Abstract
Parameter estimation constitutes a major challenge in dynamic modeling of metabolic networks. Here we examine, via computational simulations, the influence of system nonlinearity and the nature of available data on the distribution and predictive capability of identified model parameters. Simulated methionine-cycle metabolite concentration data (both with and without corresponding flux data) was inverted to identify model parameters consistent with it. Thousands of diverse parameter families were found to be consistent with the data to within moderate error, with most of the parameter values spanning over 1000-fold ranges irrespective of whether flux data was included. Due to strong correlations within the extracted parameter families, model predictions were generally reliable despite the broad ranges found for individual parameters. Inclusion of flux data, by strengthening these correlations, resulted in substantially more reliable flux predictions. These findings suggest that, despite the difficulty of extracting biochemically accurate model parameters from system level data, such data may nevertheless prove adequate for driving the development of predictive dynamic metabolic models.
Keywords: parameter identification, data inversion, biochemical modeling, metabolism, cyclic pathway
Introduction
Recent technological advances are enabling improved quantification of intracellular metabolite concentrations and fluxes (Brauer et al., 2006; Hollywood et al., 2006; Lu et al., 2006; Sauer, 2006). Constraints-based approaches to modeling metabolism, such as flux-balance analysis, have substantial power for predicting metabolic fluxes in certain organisms (Edwards et al., 2001; Schilling and Palsson, 1998; Steffen et al., 2002; Yuan et al., 2006). Despite this progress, there remains the need to also develop dynamic metabolic models which incorporate understanding of metabolite regulation to predict both metabolite concentrations and fluxes (Di Ventura et al., 2006; Gombert and Nielsen, 2000). The development of such models has been limited in part by the challenge of model parameter identification from laboratory data.
The nonlinear nature of metabolic dynamics calls for nonlinear parameter estimation (also known as data inversion) methods. Gradient based approaches, such as the Gauss-Newton method, are fast for solving convex optimization problems. However, when the inverse problem is non-convex, these approaches may fail to provide globally optimal solutions (Glowinski and Stocki, 1981; Linga et al., 2006). Global optimization methods such as simulated annealing (Eftaxias et al., 2002; Kirkpatrick et al., 1983), scatter search (Rodriguez-Fernandez et al., 2006b), evolutionary computation (Fogel, 2000; Goldberg, 1989; Moles et al., 2003; Tsuchiya and Ross, 2001), and variants (Rodriguez-Fernandez et al., 2006a), are generally more reliable in these situations. The application of global methods has been reported in a few studies modeling metabolic networks (Himmelblau et al., 1967; Moles et al., 2003; Polisetty et al., 2006).
Another challenge of bio-network model identification arises from the existence of laboratory noise. For most identification methods, the mean values of the data points are utilized to yield one solution for each unknown model parameter, and variations in the extracted parameters are subsequently estimated via linear propagation of the data noise. As a result, distributions of the identified parameters tend to look regular due to the linear error propagation (Kutalik et al., 2007), giving the impression that the mean parameter values are reliable estimates of the true values. However, the combination of data noise and system nonlinearity (hence nonlinear error propagation) can easily result in wide and highly irregular parameter distributions, where the true parameter values can be located anywhere in the distributions.
In order to address the above issues, Feng and Rabitz developed a general nonlinear method that yields not one, but a representative family of model parameters that can reproduce the bionetwork behavior to within the laboratory error ranges (Feng and Rabitz, 2004). This family then automatically forms the parameter distribution without having to perform explicit error propagations. Initial application of this method (to simulated in vitro reactions related to proofreading of tRNA loading) provided evidence that parameter distributions are better captured by this method than by traditional techniques.
In this work, we apply this method to parameter estimation for a metabolic network model that simulates the methionine cycle (Martinov et al., 2000; Reed et al., 2004). The focus is to understand the properties of the extracted parameter families (including the reliability of the predictions that they make) as a function of the input data provided. Biologically, the methionine cycle was selected because it is a representative of cyclic metabolic pathways (an important metabolic network motif), contains multiple positive and negative feedback mechanisms, and describes a physiologically and medically important system (Figure 1). Abnormalities of methionine cycle metabolism are associated with liver disease (Avila et al., 2002), cancer (Lu and Mato, 2005), neural tube defects (Refsum, 2001), Alzheimer's disease (Clarke et al., 1998), and cardiovascular disease (Mangoni and Jackson, 2002).
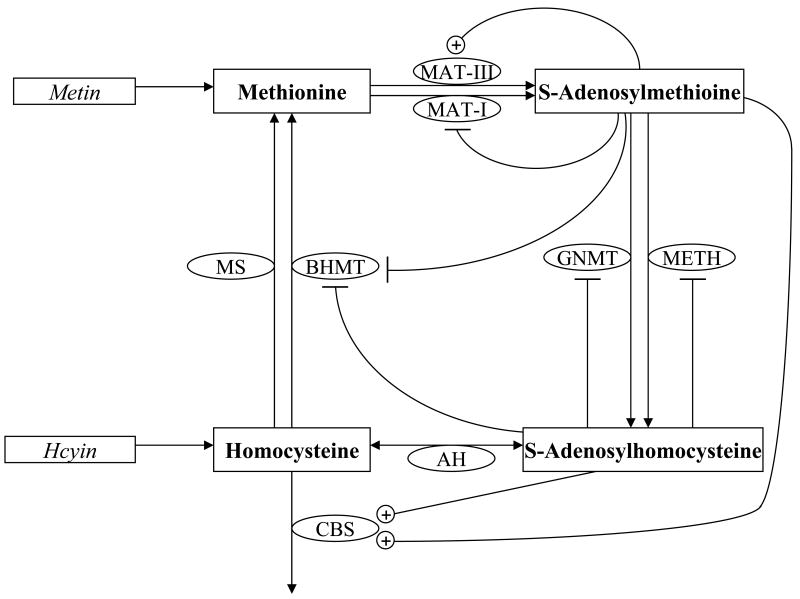
Figure 1. Methionine cycle as modeled by Reed et al (2004).
Modeled metabolites are in bold within rectangles; enzymes are in ovals; metabolite influxes are in italics. Unmodeled metabolites are not shown. The conversion of methionine (Met) to S-adenosylmethione (AdoMet) is catalyzed by either methionine adenosyl transferase I (MATI) or methionine adenosyl transferase III (MATIII), which are isozymes derived from the same polypeptide chain. The former is feedback inhibited by AdoMet, while the latter is upregulated by it in the concentration ranges used in this study. AdoMet is demethylated by either glycine N-methyltransferase (GNMT) or a general AdoMet dependent methyltransferases (METH) to give S-adenosylhomocysteine (AdoHcy). Both enzymes are feedback inhibited by AdoHcy. AdoHcy is converted to homocysteine (Hcy) by AdoHcy hydrolase. Hcy can be methylated by Met synthase (MS) or by betaine:homocysteine methyltransferase (BHMT, inhibited by AdoHcy and AdoMet) to re-form Met. Alternatively, it can combine with serine via cystathionine β-synthase (CBS, up-regulated by AdoHcy and AdoMet) to exit the cycle.
We find that exceptionally diverse and inter-correlated parameter sets exist that generate time-dependent methionine cycle behavior indistinguishable (within modest error estimates) from the published model. These parameter sets not only recapitulate the data used in their identification, they also result in similar model predictions under different perturbations. Sequential addition of flux and biochemical constraints modestly reduced the parameter ranges. Despite the remarkable diversity of the identified parameters, those extracted from concentration data generally yielded reliable concentration predictions. Addition of flux constraints into the parameter search identified parameter sets that were also flux predictive. These findings suggest that, while identification of the true (biochemically accurate) metabolic model parameter values (Teusink et al., 2000) from metabolomic data will generally be difficult owing to system nonlinearity and data noise, development of dynamic models with substantial predictive power may nevertheless prove feasible. This overall message is consistent with recent analysis of parameter sensitivities in other systems biology models (Gutenkunst et al., 2007).
Materials and Methods
Methionine cycle model
A previously published mathematical model of the methionine cycle developed initially by Martinov et al. (2000) and expanded by Reed et al. (2004) was used to generate the simulated data. The model consists of four non-linear ordinary differential equations describing the rate of change of methionine (Met), S-adenosylmethionine (AdoMet), S-adenosylhomocysteine (AdoHyc), and homocysteine (Hyc) (see Figure 1). These four metabolites interconvert via eight different enzyme catalyzed reactions:
Table 1 lists the functional forms of the flux expressions for each of the eight reactions. The formulation of the model here is identical to that of Reed et al. (2004), except for elimination of any over-parameterization. Specifically, in the expression for VMETH, the parameter KMETHm/[A] from the original publication was absorbed into VMETHmax, where [A] represents an arbitrary methylation substrate. In the case of VMS, the reaction was initially formulated as an ordered bi-bi reaction of the substrates. However, since [5mTHF] is a constant in the model, VMS was re-written as a simple, one substrate Michaelis-Menten expression. Table 2 lists the parameter values used in the revised, reference model. This model was used to generate all the data supplied to the inversion algorithm in the simulations.
Table 1.
Functional forms of rate expressions and associated parameters
| Reaction rate | Rate expression | Relevant Parameters |
|---|---|---|
| VMATI | ||
| VMATIII | ||
| VGNMT | ||
| VMET | ||
| VAH | α1, α2 | |
| VBHMT | ||
| VMS | ||
| VCBS | β1, β2 |
Table 2.
Model parameter values and search ranges
| Parameter | Published Model Value | Literature Values | Biochemical parameter range constraints |
|---|---|---|---|
| VMATImax | 561 μM/h | - | - |
| KMATIm | 41 μM | 41 μM (Sullivan and Hoffman, 1983) | 33 μM – 49 μM |
| KMATIi | 50 μM | 230 μM (Sullivan and Hoffman, 1983) | 172 μM – 287 μM |
| VMATIIImax | 22870 μM/h | - | - |
| KMATIIIm | 21.1 μM | - | - |
| VGNMTmax | 10600 μM/h | - | - |
| KGNMTm | 4500 μM | 100 μM (Heady and Kerr, 1973; Kerr, 1972) | 75 μM – 125 μM |
| KGNMTi | 20 μM | 35 μM (Kerr, 1972) | 26.2 μM – 43.7 μM |
| VMETHmax | 421 μM/h | - | - |
| α1 | 100 h-1 | - | - |
| α2 | 10 | - | - |
| β1 | 1.7 μM-1h-1 | - | - |
| β2 | 30 h-1 | - | - |
| VMSmax | 86.1 μM/h | - | - |
| KMSm,Hcy | 0.0205 μM | - | - |
| VBHMTmax | 2500 μM/h | - | - |
| KBHMTm | 12 μM | 32 μM (Garrow, 1996), 12 μM (Finkelstein et al., 1972) | 9 μM – 40 μM |
Simulated perturbations
Perturb-and-observe experiments were simulated with influxes to the system limited to methionine (Metin) and/or homocysteine (Hcyin), as described in the Results. The magnitudes of the perturbations were chosen to approach but not exceed the largest values that could be simulated by the model while retaining consistency with the thermodynamic constraints (i.e., larger perturbations resulted, due to limitations of the published models, in negative fluxes through reactions that are chemically essentially irreversible). Concentrations of ancillary compounds (not described by the ordinary differential equations of the model; e.g., 5mTHF) were assumed to be constant. The greatest change in the dynamics of the system occurs within 1 hour after each perturbation. Simulated data points were collected every 2.4 min over this 60 min period to supply the inversion algorithm with dynamically rich data for parameter identification. A total of 200 metabolite time points were used for data inversion (4 metabolites × 25 time points/perturbation × 2 perturbations). When fluxes (amounts of metabolites per unit volume enzymatically transformed per unit time; i.e., metabolite flows) were also used in the inversion, flux data for each of the five interconversions in the pathway were gathered at each time point as well. A relative error of 15% was assigned to each concentration and flux data point to simulate experimental noise. The typical reported error in metabolomic and fluxomic experimental studies ranges from ∼ 15% to > 50% (Ikeda et al., 1996; Ruijter and Visser, 1997) and sampling 25 time points per perturbation somewhat exceeds the time-resolution of the most intensive dynamic metabolomic studies to date (Brauer et al., 2006).
The inversion algorithm
A routine based on genetic algorithms (GAs) (Goldberg, 1989) was used for the model parameter identification. An initial population of the unknown parameter vectors was generated, with each parameter value randomly selected within the range of 10-2 to 105. The “fitness” score of each parameter vector in the population was determined by an objective function which measured how well the parameter vector generated the simulated data. The half of the population with the better fitness scores was retained and the rest were replaced by new parameter vectors generated through crossover and mutation of existing parameter vectors, with the total (parameter vector) population size held constant at 5000. The mating process operated as follows: two parent parameter vectors were chosen statistically with a probability derived from their respective fitness scores. Each element of each parent parameter vector had a probability Nr = 0.7 of exchanging values with the same element of the corresponding mate parameter vector (genetic recombination) and a probability Nm = 0.1 of being replaced with a random value determined by a Gaussian distribution (genetic mutation). Once the new population of parameter vectors was generated, the process of selection and mating continued until the objective function was minimized.
The objective function can take a variety of forms depending on the type of input data and the nature of the system. The objective here was to minimize the difference between data generated by the model (with the parameter values selected via the inversion algorithm) and the simulated laboratory data. The form of the objective function F used is given by Equation 1:
| (1) |
where Nt is the number of time points at which data was gathered, Neq is the number of species whose concentrations were measured, is the simulated laboratory concentration measurement for the j-th species at the i-th time point, εci,j is the laboratory error associated with the j-th species at the i-th time point (set here to 15%), is the concentration calculated by the model (using the GA-generated parameter vectors) for the j-th species at the i-th time point, Nflx is the number of system fluxes measured, is the j-th simulated laboratory flux measured at the i-th time point, is the laboratory error associated with the j-th flux at the i-th time point (set here to 15%); , the j-th system flux calculated by the model (using the candidate parameter vector) at the i-th time point; wc is a weight factor for the concentration data, and wflx is a weight factor for the flux data. The objective function was calculated by evaluating the following conditional for each data point: if the absolute difference between and falls within the error εci,j, then receives a score of 1; otherwise was scored using the continuous function that increases linearly from 1 as the deviation of from increases. The scores for each data point were summed and averaged over all time points and measured species/fluxes. For weighting factors related by wc + wflx = 1, F=1 is the minimum/optimal value. Candidate parameter sets that receive a fitness score F=1 were considered acceptable solutions. For the inversion of concentration data alone, wflx =0 and wc =1. For the inversion of both concentration and flux data, wflx =0.5 and wc =0.5.
Constraints describing properties of isolated enzymes
Biochemically constrained search ranges for select parameters were based on previously published data from in vitro kinetic studies performed for the various enzymes of the methionine cycle. The third column of Table 2 lists the constraints and relevant references from which they were derived. Here we provide a brief justification for them. The constant KMATIm for MATI was determined by Sullivan and Hoffman to be 41 ± 4 μM (1983). We selected a constraint of 41 ± 8 μM, with the additional breadth of the range selected to correct for potential in vitro versus in vivo differences in the Km. KMATIi was calculated based on a product inhibition study (Sullivan and Hoffman, 1983) which found that at [Met] = 25 μM and [AdoMet] ∼ 375 μM resulting in ∼ 50% inhibition of MATI activity. Based on the reaction rate formulation for VMATI in Table 1, this implies that the Ki for MATI is ∼ 230 μM. Experimental values but not error estimates for GNMT, KGNMTm and KGNMTi have been reported (Heady and Kerr, 1973; Kerr, 1972). Absent experimental error estimates for KMATIi, KGNMTm and KGNMTi, we assigned a constrained search range of ± 25% the nominal value. The Michaelis-Mentin constant KBHMTm for the BHMT reaction rate was independently determined by Finklestein (1972) and Garrow (1996) as 12 μM and 32 μM, respectively. We assigned a biologically constrained search range for this parameter of 9 - 40 μM (25% below the lower experimental estimate to 25% above the higher one).
Computational implementation
The ODE model of the methionine cycle was integrated using a stiff Gear integrator (Petzold and Hindmarsh, 1997). The genetic algorithm was from GAlib (Wall, 1995). The parameter distributions were generated as follows: identified parameter sets from three independent inversion runs were combined into a single data set with redundant copies of any particular identified parameter vector removed. For each parameter, the logarithmic parameter search range [-2,5] was subdivided into 50 equally sized bins of length 0.14. The frequency with which identified parameter values fell in each of the bins was determined, producing a distribution of outcomes along the range [-2,5]. Several thousands of parameter sets were identified in each run, which was sufficient to provide a converging distribution for each parameter.
Results
Diverse parameter sets reproduce simulated dynamic concentration data
The simulated concentration data shown in Figure 2 was supplied to the inversion algorithm to identify families of parameter vectors that reproduce the data. The algorithm identified over 30,000 parameter vectors consistent with the simulated concentration data within error estimates of ±15%. The resulting parameter values are plotted in the histograms of Figure 3 (blue lines; the green lines show analogous results with inclusion also of flux data and the red lines with further addition of biochemical constraints; see below). In these histograms, the Y-axis represents the fractional occurrence of particular parameter value ranges (shown on the X-axis) among the parameter vectors consistent with the simulated data. A sharp distribution in Figure 3 indicates that only a narrow range of the particular value could reproduce the simulated data of Figure 2. A spiky distribution indicates that a diversity of values are consistent with the data, but the likelihood is higher for certain parameter values. A broad, smooth distribution indicates that all (or very many) values of that parameter are acceptable.
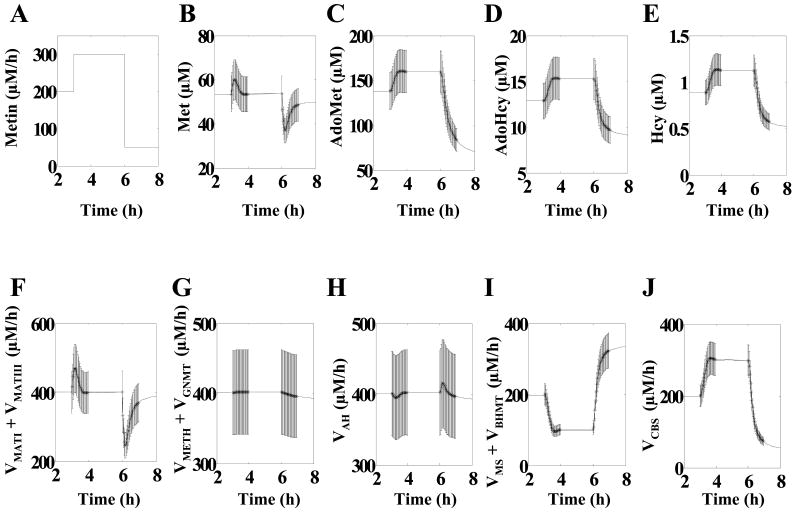
Figure 2. Simulated methionine flux perturbation and dynamic metabolic responses.
Data points used for parameter identification are surrounded by error bars of ±15% to simulate experimental noise. (A) Methionine influx (Metin). (B) Methionine concentration (Met). (C) S-Adenosylmethionine concentration (AdoMet). (D) S-Adenosylhomocysteine concentration (AdoHcy). (E) Homocysteine concentration (Hcy). (F) Flux from Met to AdoMet (VMATI + VMATIII). (G) Flux from AdoMet into AdoHcy (VMETH + VGNMT). (H) Flux between AdoHcy and Hcy (VAH). (I) Flux from Hcy to Met (VMS+VBHMT). (J) Flux from Hcy to cystathionine (VCBS).
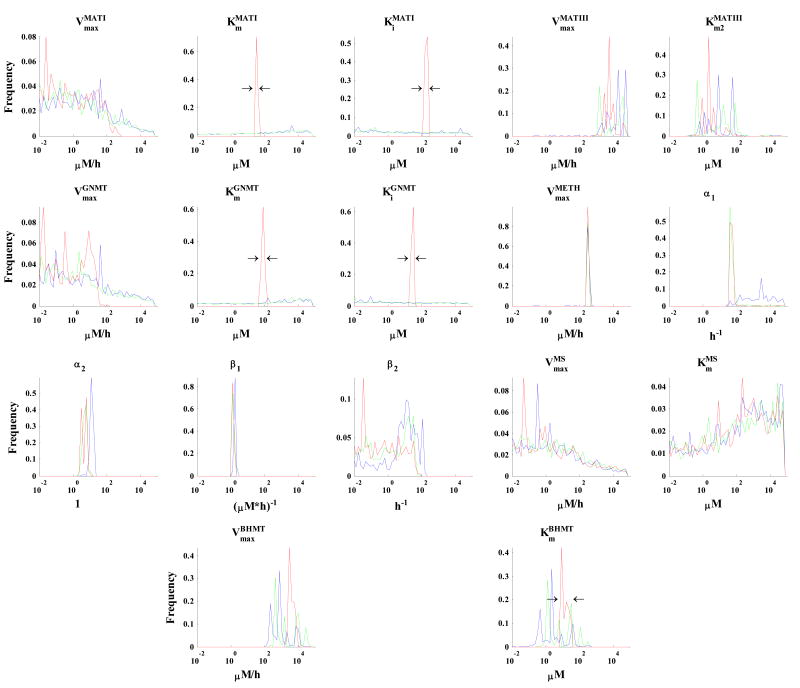
Figure 3. Histograms of parameter values that reproduce the dynamic data shown in Figure 2 within the error bars.
The results are from inversion of the simulated concentration data only (blue histograms), of both concentration and flux data (green histograms), and of both concentration and flux data with additional biochemical constraints on the search ranges for selected parameters (red histograms; constraints are shown by black arrows surrounding the red peaks for KMATIm, KMATIi, KGNMTm, KGNMTi, and KBHMTm; see Table 2 for exact ranges). For all parameters, the X-axis spans from 10-2 to 105. Global search was conducted over this 107-fold range, except for the indicated constraints mentioned above for the red histograms. The X-axis has been binned into 50 equally spaced segments of the parameter search range (on the log10 scale). The Y-axis values represent the frequency with which the identified parameter values fell within each bin.
The most notable feature of the blue lines in Figure 3 is the exceptional breadth of most of the parameter ranges, which cover the entire dynamic range of 107 for eight parameters (VMATImax, kMATIm, kMATIi, VGNMTmax, kGNMTm, kGNMTi, VMSmax, and kMSm) and exceeded a range of 103 for seven parameters (VMATIIImax, kMATIIIm2, VMETHmax, α1, β2, VBHMTmax, and kBHMTm). While some of the parameters in this latter category, such as VMETHmax, favor a smaller range of values, scattered cases demonstrate that other divergent values are acceptable. Only two parameters, α2 and β1, were identified to within a ∼ 10-fold range.
Correlations within identified parameter vectors explain the parameter value diversity
While indicating that divergent parameter vectors can produce similar kinetic behavior, the above results do not imply that random parameters selected from the identified individual parameter ranges can do so. Notably, we found that only 1 in 2×106 parameter vectors, when randomly sampled from the identified ranges, replicated the concentration data within the 15% error range. Thus, while any particular parameter can generally take on a huge diversity of values, the network behavior is conserved only for parameter vectors with highly correlated members.
To better understand the model features resulting in the remarkable diversity and correlation of feasible parameter values, we examined the roles of particular parameters in the ODE model. The parameters that exhibit the most diversity in identified values are associated with the enzymes MATI, GNMT, and MS. Each of these enzymes is involved in one of the three conversions of the methionine cycle that is catalyzed by multiple enzymes (Figure 1). Most of the identified parameter sets partition the fluxes for these conversions unequally such that MATI, GNMT, and MS have relatively minor flux contributions (with the major fluxes coming from MATIII, METH, and BHMT, respectively). Because the MATI, GNMT, and MS fluxes are small, changes in the values of the parameters associated with these reactions generally have little effect on the overall network behavior. Only parameter values which result in large fluxes through these enzymes are prohibited. Acceptably low fluxes are ensured either by keeping Vmax low or by offsetting increasing Vmax values with corresponding increases in Km. Figure 4A, 4B, and 4C show that the Vmax and Km combinations identified for MATI, MS, and GNMT indeed result in low fluxes through these enzymes: the black lines in each of these plots represents the parameter combinations which result in these enzymes producing 10% of the estimated total flux of the pathway step (and the alternative enzymes of MATIII, METH, or BHMT 90% of the flux). The vast majority of the parameter combinations for MATI, MS, and GNMT fall below this line and thus contribute <10% to the total flux. Thus, one reason for the diversity of acceptable parameter values is that, for pathway steps catalyzed by two different enzymes, if one enzyme contributes very little to the total flux, changes in the parameter values for that enzyme (as long as they do not increase its flux greatly) will have very little impact on the overall network behavior.
Figure 4. Correlations between parameter values.
Symbols indicate parameter combinations that reproduce the concentration data shown in Figure 2. MATI (A) and MATIII (D) catalyze the same reaction, as do MS (B) and BHMT (E). The conversion catalyzed by GNMT (C) is also catalyzed by METH, but no correlation plot shown for METH, as it is characterized by only a single parameter value. (A, B, C) Enzymes with minor fluxes. The solid dark lines correspond to parameter combinations producing 10% of the overall reaction flux of the reference model. For these enzymes, the observed parameter values are restricted to combinations of Vmax and Km that ensure low flux (< 10% of total). (D, E) Flux-dominant enzymes, for which Vmax and Km are roughly linearly correlated (R2 ≥ 0.95).
The parameter ranges for the flux-dominant enzymes, such as MATIII and BHMT, while better defined than for the minor flux enzymes, are nevertheless broad (most identified values fall within a 1000 to 10,000-fold range). One possible reason for such breadth could be correlated changes in two or more parameter values which offset each other to conserve the flux. For unsaturated enzymes described by Michaelis-Menten kinetics, paired increases in Vmax and Km offset, and the flux is unchanged. Both MATIII and BHMT are far from saturation in both the original model and most of the new parameter sets. As shown in Figure 4D and E, for each of these enzymes, Vmax and Km are strongly correlated (Pearson's R2 0.95 and 0.97 for MATIII and BHMT respectively). Thus, another reason for the diversity of acceptable parameter values is that, for unsaturated enzymes, correlated increases in Vmax and Km do not alter the enzymatic flux.
Identified parameter vectors generate similar concentration data predictions
We examined the reliability of the new models (using as their parameters the values identified by the inversion of the simulated concentration data) in making concentration and flux predictions for perturbations (of both methionine and homocysteine influxes, Figure 5A) different from those used for the parameter identification. The new models (grey lines), despite their high degree of parameter diversity, generally resulted in concentration predictions which closely mimicked the reference system (Figure 5B). However, certain fluxes, most notably the rate of homocysteine methylation (VMS + VBHMT), were not accurately predicted (Figure 5C). Equation (1) provides a quantitative metric of the predictive power of the new models with respect to concentration data (wc = 1,wflx = 0) and flux data (wc = 0,wflx = 1). Predictions that are accurate with ±15% error receive the optimal score of 1; worse predictions yield higher scores. The average score of the new models for concentration predictions was almost perfect (mean, 1.0043; SD, 0.0002). Conversely, the average score for flux predictions was far from perfect (23 ± 0.15). For comparison, random parameter vectors (not generated by inversion of the data shown in Figure 2) yielded scores of 280 ± 150 and 97 ± 3.3 for concentration and flux predictions, respectively. Hence, while the identified parameter sets had some predictive power for both concentrations and fluxes, they were much more accurate for concentrations.
Figure 5. Predictions of models that contain parameter vectors identified by inversion of the simulated data shown in Figure 2.
(A) Simulated methionine (Metin, black line) and homocysteine (Hcyin, blue line) influx perturbations used to generate the output shown in the other panels. (B) Metabolite concentration responses. (C, D) Homocysteine methylation reaction flux (VMS + VBHMT) response. In panels (B) – (D), blue lines show the output of the reference model; grey lines (N = 200) show the output of the models containing the newly identified parameter vectors. The parameter vectors used in (B) and (C) were identified by inversion of concentration data only, and in (D) by inversion of both concentration and flux data.
Inversion of flux data yields parameter vectors producing reliable flux predictions
Flux data from the perturbation simulation in Figure 2 (in addition to concentration data) was incorporated into the inversion procedure to further constrain the parameter search. The inversion algorithm identified >30,000 parameter vectors consistent with the simulated concentration and flux data to within 15% error (Figure 3, green). The predictive capability of the associated new models was examined using the methionine-homocysteine perturbations of Figure 5A. The concentration predictions were comparable to those of models trained on concentration data alone (see Figure 5B); however, the flux predictions were greatly improved (compare Figure 5C with Figure 5D). Quantitatively, the new models received scores for concentrations and fluxes of 1.0041 ± 0.0003 and 1.2 ±0.01, respectively.
The parameters identified using both flux and concentration data show a discernable reduction in the ranges of acceptable parameter values compared to parameters obtained from inversion of only concentration data (Figure 3). For VMATIIImax and VMETHmax, the breadth of identified distributions dropped ∼ 104 fold. This reduction in diversity results in parameter ranges that span a region of the parameter space richer in parameter sets that match the simulation data: ∼ 1 in 104 parameter vectors randomly sampled within these ranges regenerated the simulated concentration data from Figure 2 within the 15% error (compared to 1 in 2×106 parameter vectors from the broader ranges found by inversion of only concentration data).
As in the case of the parameters identified from concentration data alone, the diversity of flux and concentration-constrained parameters for MATI, GNMT, and MS, can be explained by the lax requirements on these enzymes to maintain < 10% flux contributions. Similarly, the identified parameters for MATIII and BHMT remain correlated in a manner that holds their associated enzyme fluxes constant, with the strength of the correlation enhanced by inclusion of the flux data (Pearson's R2 increase from 0.95 to 0.99 and from 0.97 to 0.998 for MATIII and BHMT respectively)
Inclusion of biochemical constraints
Even after including flux constraints, considerable diversity of identified parameters remained (Figure 3). A major source of the observed diversity arises from strong correlations between the individual parameters. This suggests that narrowing the ranges of selected parameter values (using data from literature on their in vitro biochemical properties) should also reduce the ranges of other parameters. We accordingly restricted the search ranges for certain parameters describing MATI, GNMT, and BHMT (see Table 2). As before, the inversion algorithm was supplied with concentration and flux data as in Figure 2. More than 3×104 parameter vectors that fit the simulated data well were obtained. The distributions of the resulting parameter values are shown in the red lines in Figure 3. The black arrows surrounding the plots for kMATIm, kMATIi, kGNMTm, kGNMTi, and kBHMTm indicate the biochemically-restricted boundaries of the search ranges. The biochemical restrictions on the six parameters somewhat limited the feasible ranges for the other parameters. Most notably, the distributions of both VMATImax and VGNMTmax obtained clear upper boundaries absent in the previous inversions (compare the red to the blue and green lines on Figure 3). Thus, incorporation of suitable biochemical data to restrict the parameter search may significantly reduce the diversity of the consistent parameter vectors. Interestingly, random parameter vectors selected within the parameter ranges obtained with the biochemical constraints were substantially more likely to yield models consistent with the simulation data with ∼ 1 in 103 random parameter vectors yielding results consistent with the simulated concentration data. Thus, the biochemically constrained parameter vectors lie within a region of parameter space that is especially rich in vectors yielding the desired model behavior.
Discussion
A series of recent modeling studies have made major strides towards identifying topological features of bionetworks that give rise to emergent properties such as the ability to sense and respond to environmental signals, maintain metabolic homeostasis, and form complex developmental patterns, all in a manner that is robust to environmental and genetic noise (Barkai and Leibler, 1997; Bhartiya et al., 2006; Brandman et al., 2005; Kollmann et al., 2005; Meir et al., 2002; Stelling et al., 2002; von Dassow et al., 2000; Wagner, 2005). A complement to understanding the topological features of bionetworks that lead to their qualitative properties is finding the precise parameter values which lead to specific quantitative phenotypes. This type of detailed quantitative understanding is of practical importance in that parameter values ultimately determine quantitative biochemical phenotypes such as bioreactor yields, insulin sensitivity, etc.
With a focus on the parameter identification problem and its importance in metabolic modeling, here we examined the ranges of specific model parameter values capable of producing particular quantitative behavior of a previously published methionine cycle model (Reed et al., 2004). Using a nonlinear global parameter identification algorithm, we found that remarkably broad ranges of parameter values were consistent with simulated metabolite concentration and flux data within modest error estimates. Such wide and irregular distributions are unlikely to be obtained from linear error propagation as occurs in traditional identification methods. Within these broad parameter value ranges, parameter sets consistent with published biochemical kinetics data were identified. This contrasts to previous methionine cycle models, which resorted to using kinetic constants inconsistent with biochemical literature for certain enzymes (notably for kMATIi and kGNMTm, see Table 1). The ability to rectify this deficiency points to the value of employing a global search algorithm for selecting more realistic metabolic model parameters.
The primary findings of the present work are that broad ranges of parameter values produce similar model behavior, and that models with highly divergent parameter values nevertheless can make similar quantitative predictions. Similar inferences were made recently for a diversity of systems biology models based on local sensitivity analysis of system response to parameter variations (Gutenkunst et al., 2007). Rather than using sensitivity analysis, our conclusions result directly from the wide and correlated parameter distributions identified by global inversion. Thus, they are a useful complement to those of Gutenkunst et al.
The results of both the present work and Gutenkunst et al. (2007) have clear implications for robustness: at some level, the methionine cycle model used here (Reed et al., 2004), as well as the models studied by Gutenkunst et al., are robust to parameter value variation. However, robustness generally implies similar behavior with parameter value variation in any direction, whereas Gutenkunst et al. showed that parameter value variation in only certain directions is tolerated without markedly altering the quantitative output of the model. Similarly, we find that highly divergent parameter values can produce similar quantitative model behavior, but only if certainly parameter value correlations are maintained. Thus, the breadth of the observed parameter distributions found here does not necessarily imply robustness in the traditional sense.
A basic question regarding our identification of such broad parameter value ranges via data inversion is whether similar results would be obtained with real, rather than simulated, data. The parameter identification here was aided by the fact that the model subjected to inversion had exactly the same form of the one generating the simulated data. This may not be the case in laboratory settings where there will generally be (1) unknown and/or un-modeled variables in the real network that impact the observed dynamics or (2) failure of the precise mathematical form of the flux equations to completely describe the actual chemical events occurring in the real biological system (due to complex enzymatic mechanisms, diffusion, etc.). In addition, a constraint in the present work was the need to limit the extent of perturbations to avoid non-physiological behavior of the model (e.g., thermodynamically infeasible fluxes). In experimental work, responses to more extreme perturbations might prove useful for better defining feasible parameter ranges. In addition, more extreme perturbations might highlight limitations to model predictive power.
The ability to generalize from the findings here also depends on the degree to which the methionine cycle is representative of a typical metabolic sub-network. Particular features of the methionine cycle found in some but not all other metabolic pathways include (1) the network's cyclic topology, (2) the presence of a large number of regulatory interactions, and (3) the existence of multiple enzymes catalyzing almost all of the pathway steps.
With the above caveats in mind, it is worthwhile to point out some key lessons for future modeling efforts that are suggested by the present results. One is that inclusion of additional types of data (in this case, the flux data), even when leading to only modestly better definition of the feasible parameter ranges, can nevertheless yield important gains in predictive power. Such gains may be hard to realize by simply increasing the amount of data of a given type, and support the application of multiple analytical methods in parallel experimentally.
Other lessons are suggested by the causes of the diversity of the parameter values found here. One cause was the ability of low-flux producing enzymes to adopt a broad range of parameter values while making minimal flux contributions. This suggests an approach to investigating metabolic networks in which one first experimentally identifies the flux-dominant enzymes (e.g., via gene knockout or siRNA experiments) and then focuses initial modeling efforts on them. Subsequent experiments in cells in which the flux-dominant enzymes have been knocked out could then be used to determine biochemically accurate parameters for the minor-flux enzymes.
Another cause of the parameter diversity was correlations between the flux parameters (typically Vmax and Km) for enzymes with dominant fluxes. Such correlations suggest that several of the mathematical flux formulations were over-parameterized, given the relatively narrow ranges of metabolite concentrations observed in the perturbation simulations, which precluded the enzymes from taking on the full range of Michaelis-Menten behaviors. When substrate ranges are limited (e.g., to a < 4-fold range), simple linear flux formulations (e.g., flux = k [substrate]) may be preferable to representations based on Michaelis-Menten kinetics or biochemical systems theory (Holmberg, 1982; Savageau, 1969; Savageau, 1991). Alternatively, as shown in the Results section, when in vitro kinetic data are available, they can be used to tightly constrain Km values in Michaelis-Menten formulations, eliminating the problem of dramatic swings in Vmax to accommodate widely changing values of Km.
A complementary approach to incorporation of diverse types of data is specifically tailoring of experimental perturbations and data sampling for senstitive parameter identification. This can be done through computational simulation of the extent of refinement of the feasible parameter ranges likely to be obtained with new data (Feng and Rabitz, 2006). One can imagine especially favorable results by coupling such computationally-guided experimental design to laboratory research in a closed-loop fashion.
Acknowledgments
The authors acknowledge the support of a National Science Foundation (NSF) Dynamic Data-Driven Application System–Small Multidisciplinary Research Projects (DDDAS-SMRP) grant (0540181), NSF CAREER grant (0643859), and Environmental Protection Agency STAR grant, as well as the National Institutes of Health Center for Quantitative Biology at Princeton University (P50 GM071508), the Beckman Foundation, and the American Heart Association.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Avila MA, Garcia-Trevijano ER, Martinez-Chantar ML, Latasa MU, Perez-Mato I, Martinez-Cruz LA, del Pino MMS, Corrales FJ, Mato JM. S-Adenosylmethionine revisited: its essential role in the regulation of liver function. Alcohol. 2002;27:163–167. doi: 10.1016/s0741-8329(02)00228-8. [DOI] [PubMed] [Google Scholar]
- Barkai N, Leibler S. Robustness in simple biochemical networks. Nature. 1997;387:913–917. doi: 10.1038/43199. [DOI] [PubMed] [Google Scholar]
- Bhartiya S, Chaudhary N, Venkatesh KV, Doyle FJ. Multiple feedback loop design in the tryptophan regulatory network of Escherichia coli suggests a paradigm for robust regulation of processes in series. Journal of the Royal Society Interface. 2006;3:383–391. doi: 10.1098/rsif.2005.0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandman O, Ferrett JE, Li R, Meyer T. Interlinked fast and slow positive feedback loops drive reliable cell decisions. Science. 2005;310:496–498. doi: 10.1126/science.1113834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauer MJ, Yuan J, Bennett BD, Lu WY, Kimball E, Botstein D, Rabinowitz JD. Conservation of the metabolomic response to starvation across two divergent microbes. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:19302–19307. doi: 10.1073/pnas.0609508103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke R, Smith AD, Jobst KA, Refsum H, Sutton L, Ueland PM. Folate, vitamin B-12, and serum total homocysteine levels in confirmed Alzheimer disease. Archives of Neurology. 1998;55:1449–1455. doi: 10.1001/archneur.55.11.1449. [DOI] [PubMed] [Google Scholar]
- Di Ventura B, Lemerle C, Michalodimitrakis K, Serrano L. From in vivo to in silico biology and back. Nature. 2006;443:527–533. doi: 10.1038/nature05127. [DOI] [PubMed] [Google Scholar]
- Edwards JS, Ibarra RU, Palsson BO. In silico predictions of Escherichia coli metabolic capabilities are consistent with experimental data. Nature Biotechnology. 2001;19:125–130. doi: 10.1038/84379. [DOI] [PubMed] [Google Scholar]
- Eftaxias A, Font J, Fortuny A, Fabregat A, Stuber F. Nonlinear kinetic parameter estimation using simulated annealing. Computers & Chemical Engineering. 2002;26:1725–1733. [Google Scholar]
- Feng XJ, Rabitz H. Optimal identification of biochemical reaction networks. Biophysical Journal. 2004;86:1270–1281. doi: 10.1016/S0006-3495(04)74201-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng XJ, Rabitz H, Turinici G, Le Bris C. A closed-loop identification protocol for nonlinear dynamical systems. Journal of Physical Chemistry A. 2006;110:7755–7762. doi: 10.1021/jp056189o. [DOI] [PubMed] [Google Scholar]
- Finkelstein JD, Harris BJ, Kyle WE. Methionine Metabolism in Mammals - Kinetic Study of Betaine-Homocysteine Methyltransferase. Archives of Biochemistry and Biophysics. 1972;153:320–4. doi: 10.1016/0003-9861(72)90451-1. [DOI] [PubMed] [Google Scholar]
- Fogel DB. What is evolutionary computation? IEEE Spectrum. 2000;37:26–32. [Google Scholar]
- Garrow TA. Purification, kinetic properties, and cDNA cloning of mammalian betaine-homocysteine methyltransferase. Journal of Biological Chemistry. 1996;271:22831–22838. doi: 10.1074/jbc.271.37.22831. [DOI] [PubMed] [Google Scholar]
- Glowinski J, Stocki J. Estimation of Kinetic-Parameters - Initial Guess Generation Method. Aiche Journal. 1981;27:1041–1043. [Google Scholar]
- Goldberg DE. Genetic algorithms in search, optimization, and machine learning. Addison-Wesley Pub. Co.; Reading, Mass: 1989. [Google Scholar]
- Gombert AK, Nielsen J. Mathematical modelling of metabolism. Current Opinion in Biotechnology. 2000;11:180–186. doi: 10.1016/s0958-1669(00)00079-3. [DOI] [PubMed] [Google Scholar]
- Gutenkunst RN, Waterfall JJ, Casey FP, Brown KS, Myers CR, Sethna JP. Universally sloppy parameter sensitivities in systems biology models. PLoS Comput Biol. 2007;3:e189. doi: 10.1371/journal.pcbi.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heady JE, Kerr SJ. Purification and Characterization of Glycine N-Methyltransferase. Journal of Biological Chemistry. 1973;248:69–72. [PubMed] [Google Scholar]
- Himmelblau DM, Jones CR, Bischoff KB. Determination of Rate Constants for Complex Kinetics Models. Industrial & Engineering Chemistry Fundamentals. 1967;6:539–543. [Google Scholar]
- Hollywood K, Brison DR, Goodacre R. Metabolomics: Current technologies and future trends. Proteomics. 2006;6:4716–4723. doi: 10.1002/pmic.200600106. [DOI] [PubMed] [Google Scholar]
- Holmberg A. On the Practical Identifiability of Microbial-Growth Models Incorporating Michaelis-Menten Type Nonlinearities. Mathematical Biosciences. 1982;62:23–43. [Google Scholar]
- Ikeda TP, Shauger AE, Kustu S. Salmonella typhimurium apparently perceives external nitrogen limitation as internal glutamine limitation. Journal of Molecular Biology. 1996;259:589–607. doi: 10.1006/jmbi.1996.0342. [DOI] [PubMed] [Google Scholar]
- Kerr SJ. Competing Methyltransferase Systems. Journal of Biological Chemistry. 1972;247:4248–&. [PubMed] [Google Scholar]
- Kirkpatrick S, Gelatt CD, Vecchi MP. Optimization by Simulated Annealing. Science. 1983;220:671–680. doi: 10.1126/science.220.4598.671. [DOI] [PubMed] [Google Scholar]
- Kollmann M, Lovdok L, Bartholome K, Timmer J, Sourjik V. Design principles of a bacterial signalling network. Nature. 2005;438:504–507. doi: 10.1038/nature04228. [DOI] [PubMed] [Google Scholar]
- Kutalik Z, Tucker W, Moulton V. S-system parameter estimation for noisy metabolic profiles using Newton-flow analysis. IET Systems Biology. 2007;1:174–180. doi: 10.1049/iet-syb:20060064. [DOI] [PubMed] [Google Scholar]
- Linga P, Al-Saifi N, Englezos P. Comparison of the Luus-Jaakola optimization and Gauss-Newton methods for parameter estimation in ordinary differential equation models. Industrial & Engineering Chemistry Research. 2006;45:4716–4725. [Google Scholar]
- Lu SC, Mato JM. Role of methionine adenosyltransferase and S-adenosylmethionine in alcohol-associated liver cancer. Alcohol. 2005;35:227–234. doi: 10.1016/j.alcohol.2005.03.011. [DOI] [PubMed] [Google Scholar]
- Lu WY, Kimball E, Rabinowitz JD. A high-performance liquid chromatography-tandem mass spectrometry method for quantitation of nitrogen-containing intracellular metabolites. Journal of the American Society for Mass Spectrometry. 2006;17:37–50. doi: 10.1016/j.jasms.2005.09.001. [DOI] [PubMed] [Google Scholar]
- Mangoni AA, Jackson SHD. Homocysteine and cardiovascular disease: Current evidence and future prospects. American Journal of Medicine. 2002;112:556–565. doi: 10.1016/s0002-9343(02)01021-5. [DOI] [PubMed] [Google Scholar]
- Martinov MV, Vitvitsky VM, Mosharov EV, Banerjee R, Ataullakhanov FI. A substrate switch: A new mode of regulation in the methionine metabolic pathway. Journal of Theoretical Biology. 2000;204:521–532. doi: 10.1006/jtbi.2000.2035. [DOI] [PubMed] [Google Scholar]
- Meir E, von Dassow G, Munro E, Odell GM. Robustness, flexibility, and the role of lateral inhibition in the neurogenic network. Current Biology. 2002;12:778–+. doi: 10.1016/s0960-9822(02)00839-4. [DOI] [PubMed] [Google Scholar]
- Moles CG, Mendes P, Banga JR. Parameter estimation in biochemical pathways: A comparison of global optimization methods. Genome Research. 2003;13:2467–2474. doi: 10.1101/gr.1262503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petzold LR, Hindmarsh AC. Livermore Solver for Ordinary Differential Equations. Lawrence Livermore National Laboratory; 1997. [Google Scholar]
- Polisetty P, Voit E, Gatzke E. Identification of metabolic system parameters using global optimization methods. Theoretical Biology and Medical Modelling. 2006;3:4. doi: 10.1186/1742-4682-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed MC, Nijhout HF, Sparks R, Ulrich CM. A mathematical model of the methionine cycle. Journal of Theoretical Biology. 2004;226:33–43. doi: 10.1016/j.jtbi.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Refsum H. Folate, vitamin B12 and homocysteine in relation to birth defects and pregnancy outcome. British Journal of Nutrition. 2001;85:S109–S113. [PubMed] [Google Scholar]
- Rodriguez-Fernandez M, Mendes P, Banga JR. A hybrid approach for efficient and robust parameter estimation in biochemical pathways. Biosystems. 2006a;83:248–265. doi: 10.1016/j.biosystems.2005.06.016. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Fernandez M, Egea JA, Banga JR. Novel metaheuristic for parameter estimation in nonlinear dynamic biological systems. Bmc Bioinformatics. 2006b;7 doi: 10.1186/1471-2105-7-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruijter GJG, Visser J. Determination of intermediary metabolites in Aspergillus niger (vol 25, pg 295, 1996) Journal of Microbiological Methods. 1997;30:171–171. [Google Scholar]
- Sauer U. Metabolic networks in motion: C-13-based flux analysis. Molecular Systems Biology. 2006 doi: 10.1038/msb4100109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savageau MA. Biochemical Systems Analysis .1. Some Mathematical Properties of Rate Law for Component Enzymatic Reactions. Journal of Theoretical Biology. 1969;25:365–&. doi: 10.1016/s0022-5193(69)80026-3. [DOI] [PubMed] [Google Scholar]
- Savageau MA. Biochemical Systems-Theory - Operational Differences among Variant Representations and Their Significance. Journal of Theoretical Biology. 1991;151:509–530. doi: 10.1016/s0022-5193(05)80367-4. [DOI] [PubMed] [Google Scholar]
- Schilling CH, Palsson BO. The underlying pathway structure of biochemical reaction networks. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:4193–4198. doi: 10.1073/pnas.95.8.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen M, Petti A, Aach J, D'Haeseleer P, Church G. Automated modelling of signal transduction networks. Bmc Bioinformatics. 2002;3 doi: 10.1186/1471-2105-3-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelling J, Klamt S, Bettenbrock K, Schuster S, Gilles ED. Metabolic network structure determines key aspects of functionality and regulation. Nature. 2002;420:190–193. doi: 10.1038/nature01166. [DOI] [PubMed] [Google Scholar]
- Sullivan DM, Hoffman JL. Fractionation and Kinetic-Properties of Rat-Liver and Kidney Methionine Adenosyltransferase Isozymes. Biochemistry. 1983;22:1636–1641. doi: 10.1021/bi00276a017. [DOI] [PubMed] [Google Scholar]
- Tsuchiya M, Ross J. Application of genetic algorithm to chemical kinetics: Systematic determination of reaction mechanism and rate coefficients for a complex reaction network. Journal of Physical Chemistry A. 2001;105:4052–4058. [Google Scholar]
- von Dassow G, Meir E, Munro EM, Odell GM. The segment polarity network is a robust development module. Nature. 2000;406:188–192. doi: 10.1038/35018085. [DOI] [PubMed] [Google Scholar]
- Wagner A. Circuit topology and the evolution of robustness in two-gene circadian oscillators. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:11775–11780. doi: 10.1073/pnas.0501094102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall M. The GAlib Genetic Algorithm Package (copyright 1995-1996 Massachusetts Institute of Technology; copyright 1996-1999 Matthew Wall) 1995 Available at http://lancet.mit.edu/ga.
- Yuan J, Fowler WU, Kimball E, Lu WY, Rabinowitz JD. Kinetic flux profiling of nitrogen assimilation in Escherichia coli. Nature Chemical Biology. 2006;2:529–530. doi: 10.1038/nchembio816. [DOI] [PubMed] [Google Scholar]