Menopausal | |||
| Post | 17 (23%) | 8 (28%) | .6 |
| Pre | 58 (77%) | 21 (72%) | |
| Unknown | 6 | ||
| Total | 81 | 29 | |
DM indicates distal metastasis; ER, estrogen receptor; LR, local recurrence; PGR, progesterone receptor.
For consistency with our rating system, HERV-K-T47D-RT-positive expression was an AAI value above the cutoff of 1.2.
χ2 test.
Discussion
After years of intensive search for retroviral-related factor(s) involved in human breast cancer, different clues were found to show retroviral involvements in the disease. However, viral etiology was not convincingly proven. Most of the studies prove the existence of DNA that encodes for ORFs of retroviral genes in human breast cancer cell lines [13,17,18]. Few studies have shown the over-expression of mRNA species encoding for the retroviral Env protein in human breast cancer cell lines and biopsies [21,23]. However, as far as we know, there are no reports showing the selective expression of the Pol and Gag viral proteins in human breast cancer.
In this work, we have isolated a gene that encodes the HERV-K-RT from the human breast carcinoma cell line T47D. The putative sequence of the RT encoded by this new gene possesses high identity (96%) with a sequence of the pol gene product of HERV-K10, the first full-length provirus also isolated from T47D cells [13]. The sequence of the HERV-K-T47D-RT protein possesses high identity (97%) with the sequence of the product of pol isolated from particles released from T47D cells [18] and only 71% identity with the sequence encoded by the comparable gene isolated from similar particles that belongs to other HERV-K subfamily [17]. Phylogenetic analysis showed that the HERV-K-T47D-RT, isolated by us in the present study, belongs to the HML-2 subfamily as the HERV-K10 (also expressed in T47D cells) and is closely related to HERV-K sequences. It seems that, in T47D cells, there are several HERV-K (HML-2) sequences distinctive from each other by only a few nucleotides. It is estimated that the HERV-K (HML-2) integration event occurred approximately 1.2 Ma (based on minimal difference between the LTRs) and thus is considered an evolutionary young HERV-K family [9]. This may be the cause for the conserved active genes in this family compared to other HERVs. Another possibility is that these genes have an essential function for the cell's normal life cycle, and therefore they remained functional during evolution.
The recent dramatic progress in real-time PCR techniques enables investigators to learn more about viral genes' expression in human breast cancer, but the interpretation of these results can be sometimes deceiving. mRNA expression does not necessarily lead to active protein expression. Hence, it is essential to establish the biologic significance of an mRNA presence by showing that it was translated into a protein and by further confirming that the protein is biologically active and has an enzymatic activity. We have shown here that the HERV-K-T47D-RT is expressed in different human breast cancer cell lines but not in the normal human breast cell line HB2. These results support the assumption that the HERV-K-T47D-RT is specific to human breast cancer. However, only in T47D cells the HERV-K-T47D-RT protein was shown to be induced by the steroid hormones estrogen and progesterone. Our results highly correlate with a work that examined the mRNA expression of HERV-K-env in different hormonally induced breast cancer cell lines [22]. Because all tested breast cancer cell lines express the HERV-K-T47D-RT, albeit some express this protein constitutively, hormonal induction probably depends on the presence of receptors to both steroid hormones tested. The T47D cells express low levels of estrogen receptors and high levels of progesterone receptors [24], whereas in MCF7, this ratio is reversed such that estrogen receptors levels are high [45] and the progesterone receptor levels are low [46]. Thus, it is not surprising that the pattern of hormonal regulations in these two cell lines is likely to be different. The common assumption is that the hormonal regulation is gained by the hormone receptor binding to specific response elements in the viral LTR promoter [47]. In the MDA-MB-231 and BT549 cells that do not express steroid receptors, there is a constitutively high expression of the HERV-K-T47D-RT. This observation may result from the loss of hormonal sensitivity that accompanies malignant conversion and causes unregulated constitutive expression of the HERV-K-RT.
Although it was previously shown by our group and others [12,18,48] that the particles released from T47D cells exhibit an RT-related activity, to the best of our knowledge, so far there is no report connecting the relationship between the enzymatic activities and the HERV-K-RT expression. The only HERV-K-RT expressed was a recombinant protein with low in vitro polymerase enzymatic activity [20], similar to the HERV-K-T47D-RT studied herein. This low activity of the recombinant RT may result from the requirements for additional mammalian cellular cofactors that are missing from the in vitro reactions (such as retroviral NC protein, known to induce RT activity) or from possible posttranslational modifications that take place only in mammalian cells and not in bacteria. It is less likely that minor bacterial contaminants inhibit the RT activity; as we have already expressed, there were many retroviral RTs and all were very active after similar purification procedures [28,49–51]. In addition, we show here that over-expression of YFP-HERV-K-T47D-RT chimeric protein in human cell lines leads to a dramatic increase in the cytoplasmic RT-associated DNA polymerase activity. This activity was proved to be specific to the YFP-HERV-K-T47D-RT enzyme, because mutation in the YIDD active site abolished this activity. Furthermore, this conclusion is strongly supported by the finding that the intracellular T47D endogenous RT-related activity was significantly decreased because of a specific RNA interference. These results clearly indicate that the HERV-K-T47D-RT protein is expressed and is enzymatically active in the human breast cancer cell line T47D. Over-expression of YFP HERV-K-T47D-RT and down-regulation of the endogenous HERV-K-T47D-RT did not have any obvious effect neither on the transformed breast cancer cells morphology nor on their growth rate. These results indicate that other viral proteins might be involved in the tumorigenic process. We have also shown that the retroviral-like particles released from hormonally induced T47D cells contain the HERV-K-T47D-RT protein (data not shown). Furthermore, a 20-fold induction of the RT-dependent DNA polymerase activity was observed in these particles released from T47D cells treated with estrogen and progesterone. Because RT catalyzes a vital step in the retroviral life cycle, the presence of such enzymatically active RT in retroviral-like particles may suggest that these particles are infectious. Although no infectivity of HERV-K particles was reported so far [47,52], recent studies deduce that there is a possibility that HERV-K (HML-2) particles are infective in cultures cells [53–56].
Based on the examination of the insertion polymorphism levels, it was suggested that the HERV-K (HML-2) family are active in present-day humans [53]. Active elements are likely to have been inserted quite recently in human evolution and are therefore present at low allele frequencies. These active elements may lead to the development of malignancies. Because the HERV-K-T47D-RT enzyme, isolated in this study, belongs to the same HML-2 subfamily, the findings described herein may give rise to questions about a possible infectivity of the HERV-K-T47D-RT-containing particles. According to another hypothesis, the enzymatically active HERV-K-RTmay play also a role in retrotransposition. An active RT in cancer cells reinforces the hypothesis that, like MMTV, HERV-K can retrotranspose endogenous oncogenes or into tumor suppressor genes and thus promote tumorigenesis [8]. There are evidences showing that retrotransposons of HERV-K origin are responsible for polymorphism in the length of the complement genes C3 [57] and C4 [58]. HERV-K sequences were also located very close to the BRCA1 breast predisposition gene [59] and close to the glucose-6-phosphate dehydrogenase (G6PD) gene [60]. A third hypothesis suggests that the active RT enzyme generates pseudogenes by reverse transcription and reintegration of cellular mRNA. Some works suggested that the over-expression of different RTs could lead to an increase in cDNAs and pseudogenes levels [61–63].
The presence of HERV-K-T47D-RT was further tested in human breast tumors. We have shown that the HERV-K-RT protein is expressed in 26% of breast carcinoma cases tested (28/110) and in 18% of adjacent normal tissue tested (15/85). The HERV-K-RTexpression is highly associated with the overall survival's poor prognosis. The results implicate that the HERV-K virus and its active RT enzyme may be associated with human breast carcinogenesis. The expression of HERV-K-RT in the normal tissue adjacent to the tumor may indicate that HERV-K is expressed very early in the tumorigenic process. These data correlate with the previously published results showing that HERV-K ENV is expressed in 45% of the breast tumors and in 18% of the adjacent normal tissue [21]. However, we must emphasize that this association is not necessarily an evidence of causation. As tailoring personal patient treatment becomes more customary in breast carcinoma treatment, there is a growing necessity in finding additional biologic markers for determining suitable treatment. A novel marker such as HERV-K-T47D-RT can have a great significance because of its warning of poor prognosis that is independent of other tumor markers.
Anti-RT drugs are vastly used in the treatment of retroviral diseases (most notably AIDS, caused by HIV). In addition, RT inhibitors have been proposed in differentiation therapy for cancer [64]. Several reviews report epidemiological data that support the conclusion that cancer incidence is reduced in HAART-treated HIV-AIDS patients [65,66]. These data indicate that anti-RT drugs might also help in preventing or treating breast cancers expressing an RT marker. To conclude, the findings presented herein are consistent with T47D-RT as a novel independent prognostic marker for breast cancer and provide evidence for the possible involvement of endogenous retrovirus in human breast carcinoma.
Supplementary Material
Acknowledgments
The authors thank R. Tonjes for his generous gifts of the plasmid. The authors also thank Leonid Mitelman (Interdepartmental Core Facility at the Sackler School of Medicine) for his excellent assistance with the CLSM, Mati Bornshtein (Department of Cell Research and Immunology at the George S. Weiss Faculty of Life Science) for her outstanding assistance with cell culture growing, and Shira Reitkoff (Department of Human Microbiology at the Sackler School of Medicine) for her assistance in this work.
Abbreviations
- HERV
human endogenous retrovirus
- RT
reverse transcriptase
- YFP
yellow fluorescence protein
Footnotes
This work was partially supported by a research grant of the Breast Cancer Research Foundation.
This article refers to supplementary materials, which are designated by Figures W1 and W2 and are available online at www.neoplasia.com.
This work was done in partial fulfillment of the requirements for the Ph.D. degree of Maya Golan, Sackler Faculty of Medicine, Tel-Aviv University, Tel-Aviv, Israel.
References
- 1.Herbst RS, Bajorin DF, Bleiberg H, Blum D, Hao D, Johnson BE, Ozols RF, Demetri GD, Ganz PA, Kris MG, et al. Clinical Cancer Advances 2005: major research advances in cancer treatment, prevention, and screening—a report from the American Society of Clinical Oncology. J Clin Oncol. 2006;24:190–205. doi: 10.1200/JCO.2005.04.8678. [DOI] [PubMed] [Google Scholar]
- 2.McPherson K, Steel CM, Dixon JM. ABC of breast diseases. Breast cancer—epidemiology, risk factors and genetics. BMJ. 2000;321:624–628. doi: 10.1136/bmj.321.7261.624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kelsey JL, Bernstein L. Epidemiology and prevention of breast cancer. Annu Rev Public Health. 1996;17:47–67. doi: 10.1146/annurev.pu.17.050196.000403. [DOI] [PubMed] [Google Scholar]
- 4.Hulka BS, Moorman PG. Breast cancer: hormones and other risk factors. Maturitas. 2001;38:103–113. doi: 10.1016/s0378-5122(00)00196-1. discussion 113–106. [DOI] [PubMed] [Google Scholar]
- 5.Bittner J. Some possible effect of nursing on the mammary tumor incidence. Science. 1936;84:62. doi: 10.1126/science.84.2172.162. [DOI] [PubMed] [Google Scholar]
- 6.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 7.Larsson E, Kato N, Cohen M. Human endogenous proviruses. Curr Top Microbiol Immunol. 1989;148:115–132. doi: 10.1007/978-3-642-74700-7_4. [DOI] [PubMed] [Google Scholar]
- 8.Leib-Mosch C, Brack-Werner R, Werner T, Bachmann M, Faff O, Erfle V, Hehlmann R. Endogenous retroviral elements in human DNA. Cancer Res. 1990;50:5636S–5642S. [PubMed] [Google Scholar]
- 9.Mayer J, Sauter M, Racz A, Scherer D, Mueller-Lantzsch N, Meese E. An almost-intact human endogenous retrovirus K on human chromosome 7. Nat Genet. 1999;21:257–258. doi: 10.1038/6766. [DOI] [PubMed] [Google Scholar]
- 10.Tonjes RR, Czauderna F, Kurth R. Genome-wide screening, cloning, chromosomal assignment, and expression of full-length human endogenous retrovirus type K. J Virol. 1999;73:9187–9195. doi: 10.1128/jvi.73.11.9187-9195.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lower R, Lower J, Kurth R. The viruses in all of us: characteristics and biological significance of human endogenous retrovirus sequences. Proc Natl Acad Sci USA. 1996;93:5177–5184. doi: 10.1073/pnas.93.11.5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keydar I, Ohno T, Nayak R, Sweet R, Simoni F, Weiss F, Karby S, Mesa-Tejada R, Spiegelman S. Properties of retrovirus-like particles produced by a human breast carcinoma cell line: immunological relationship with mouse mammary tumor virus proteins. Proc Natl Acad Sci USA. 1984;81:4188–4192. doi: 10.1073/pnas.81.13.4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ono M, Kawakami M, Ushikubo H. Stimulation of expression of the human endogenous retrovirus genome by female steroid hormones in human breast cancer cell line T47D. J Virol. 1987;61:2059–2062. doi: 10.1128/jvi.61.6.2059-2062.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keydar I, Selzer G, Chaitchik S, Hareuveni M, Karby S, Hizi A. A viral antigen as a marker for the prognosis of human breast cancer. Eur J Cancer Clin Oncol. 1982;18:1321–1328. doi: 10.1016/0277-5379(82)90136-5. [DOI] [PubMed] [Google Scholar]
- 15.Segev N, Hizi A, Kirenberg F, Keydar I. Characterization of a protein, released by the T47D cell line, immunologically related to the major envelope protein of mouse mammary tumor virus. Proc Natl Acad Sci USA. 1985;82:1531–1535. doi: 10.1073/pnas.82.5.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Medstrand P, Blomberg J. Characterization of novel reverse transcriptase encoding human endogenous retroviral sequences similar to type A and type B retroviruses: differential transcription in normal human tissues. J Virol. 1993;67:6778–6787. doi: 10.1128/jvi.67.11.6778-6787.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seifarth W, Skladny H, Krieg-Schneider F, Reichert A, Hehlmann R, Leib-Mosch C. Retrovirus-like particles released from the human breast cancer cell line T47-D display type B- and C-related endogenous retroviral sequences. J Virol. 1995;69:6408–6416. doi: 10.1128/jvi.69.10.6408-6416.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patience C, Simpson GR, Colletta AA, Welch HM, Weiss RA, Boyd MT. Human endogenous retrovirus expression and reverse transcriptase activity in the T47D mammary carcinoma cell line. J Virol. 1996;70:2654–2657. doi: 10.1128/jvi.70.4.2654-2657.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seifarth W, Baust C, Murr A, Skladny H, Krieg-Schneider F, Blusch J, Werner T, Hehlmann R, Leib-Mosch C. Proviral structure, chromosomal location, and expression of HERV-K-T47D, a novel human endogenous retrovirus derived from T47D particles. J Virol. 1998;72:8384–8391. doi: 10.1128/jvi.72.10.8384-8391.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berkhout B, Jebbink M, Zsiros J. Identification of an active reverse transcriptase enzyme encoded by a human endogenous HERV-K retrovirus. J Virol. 1999;73:2365–2375. doi: 10.1128/jvi.73.3.2365-2375.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang-Johanning F, Frost AR, Johanning GL, Khazaeli MB, LoBuglio AF, Shaw DR, Strong TV. Expression of human endogenous retrovirus K envelope transcripts in human breast cancer. Clin Cancer Res. 2001;7:1553–1560. [PubMed] [Google Scholar]
- 22.Wang-Johanning F, Frost AR, Jian B, Epp L, Lu DW, Johanning GL. Quantitation of HERV-K env gene expression and splicing in human breast cancer. Oncogene. 2003;22:1528–1535. doi: 10.1038/sj.onc.1206241. [DOI] [PubMed] [Google Scholar]
- 23.Liu B, Wang Y, Melana SM, Pelisson I, Najfeld V, Holland JF, Pogo BG. Identification of a proviral structure in human breast cancer. Cancer Res. 2001;61:1754–1759. [PubMed] [Google Scholar]
- 24.Keydar I, Chen L, Karby S, Weiss FR, Delarea J, Radu M, Chaitcik S, Brenner HJ. Establishment and characterization of a cell line of human breast carcinoma origin. Eur J Cancer. 1979;15:659–670. doi: 10.1016/0014-2964(79)90139-7. [DOI] [PubMed] [Google Scholar]
- 25.Owens RB, Hackett AJ. Tissue culture studies of mouse mammary tumor cells and associated viruses. J Natl Cancer Inst. 1972;49:1321–1332. [PubMed] [Google Scholar]
- 26.Berdichevsky F, Alford D, D'Souza B, Taylor-Papadimitriou J. Branching morphogenesis of human mammary epithelial cells in collagen gels. J Cell Sci. 1994;107(Pt 12):3557–3568. doi: 10.1242/jcs.107.12.3557. [DOI] [PubMed] [Google Scholar]
- 27.Tonjes RR, Boller K, Limbach C, Lugert R, Kurth R. Characterization of human endogenous retrovirus type K virus-like particles generated from recombinant baculoviruses. Virology. 1997;233:280–291. doi: 10.1006/viro.1997.8614. [DOI] [PubMed] [Google Scholar]
- 28.Taube R, Loya S, Avidan O, Perach M, Hizi A. Reverse transcriptase of mouse mammary tumour virus: expression in bacteria, purification and biochemical characterization. Biochem J. 1998;329:579–587. doi: 10.1042/bj3290579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harlow E, Lane D. Antibodies — A Laboratory Manual. New York, NY: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 30.Sambrook A, Fritsch EF, Maniatis T. Molecular Cloning. A Laboratory Manual. New York, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 31.Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 32.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 33.Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsarfaty I, Alvord WG, Resau JH, Altstock RT, Lidereau R, Bieche I, Bertrand F, Horev J, Klabansky RL, Keydar I, et al. Alteration of met protooncogene product expression and prognosis in breast carcinomas. Anal Quant Cytol Histol. 1999;21:397–408. [PubMed] [Google Scholar]
- 35.Tsarfaty I, Resau JH, Rulong S, Keydar I, Faletto DL, Vande Woude GF. The met proto-oncogene receptor and lumen formation. Science. 1992;257:1258–1261. doi: 10.1126/science.1387731. [DOI] [PubMed] [Google Scholar]
- 36.Shaharabany M, Rice NR, Hizi A. Expression and mutational analysis of the reverse transcriptase of the lentivirus equine infectious anemia virus. Biochem Biophys Res Commun. 1993;196:914–920. doi: 10.1006/bbrc.1993.2336. [DOI] [PubMed] [Google Scholar]
- 37.Kaplan EL, Meier P. Nonparametric estimation from incomplete observations. J Am Stat Soc. 1958;53:457–481. [Google Scholar]
- 38.Altstock RT, Stein GY, Resau JH, Tsarfaty I. Algorithms for quantitation of protein expression variation in normal versus tumor tissue as a prognostic factor in cancer: met oncogene expression, and breast cancer as a model. Cytometry. 2000;41:155–165. [PubMed] [Google Scholar]
- 39.Lower R, Boller K, Hasenmaier B, Korbmacher C, Muller-Lantzsch N, Lower J, Kurth R. Identification of human endogenous retroviruses with complex mRNA expression and particle formation. Proc Natl Acad Sci USA. 1993;90:4480–4484. doi: 10.1073/pnas.90.10.4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiong Y, Eickbush TH. Similarity of reverse transcriptase-like sequences of viruses, transposable elements, and mitochondrial introns. Mol Biol Evol. 1988;5:675–690. doi: 10.1093/oxfordjournals.molbev.a040521. [DOI] [PubMed] [Google Scholar]
- 41.Awad R, Medstrand P, Yin H, Andersson M-L, Blomberg J. Open reading frames and associated enzymatic activities in the polymerase gene of human endogenous retroviral HERV-K(HML-2) sequences. Uppsala, Sweden: Department of Medical Sciences. Upsala University; 2003. [Google Scholar]
- 42.Sharma PL, Nurpeisov V, Schinazi RF. Retrovirus reverse transcriptases containing a modified YXDD motif. Antivir Chem Chemother. 2005;16:169–182. doi: 10.1177/095632020501600303. [DOI] [PubMed] [Google Scholar]
- 43.Harris D, Yadav PN, Pandey VN. Loss of polymerase activity due Tyr to Phe substitution in the YMDD motif of human immunodeficiency virus type-1 reverse transcriptase is compensated by Met to Val substitution within the same motif. Biochemistry. 1998;37:9630–9640. doi: 10.1021/bi980549z. [DOI] [PubMed] [Google Scholar]
- 44.Kaushik N, Singh K, Alluru I, Modak MJ. Tyrosine 222, a member of the YXDD motif of MuLV RT, is catalytically essential and is a major component of the fidelity center. Biochemistry. 1999;38:2617–2627. doi: 10.1021/bi9824285. [DOI] [PubMed] [Google Scholar]
- 45.Brooks SC, Locke ER, Soule HD. Estrogen receptor in a human cell line (MCF-7) from breast carcinoma. J Biol Chem. 1973;248:6251–6253. [PubMed] [Google Scholar]
- 46.Cassanelli S, Louis J, Seigneurin D. Progesterone receptor heterogeneity in MCF-7 cell subclones is related to clonal origin and kinetics data. Tumour Biol. 1995;16:222–229. doi: 10.1159/000217939. [DOI] [PubMed] [Google Scholar]
- 47.Ono M. Molecular cloning and long terminal repeat sequences of human endogenous retrovirus genes related to types A and B retrovirus genes. Virol. 1986;58:937–944. doi: 10.1128/jvi.58.3.937-944.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Faff O, Murray AB, Schmidt J, Leib-Mosch C, Erfle V, Hehlmann R. Retrovirus-like particles from the human T47D cell line are related to mouse mammary tumour virus and are of human endogenous origin. J Gen Virol. 1992;73(Pt 5):1087–1097. doi: 10.1099/0022-1317-73-5-1087. [DOI] [PubMed] [Google Scholar]
- 49.Avidan O, Loya S, Tonjes RR, Sevilya Z, Hizi A. Expression and characterization of a recombinant novel reverse transcriptase of a porcine endogenous retrovirus. Virology. 2003;307:341–357. doi: 10.1016/s0042-6822(02)00131-9. [DOI] [PubMed] [Google Scholar]
- 50.Avidan O, Bochner R, Hizi A. The catalytic properties of the recombinant reverse transcriptase of bovine immunodeficiency virus. Virology. 2006;351:42–57. doi: 10.1016/j.virol.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 51.Perach M, Hizi A. Catalytic features of the recombinant reverse transcriptase of bovine leukemia virus expressed in bacteria. Virology. 1999;259:176–189. doi: 10.1006/viro.1999.9761. [DOI] [PubMed] [Google Scholar]
- 52.Lower J, Wondrak EM, Kurth R. Genome analysis and reverse transcriptase activity of human teratocarcinoma-derived retroviruses. J Gen Virol. 1987;68(Pt 11):2807–2815. doi: 10.1099/0022-1317-68-11-2807. [DOI] [PubMed] [Google Scholar]
- 53.Belshaw R, Dawson AL, Woolven-Allen J, Redding J, Burt A, Tristem M. Genomewide screening reveals high levels of insertional polymorphism in the human endogenous retrovirus family HERV-K(HML2): implications for present-day activity. J Virol. 2005;79:12507–12514. doi: 10.1128/JVI.79.19.12507-12514.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Belshaw R, Pereira V, Katzourakis A, Talbot G, Paces J, Burt A, Tristem M. Long-term reinfection of the human genome by endogenous retroviruses. Proc Natl Acad Sci USA. 2004;101:4894–4899. doi: 10.1073/pnas.0307800101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Turner G, Barbulescu M, Su M, Jensen-Seaman MI, Kidd KK, Lenz J. Insertional polymorphisms of full-length endogenous retroviruses in humans. Curr Biol. 2001;11:1531–1535. doi: 10.1016/s0960-9822(01)00455-9. [DOI] [PubMed] [Google Scholar]
- 56.Macfarlane C, Simmonds P. Allelic variation of HERV-K(HML-2) endogenous retroviral elements in human populations. J Mol Evol. 2004;59:642–656. doi: 10.1007/s00239-004-2656-1. [DOI] [PubMed] [Google Scholar]
- 57.Zhu ZB, Hsieh SL, Bentley DR, Campbell RD, Volanakis JE. A variable number of tandem repeats locus within the human complement C2 gene is associated with a retroposon derived from a human endogenous retrovirus. J Exp Med. 1992;175:1783–1787. doi: 10.1084/jem.175.6.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dangel AW, Mendoza AR, Baker BJ, Daniel CM, Carroll MC, Wu LC, Yu CY. The dichotomous size variation of human complement C4 genes is mediated by a novel family of endogenous retroviruses, which also establishes species-specific genomic patterns among Old World primates. Immunogenetics. 1994;40:425–436. doi: 10.1007/BF00177825. [DOI] [PubMed] [Google Scholar]
- 59.Jones KA, Black DM, Brown MA, Griffiths BL, Nicolai HM, Chambers JA, Bonjardim M, Xu CF, Boyd M, McFarlane R, et al. The detailed characterisation of a 400 kb cosmid walk in the BRCA1 region: identification and localisation of 10 genes including a dual-specificity phosphatase. Hum Mol Genet. 1994;3:1927–1934. [PubMed] [Google Scholar]
- 60.Sedlacek Z, Korn B, Konecki DS, Siebenhaar R, Coy JF, Kioschis P, Poustka A. Construction of a transcription map of a 300 kb region around the human G6PD locus by direct cDNA selection. Hum Mol Genet. 1993;2:1865–1869. doi: 10.1093/hmg/2.11.1865. [DOI] [PubMed] [Google Scholar]
- 61.Levine KL, Steiner B, Johnson K, Aronoff R, Quinton TJ, Linial ML. Unusual features of integrated cDNAs generated by infection with genome-free retroviruses. Mol Cell Biol. 1990;10:1891–1900. doi: 10.1128/mcb.10.5.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Maestre J, Tchenio T, Dhellin O, Heidmann T. mRNA retroposition in human cells: processed pseudogene formation. EMBO J. 1995;14:6333–6338. doi: 10.1002/j.1460-2075.1995.tb00324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tchenio T, Segal-Bendirdjian E, Heidmann T. Generation of processed pseudogenes in murine cells. EMBO J. 1993;12:1487–1497. doi: 10.1002/j.1460-2075.1993.tb05792.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sinibaldi-Vallebona P, Lavia P, Garaci E, Spadafora C. A role for endogenous reverse transcriptase in tumorigenesis and as a target in differentiating cancer therapy. Genes Chromosomes Cancer. 2006;45:1–10. doi: 10.1002/gcc.20266. [DOI] [PubMed] [Google Scholar]
- 65.Rabkin CS. AIDS and cancer in the era of highly active antiretroviral therapy (HAART) Eur J Cancer. 2001;37:1316–1319. doi: 10.1016/s0959-8049(01)00104-6. [DOI] [PubMed] [Google Scholar]
- 66.Laurence J. Drug resistance among HIV and its “friends”. AIDS Read. 2003;13:103, 107. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.