Abstract
In Saccharomyces cerevisiae, Rad52 protein plays an essential role in the repair of DNA double-stranded breaks (DSBs). Rad52 and its orthologs possess the unique capacity to anneal single-stranded DNA (ssDNA) complexed with its cognate ssDNA-binding protein, RPA. This annealing activity is used in multiple mechanisms of DSB repair: single-stranded annealing, synthesis-dependent strand annealing, and cross-over formation. Here we report that the S. cerevisiae DNA strand exchange protein, Rad51, prevents Rad52-mediated annealing of complementary ssDNA. Efficient inhibition is ATP-dependent and involves a specific interaction between Rad51 and Rad52. Free Rad51 can limit DNA annealing by Rad52, but the Rad51 nucleoprotein filament is even more effective. We also discovered that the budding yeast Rad52 paralog, Rad59 protein, partially restores Rad52-dependent DNA annealing in the presence of Rad51, suggesting that Rad52 and Rad59 function coordinately to enhance recombinational DNA repair either by directing the processed DSBs to repair by DNA strand annealing or by promoting second end capture to form a double Holliday junction. This regulation of Rad52-mediated annealing suggests a control function for Rad51 in deciding the recombination path taken for a processed DNA break; the ssDNA can be directed to either Rad51-mediated DNA strand invasion or to Rad52-mediated DNA annealing. This channeling determines the nature of the subsequent repair process and is consistent with the observed competition between these pathways in vivo.
In Saccharomyces cerevisiae, the repair of DNA double-stranded breaks (DSBs)3 is accomplished primarily by homologous recombination. Genes from the RAD52 epistasis group, including RAD50, RAD51, RAD52, RAD54, RAD55-57, RAD59, MRE11, XRS2, and RFA1, are responsible for this recombination-dependent DSB repair (1, 2). To repair a DSB, the DNA end is first processed to produce a 3′ single-stranded tailed duplex DNA. The ssDNA is then channeled into one of the many recombinational pathways, which can be further categorized into RAD51-dependent and -independent pathways.
RAD51-dependent recombination requires functions of RAD51, RAD52, RAD54, RAD55, RAD57, and RFA1 for efficient DNA repair (1). The central step of this pathway involves DNA strand invasion of homologous duplex DNA by the processed DSB complexed with Rad51 protein (3). DNA replication from the invading 3′-end replaces the genetic information missing from the broken dsDNA, and subsequent DNA pairing and resolution steps restore DNA integrity.
RAD51-independent recombination requires RAD52 (4) and is enhanced by RAD59 (5-7). In addition to the recombination genes MRE11, RAD50, XRS2, RAD52, and RAD59 (6, 8), this pathway also depends on MSH2, MSH3 (6, 9), RAD1, and RAD10 (10, 11). This RAD51-independent recombination is most easily assayed as DSB repair occurring between directly repeated sequences, by a mechanism termed single-stranded annealing (SSA) (8, 12). As implied, the central step of SSA is annealing between complementary single-stranded DNA (ssDNA) on either side of the DSB, followed by removal of heterologous tails and ligation of the nicks (1). In addition, RAD51-independent recombination can mediate gene conversion with or without cross-overs (5, 13), presumably via break-induced replication coupled to SSA (14).
Genetic studies show that the loss of either the RAD51-dependent or the RAD51-independent pathway only moderately compromises recombinational repair efficiency, whereas the loss of both pathways severely impairs repair (5, 15, 16). Those studies showed a compensatory increase in utilization of one pathway upon elimination of the other and suggested competition between the two pathways. Moreover, examination of RAD52-RAD59-dependent recombination revealed that deletion of RAD51, RAD54, RAD55, or RAD57 results in an elevated utilization of SSA (5, 7, 8, 17, 18); whereas deletion of SRS2, an inhibitor of the RAD51-dependent pathway, decreases SSA efficiency (6, 17). Both results indicate that RAD51-dependent recombination inhibits RAD52-RAD59-dependent recombination.
The Rad52 protein possesses two distinct biochemical functions: the abilities to mediate formation of the Rad51-ssDNA nucleoprotein filament (19-21) and to catalyze ssDNA annealing in the presence of RPA (replication protein A) (22, 23). The nucleoprotein complexes that participate in, and result from, each of these biochemical processes are different. The Rad51-ssDNA filament is a dynamic complex that forms after Rad51 transiently displaces both RPA and Rad52 from ssDNA (24) and that is needed for the homology search, DNA strand invasion, and DNA strand exchange steps essential in the RAD51-dependent pathways. On the other hand, annealing of complementary ssDNA requires the preservation of the Rad52-ssDNA complex, at least on a steady-state basis, rather than the Rad51 nucleoprotein filament.
RAD59 displays a wide range of behavior in RAD51-dependent recombination. In the absence of RAD59, heteroallelic recombination between interchromosomal homologous sequences can increase (16), slightly decrease (7), or remain unaffected (25). Because recombination in most assay systems is determined by both RAD51-dependent and -independent mechanisms and because deletion of RAD59 has a greater impact on the RAD51-independent pathways, its effect on the RAD51-dependent pathways is often not obvious. However, there are reasons to believe that RAD59 plays a role in the RAD51-dependent pathways. First, the last steps of the RAD51-dependent synthesis-dependent strand annealing (SDSA) pathway, which consist of DNA strand annealing, removal of heterologous or over-replicated 3′-ssDNA tails, and gap sealing, are noticeably similar to those of the SSA pathway (1, 2, 26). Proteins that are important for removal of heterologous tails in SSA, the Rad1-Rad10 endonuclease and the Msh2-Msh3 mismatch repair complex, are also important for removal of nonhomologous sequences during mating type switching, an SDSA-type of recombination (27). Second, in a strain defective for the flap endonuclease Rad27, which functions in Okazaki fragment maturation, all of the proteins of the RAD52 epistasis group, i.e. both Rad51 and Rad59, are required for viability (28-30). Third, in the rad52-R70K background where the functions of RAD52 are partially compromised, deletion of RAD59 causes a synergistic reduction in sporulation efficiency, spore viability, and mating type switch (16), all of which are dependent on RAD51. Finally, Rad59 interacts with Rad51 indirectly through Rad52 in immunoprecipitation experiments (31). Based on these observations, it was proposed that Rad59 augments the activity of Rad52 protein in both the RAD51-dependent and -independent pathways (5, 16, 31, 32).
In this report, we demonstrate that Rad51 inhibits Rad52-dependent DNA annealing in vitro not simply by indirect competition for ssDNA, but through a direct interaction with Rad52 itself. However, by working in concert with Rad52, Rad59 partially alleviates the inhibitory effect of Rad51 on DNA annealing. We suggest that this counteracting regulation of Rad52-dependent DNA annealing function by Rad51 and Rad59 determines the biochemical steps that are used for DSB recombinational repair and may serve to enhance the fidelity of second end capture by annealing.
EXPERIMENTAL PROCEDURES
DNA Substrates—Oligonucleotides were purchased from Operon and were purified using denaturing polyacrylamide gel electrophoresis (7 m urea, 9% polyacrylamide, 19:1 cross-linking in 1× Tris-borate-EDTA (TBE) buffer (89 mm Tris borate, pH 8.3, 2 mm EDTA)). Sequences for the 48-nucleotide annealing substrates ssDNA (W) and ssDNA (C) are 5′-GCAATTAAGCTCTAAGCCATCCGCAAAAATGACCTCTTATCAAAAGGA-3′ and 5′-TCCTTTTGATAAGAGGTCATTTTTGCGGATGGCTTAGAGCTTAATTGC-3′, respectively. Concentration of ssDNA (W) and ssDNA (C) was determined using the extinction coefficient of 1 × 104 and 9.6 × 103 m-1·cm-1, respectively. Sequences for the 100-nucleotide heterologous ssDNA (PB78) and its complementary strand (PB77) are 5′-TGGCCTGCAACGCGGGCATCCCGATGCCGCCGGAAGCGAGAAGAATCATAATGGGGAAGGCCACCAGCCTCGCGTCGCGAACGCCAGCAAGACGTAGCCC-3′ and 5′-GGGCTACGTCTTGCTGGCGTTCGCGACGCGAGGCTGGTGGCCTTCCCCATTATGATTCTTCTCGCTTCCGGCGGCATCGGGATGCCCGCGTTGCAGGCCA-3′, respectively. The concentrations of PB78 and PB77 were determined using the extinction coefficient of 9.7× 103 and 8.9 × 103 m-1·cm-1, respectively. To generate the heterologous dsDNA, two complementary oligonucleotides of equal concentration were mixed together in 10 mm Tris-HCl (pH 7.5), 50 mm NaCl, 10 mm MgCl2, and 1 mm dithiothreitol. The mixture was heated at 100 °C for 5 min and then allowed to cool to room temperature slowly over a 2-h period.
Proteins—Rad51 (33) and RPA (66) were purified as described. Rad52 was purified as described (19) except that the Superose-12 column was substituted with a Superdex-200 column (GE Healthcare). C-terminally His6-tagged Rad59 was purified as described (37) except that a purification step over ssDNA cellulose was added. In brief, the Rad59 that eluted from Ni2+-charged chelating Sepharose was dialyzed against MDEG buffer (25 mm K-MES, pH 6.5, 1 mm dithiothreitol, 1 mm EDTA, and 10% glycerol) containing 100 mm NaCl. The protein was loaded onto an ssDNA-cellulose column, washed with 200 mm NaCl in MDEG, and eluted in the 300 and 400 mm NaCl steps. This fraction was then dialyzed and subjected to Q-Sepharose and heparin-Sepharose chromatography as described previously. Because the ssDNA-cellulose step purifies on the basis of ssDNA affinity, the specific activity of Rad59 is ∼2-fold greater than the previous preparation. Escherichia coli RecA was purified as described (67) and was provided by Dr. Roberto Galletto in our laboratory. E. coli SSB protein was purified as described (68). Human Rad51 (hRad51) protein was purified as described (69) and was provided by Dr. Anthony Forget in our laboratory. T4 polynucleotide kinase was purchased from New England Biolabs. Proteinase K was purchased from Roche Applied Science.
DNA Annealing—Unless otherwise indicated, DNA annealing reactions were carried out at 30 °C essentially as described (23) using complementary oligonucleotide 25 (ssDNA (W)) and oligonucleotide 26 (ssDNA (C)) (200 nm each); oligonucleotide 25 was labeled at 5′-end. In the control reaction where a protein was omitted, an equal amount of corresponding protein storage buffer was added instead. Where Rad59 is present, Rad52 and Rad59 were preincubated on ice for at least 15 min before adding to the reaction. For reactions without RPA, ssDNA (W) was first incubated with Rad51 (67 nm) in DNA annealing buffer (30 mm Tris OAc, pH 7.5, 5 mm Mg(OAc)2, and 1 mm dithiothreitol) in the presence of 1 mm ATP at 30 °C for 5 min. Rad52 (20 nm) was added and incubated for another 5 min before addition of ssDNA (C) to initiate the reaction. For reactions containing RPA, the two oligonucleotides were first incubated with RPA (30 nm) for 5 min in DNA annealing buffer supplemented with 1 mm ATP prior to the addition of Rad51 (134 nm). After incubation for 5 min, Rad52 (40 nm) and/or Rad59 (80 nm) were added to initiate the reaction. For each time point, an aliquot was withdrawn into 0.8 volume of stop buffer (1.5% SDS, 10 mm EDTA, 4 mg/ml proteinase K, and 10 μm unlabeled ssDNA (W)) and was incubated for another 15 min at 30 °C. The samples were analyzed by native polyacrylamide gel electrophoresis (either 6% in 1× TBE or 10% in 0.5× TBE buffer; cross-linking ratio 19:1), and the gels were dried onto DEAE paper (Whatman DE81). The extent of DNA annealing was visualized and quantified using a Storm 860 system (Molecular Dynamics) or a Personal FX phosphorimaging device (Bio-Rad).
Pulldown Assay—Protein-protein interactions between Rad51 and Rad59 were studied using pulldown assays with Ni-NTA magnetic beads (Qiagen) as described (34). Unless otherwise indicated, the reaction mixtures contained 1 μm each of Rad51 and Rad59 proteins in interaction buffer (50 mm Tris-HCl, pH 7.5, 50 mm NaCl, 30 mm imidazole, 0.2% Triton X-100). The reactions were preincubated for 15 min at 37 °C before Ni-NTA magnetic beads were added to final concentration of 1%. The beads were separated from the solution phase using a Qiagen “12-Tube Magnet”. Aliquots (20 μl) from each assay mixture containing unbound proteins were analyzed by 11% SDS-PAGE. The beads were washed three times with 300 μl of interaction buffer to remove unbound proteins. Proteins bound to the beads were eluted with two washes of 20 μl of elution buffer (150 mm Tris-HCl, pH 7.5, 100 mm NaCl, and 300 mm imidazole) and were analyzed using 11% SDS-PAGE.
RESULTS
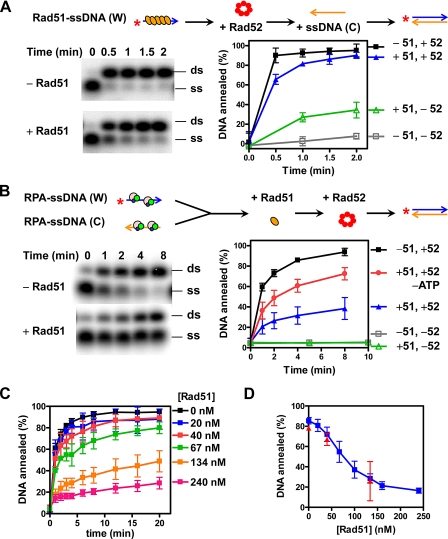
Rad51 Inhibits Rad52-promoted DNA Annealing of ssDNA Complexed with RPA—Because genetic analyses suggested that Rad51 may block RAD52-RAD59-mediated recombinational repair, we determined the effect of Rad51 on Rad52-promoted DNA annealing in vitro. We used complementary oligonucleotide substrates and analyzed annealing by native polyacrylamide gel electrophoresis (23). First, we examined DNA annealing in the absence of RPA. We observed that Rad51 itself also promotes DNA annealing, but much more slowly than Rad52-mediated DNA annealing (Fig. 1A); in addition, we noticed that the presence of Rad51 slightly decreased the rate of Rad52-mediated DNA annealing. In the absence of ATP, Rad51 was unable to anneal DNA or inhibit Rad52-mediated annealing (data not shown). Because ATP binding is essential for the binding of Rad51 to ssDNA (33), we tentatively concluded that both of these activities reflected a property of the Rad51 nucleoprotein filament.
FIGURE 1.
Rad51 inhibits Rad52-mediated DNA annealing. The reactions were conducted as described under “Experimental Procedures” and are illustrated schematically at the top of the figure. Rad51 inhibits Rad52-mediated annealing of complementary ssDNA (A) and RPA-ssDNA complexes (B). A representative gel of Rad52-promoted DNA annealing is shown in the left panel, and the quantification is shown in the right panel. The reactions contained Rad52 alone (-51, +52; black squares), both Rad51 and Rad52 (+51, +52; blue triangles), neither Rad51 nor Rad52 (-51, -52; gray squares), and Rad51 alone (+51, -52; green triangles). ATP was omitted from the experiment represented by red circles. The results are the averages obtained from at least three independent experiments, and the error bars represent one standard deviation (where absent, the error bars are smaller than the symbol). C, time course of Rad52-mediated DNA annealing as a function of Rad51 concentration (gels not shown). The extent of DNA annealing at 4 min is plotted versus the Rad51 concentration and is shown in D as blue squares; the red triangles are from experiments where Rad51 and Rad52 were incubated together and added simultaneously. The results are the averages obtained from at least two independent experiments, and the error bars represent the variation.
RPA plays an important role in recombinational DSB repair; therefore, it was included in all reactions hereafter. We observed that RPA blocked both spontaneous and Rad51-promoted annealing (Fig. 1B, gray squares and green triangles, respectively) but not Rad52-promoted DNA annealing (black squares) (22, 23), showing that Rad51 does not displace RPA from these short ssDNA oligonucleotides in these experiments. When Rad52-promoted annealing of the RPA-ssDNA complex was examined in the presence of Rad51, the rate of annealing was reduced (blue triangles). Because we anticipated that the binding of Rad51 to ssDNA could inhibit its annealing by Rad52, we also examined annealing in the absence of ATP. Omitting ATP in the reaction greatly, but not completely, alleviated the inhibition (red circles), suggesting that ATP and/or ssDNA binding by Rad51 is needed for the inhibitory effect.
The reduction of DNA annealing was dependent on Rad51 concentration (Fig. 1, C and D, blue squares). As the concentration of Rad51 was increased, the rate of DNA annealing progressively decreased. Where the rate was not inhibited severely (≤67 nm), the amount of dsDNA was nonetheless approaching completion, showing that only the rate of annealing was reduced but not the yield of product. The apparent affinity (Kd) of Rad51 for Rad52 as defined by this assay is ∼100 nm.To eliminate the possibility that Rad51 was displacing RPA from the ssDNA during the 5-min incubation step prior to Rad52 addition and that it was directly competing with Rad52 for ssDNA binding, Rad51 and Rad52 were preincubated and added together to initiate the reaction; the same inhibitory effect was observed (Fig. 1D, red triangles). Finally, the inhibitory effect of Rad51 was the same with a different pair of oligonucleotide substrates of unrelated sequence or plasmid-length ssDNA (data not shown), showing that the effect of Rad51 on Rad52-promoted DNA annealing is not sequence-specific. Thus, we conclude that Rad51 inhibits the DNA annealing activity of Rad52; however, these experiments do not define the mechanism of inhibition, which is addressed in the following sections.
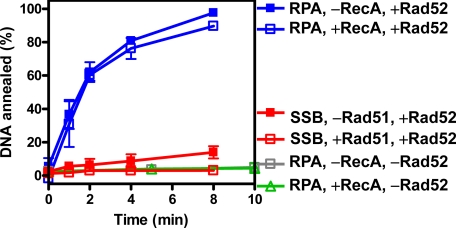
Inhibition of Rad52-mediated DNA Annealing by Rad51 Is Species-specific—Both DNA annealing promoted by Rad52 and RPA, and DNA strand exchange promoted by Rad51, Rad52, and RPA require species-specific interactions (19, 23, 24). To determine whether Rad51 inhibited Rad52-mediated DNA annealing through species-specific interactions, the reactions were carried out with Rad51 or RPA replaced by their corresponding counterparts from E. coli, RecA, or SSB proteins (Fig. 2).
FIGURE 2.
Inhibition of Rad52-dependent DNA annealing by Rad51 is species-specific. The reactions were carried out as described under “Experimental Procedures” except RecA (134 nm, blue lines) or SSB (50 nm, red lines) was substituted for Rad51 or RPA, respectively. Control reactions containing RPA only or RPA and RecA are shown as gray squares and green triangles, respectively. The results are the averages obtained from two independent experiments, and the error bars represent the variation.
RecA did not inhibit Rad52-mediated annealing of RPA-coated ssDNA (Fig. 2, blue lines, compare filled and open squares). The annealing observed was not being promoted by the RecA, because RPA blocked RecA-mediated annealing completely (green triangles). To eliminate the concern that RecA may not be fully active at 30 °C, the experiments were repeated at 37 °C, and the same results were obtained (data not shown). Therefore, RecA does not repress Rad52-mediated DNA annealing. These results mirror our previous observations that RPA inhibits RecA nucleoprotein filament formation and that Rad52 does not enable RecA to overcome this inhibition (24).
SSB does not interact with Rad52, nor does it fully permit Rad52-mediated DNA annealing (23). When SSB was substituted for RPA, a slow but measurable rate of ssDNA annealing was detected (Fig. 2, red filled squares). However, in contrast to the results with RecA, this residual level of DNA annealing was blocked by Rad51 to the background level (Fig. 2, red open squares). Previously, we established that Rad52 does not facilitate Rad51 nucleoprotein filament formation on SSB-coated ssDNA (24); hence, the inhibition of DNA annealing that we see here (in the presence of SSB) is not due to Rad51 nucleoprotein filament formation. Therefore, efficient DNA annealing by Rad52 is dependent on the specific interaction between Rad52 and RPA, whereas inhibition by Rad51 requires Rad52 but not RPA. Therefore, our collective results show that Rad51 does not block Rad52-promoted DNA annealing by indirect competition via displacement of RPA (or Rad52) from ssDNA; instead, Rad51 controls DNA annealing via direct species-specific interactions with Rad52.
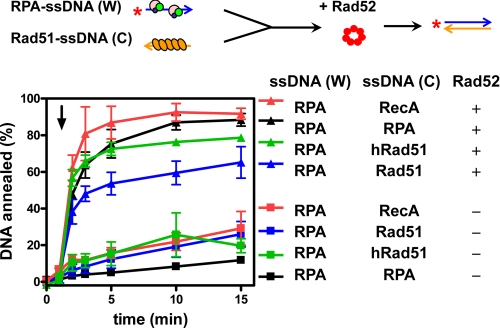
Rad52 Promotes DNA Annealing between RPA-ssDNA and Rad51-ssDNA Complexes, but at a Reduced Rate—In the previous experiments, DNA annealing was examined using only RPA-coated ssDNA. However, it is possible that one of the ssDNA substrates for DNA annealing in vivo is bound by Rad51 protein. To mimic such a potential physiological reaction, we carried out DNA annealing with RPA bound to one of the complementary ssDNA substrates, and Rad51 bound to the other (Fig. 3). For these reactions, RPA-ssDNA and Rad51-ssDNA complexes were preassembled separately, and then Rad52 was added (triangles); in all cases, DNA annealing was accelerated relative to the Rad52-free control (squares). However, DNA annealing was the slowest when yeast Rad51 was present (blue triangles), faster with human Rad51 (green triangles), and fastest with either RecA or RPA (red or black triangles, respectively). These results are consistent with our results in the previous section showing that the cognate Rad51 is the most potent inhibitor of Rad52, and they further establish that that the closely related human Rad51 is capable of a weaker but significant inhibition. These hierarchy of inhibition is consistent with our previous studies with the RecA-loading domain of E. coli RecBCD enzyme (34), which established that interaction with the cognate DNA strand exchange protein, RecA, was the strongest, but other prokaryotic and eukaryotic DNA strand exchange proteins bound to the RecA-loading domain with reduced affinities that paralleled phylogenetic distance.
FIGURE 3.
Rad52-promoted DNA annealing between RPA-ssDNA complexes and Rad51-ssDNA filament is reduced. Protein-ssDNA complexes were formed by separately incubating RPA (30 nm) with 5′-32P-ssDNA (W) (400 nm), and the protein indicated with ssDNA (C) (400 nm) in DNA annealing buffer with 1 mm ATP for 5 min. Annealing was initiated by mixing equal volumes of the two complexes, and then Rad52 was added at 1 min (triangles). The reactions lacking Rad52 are shown as squares. The reactions containing either E. coli RecA (134 nm), RPA (30 nm), human Rad51 (134 nm), and yeast Rad51 (134 nm) are represented by the red, black, green, and blue symbols, respectively. The results are the averages obtained from at least two independent experiments, and the error bars represent reaction variation.
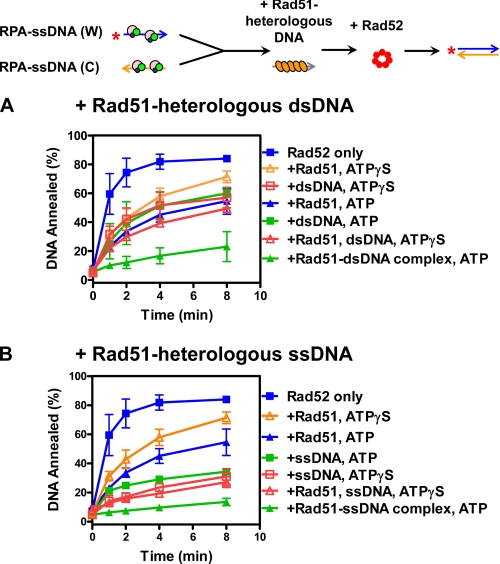
The Rad51 Nucleoprotein Filament Is the Strongest Inhibitor of Rad52-mediated DNA Annealing—The reduction of Rad52-mediated DNA annealing by Rad51 protein in the presence of ATP described above (Fig. 1) could be easily explained if Rad51 simply displaced Rad52 (and RPA) from the ssDNA to form a Rad51-ssDNA filament (24). In doing so, Rad52 would be excluded from the ssDNA and, consequently, DNA annealing by Rad51 would be inhibited by RPA (Fig. 1). However, we noticed that even though Rad52 could anneal DNA when one partner was complexed with Rad51 (Fig. 3), the rate of annealing was slower than when an equivalent amount of free Rad51 was present. This finding suggested to us that Rad51-ssDNA might be a more potent inhibitor of Rad52 function. To test this possibility, the annealing of RPA-ssDNA complexes was re-examined, but this time in the presence of Rad51 nucleoprotein filaments that were assembled on DNA (either dsDNA or ssDNA) that was heterologous to the annealing substrates and added in trans (Fig. 4).
FIGURE 4.
The Rad51 nucleoprotein complex is the most effective inhibitor of Rad52-mediated DNA annealing. The experiments were carried out as described under “Experimental Procedures” except that in A Rad51 was preassembled on heterologous dsDNA (400 nm) at 37 °C for 5 min before addition to the RPA-ssDNA complexes. DNA was 100 bp in length. B, experiments were carried out as in A except that Rad51 was preassembled on heterologous ssDNA (400 nm). DNA was 100-nucleotides in length. In both panels, Rad52 only (blue squares) and with Rad51 (blue triangles), heterologous DNA (green squares), and Rad51-DNA complex (green triangles) with ATP are shown. The experiments carried out in the presence of ATPγS in lieu of ATP are represented by the open symbols: +Rad51, orange triangles; +heterologous DNA, red squares; and +Rad51 and DNA (both), red triangles. The results are the averages obtained from at least two independent experiments, and the error bars represent the variation (where absent, the error bars are smaller than the symbol).
As surmised, the heterologous Rad51 nucleoprotein filament added in trans inhibited Rad52-mediated DNA annealing between the two complementary RPA-ssDNA complexes considerably more effectively than an equivalent concentration of free Rad51. In the case of dsDNA, a Rad51 nucleoprotein filament assembled on a 100-bp duplex (Fig. 4A, green triangles) was at least 4-fold more potent than the free Rad51 (blue triangles). Although the free dsDNA reduced the annealing effectiveness of Rad52, because of direct competition and binding of Rad52, inhibition by the Rad51 nucleoprotein filament was greater than that of either the Rad51 alone or the dsDNA alone (green squares). Thus, inhibition by the trans heteroduplex Rad51-DNA complex eliminates the possibility that Rad51 is simply displacing Rad52 from the complementary oligonucleotides.
Rad51 nucleoprotein filaments assembled on heterologous ssDNA (Fig. 4B, green triangles) behaved similarly to the filaments assembled on dsDNA. However, not unexpectedly, the free ssDNA was a more potent inhibitor of Rad52 activity than free dsDNA; both Rad52 and RPA bind more tightly to ssDNA than to dsDNA, making the ssDNA an effective titrator of these proteins. Nevertheless, when compared with the Rad51 alone (blue triangles) or the ssDNA alone controls (green squares), the ATP-Rad51-ssDNA complex is the most effective inhibitor of DNA annealing. Thus, sequestration of Rad52 by free ssDNA is insufficient to explain the inhibition by the Rad51 nucleoprotein complexes and is fully consistent with inhibition being exerted by the Rad51-complex itself.
To further confirm these observations, experiments were conducted in the presence of ATPγS instead of ATP (Fig. 4, A and B, orange triangles and red squares and triangles); ATPγSis a nonhydrolyzable ATP analog that compromises the ssDNA binding ability of Rad51 (33, 35, 36). In the presence of ATPγS, inhibition by mixtures of Rad51 and DNA showed no difference from that observed by the free DNA, within experimental errors, consistent with the expectation that Rad51-DNA complexes did not form. Therefore, the repressive effect of Rad51 protein does not rely on complex formation with the complementary ssDNA. Furthermore, these experiments using Rad51 nucleoprotein filaments in trans show that inhibition cannot be attributed to a simple competition with Rad52 for the ssDNA substrates that will be annealing. Thus, the Rad51 nucleoprotein filament is the most effective and direct controller of Rad52 annealing function.
Rad59 Alleviates the Inhibitory Effect of Rad51 in Rad52-promoted Annealing of RPA-ssDNA Complexes—Deletion of RAD59 results in a significant reduction in SSA efficiency in vivo (6, 7, 32), but only a moderate stimulating effect of Rad59 protein was observed in Rad52-mediated DNA annealing in vitro (37). Therefore, we examined whether Rad59 could help Rad52 overcome the inhibitory effect of Rad51. These experiments were conducted in the absence of heterologous DNA to avoid the complexities in interpretation that would be associated with sequestration of either Rad59 or Rad52 by the heterologous DNA. Fig. 5 shows that free Rad51 increased the time required to reach 50% annealing (the half-time) for Rad52-mediated DNA annealing by ∼16-fold (black open squares, ∼1 min, versus blue open triangles, ∼16 min). When Rad59 was added to the Rad51-inhibited reaction, the rate of DNA annealing increased ∼2-fold; the reaction half-time deceased to ∼7 min (blue solid triangles). There was no further stimulation when the concentration of Rad59 was doubled (blue diamonds). However, when the Rad59 concentration was reduced by one-half, no stimulation was observed (data not shown). Therefore, Rad59 exerted its maximal effect when present at a 1:1 ratio with Rad52. In the absence of Rad51, Rad59 had no detectable effect on DNA annealing mediated by Rad52 (black squares). This result is also consistent with our previous findings that Rad59 did not stimulate Rad52 when reaction conditions were optimal (37) and that stimulation by Rad59 is revealed only when Rad52 functions at suboptimal conditions. Rad59 alone promoted only a low level of annealing of RPA-complexed ssDNA (green filled circles), as reported previously (37), but the residual annealing was also repressed by Rad51 (green open circles). Thus, although also subject to repression itself, Rad59 mitigates the inhibition of Rad52-mediated annealing of RPA-ssDNA that is effected by interaction with Rad51.
FIGURE 5.
Rad59 alleviates the inhibitory effect of Rad51 in Rad52-mediated DNA annealing. Experiments were carried out as described under “Experimental Procedures.” RPA-complexed ssDNA was preincubated without or with Rad51 (134 nm), and the reactions were initiated by the addition of Rad52 (40 nm) and/or Rad59 (40 or 80 nm). The reactions either lacking or containing Rad51 are shown in black and blue, respectively. The reaction containing Rad52 alone (black open squares), Rad51 followed by Rad52 (blue open triangles), Rad51 followed by Rad52 and 40 nm Rad59 (blue solid triangles), and Rad51 followed by Rad52 and 80 nm Rad59 (blue diamonds) are shown. The results are the averages obtained from at least three independent experiments, and the error bars represent one standard deviation. Control reactions containing Rad59 only (80 nm; green solid circles), Rad51 and Rad59 (green open circles), and Rad52 and Rad59 (black solid squares) are shown. The results are the averages obtained from at least two independent experiments, and the error bars represent the variation (where absent, the error bars are smaller than the symbol).
Rad51 and Rad59 Weakly Interact with Each Other in Vitro—We observed that Rad59-mediated DNA annealing is inhibited by Rad51, either in the presence (Fig. 5) or absence of RPA,4 and that Rad59 could partially overcome the inhibitory effect of Rad51 in Rad52-mediated DNA annealing (Fig. 5). Even though an interaction between Rad51 and Rad59 proteins was not detected in vivo previously (31), our observations prompted us to suspect that these two proteins might nevertheless interact weakly.
To determine whether purified Rad51 and Rad59, in the absence of DNA, interact with each other, a pulldown assay using Ni-NTA magnetic beads was performed, taking advantage of the hexahistidine tag at the C terminus of Rad59 protein (Fig. 6). An excess amount of both proteins relative to bead capacity was used. After extensive washing, proteins that were still bound to the beads were eluted with imidazole. The eluted proteins (lanes 5-8) and unbound proteins (lanes 1-4) were then analyzed using SDS-PAGE followed by Coomassie Blue staining. No Rad51 was retained on the beads when Rad59 was omitted (lane 5). However, in the presence of Rad59, Rad51 was detected in the bound fraction (lanes 7 and 8), indicating complex formation. The interaction was not strong, because only 13% of Rad51 relative to the Rad59 was pulled down (lane 7), and this interaction was further weakened (to 5%) by increasing NaCl concentration to 300 mm (lane 8). However, our result is comparable with the report that human Rad51 and the N terminus of human Rad52 (which is the domain that is similar to Rad59) also interact weakly, showing only ∼10% of level of complex formation that was measured for Rad51 and the full-length Rad52 (38).
FIGURE 6.
Rad51 directly interacts with Rad59 protein in vitro. Pulldown experiments using magnetic Ni-NTA beads were carried out in the absence of DNA as described under “Experimental Procedures.” Proteins that remained free in the solution are shown in the left panel (unbound). Proteins retained on the beads were eluted by 300 mm imidazole and are shown in the right panel (bound). NaCl concentration in the initial binding reaction is indicated at the top.
DISCUSSION
In this report, we establish that Rad51 controls an important function of Rad52 by blocking Rad52-mediated DNA annealing. Rad51 acts via a direct interaction with Rad52 rather than by simply displacing Rad52 from the complementary ssDNA. Inhibition requires a species-specific interaction between Rad51 and Rad52 and is much more pronounced for the ATP-Rad51-DNA nucleoprotein complex. Interestingly, we also discovered that Rad59 can partially overcome the inhibitory effect of Rad51, likely because of the ability of Rad59 to interact with Rad52 and possibly Rad51. Our results are consistent with genetic findings that the RAD52, RAD59-dependent SSA is repressed in a RAD51-dependent manner (5, 7, 8). Furthermore, our observations offer insight into the need for both Rad52 and Rad59 in RAD51-dependent recombination at post-synaptic steps. Collectively, these findings suggest that Rad51 controls recombination outcome by a previously unappreciated mechanism. They show that free Rad51 can exert a global control by interacting with Rad52 to limit the overall level of DNA annealing by Rad52 and suggest that the Rad51 nucleoprotein filament may exerts a more local control by repressing Rad52-mediated DNA annealing in its vicinity, e.g. at sites such as single-ended joint molecules, as discussed below.
Rad51 Inhibits Rad52-mediated DNA Annealing by Directly Interacting with Rad52—Rad51 impedes Rad52-promoted annealing of complementary ssDNA, regardless of whether the ssDNA substrates are naked, RPA-complexed, or complexed with RPA and Rad51 on either strand. One function of Rad52 is to accelerate Rad51-dependent displacement of RPA from ssDNA and facilitate Rad51 nucleoprotein filament assembly (19-21, 24). Nucleoprotein filament formation by Rad51 could have been sufficient to block Rad52-mediated DNA annealing. However, a number of lines of experimental evidence show that displacement of RPA and Rad52 from the ssDNA by Rad51 cannot explain the mechanism by which Rad51 acts. Compared with Rad52-mediated DNA annealing, which is complete within a few minutes, Rad51 nucleoprotein filament assembly is relatively slow, taking at least 5-10 min even in the presence of Rad52 to displace RPA from ssDNA completely (24); thus, the displacement possibility is too slow to explain the inhibition we see here. Moreover, because the Rad51 filament also can promote DNA annealing, displacement of Rad52 or RPA to form Rad51 nucleoprotein filaments should have resulted in rapid DNA annealing. However, the most compelling evidence is that the preassembly of Rad51 nucleoprotein filaments on heterologous DNA in trans also inhibited Rad52-promoted DNA annealing, showing directly that a Rad51-DNA complex formed in trans is sufficient to control Rad52 annealing function. Our collective results therefore show that Rad51 inhibits the DNA annealing activity of Rad52 by interacting directly with the Rad52-ssDNA complex. The precise mechanism of inhibition remains to be determined, but the binding of Rad51 may modify the oligomeric state of Rad52, or it may hinder the binding of a Rad52-ssDNA complex to complementary ssDNA, thereby blocking DNA annealing.
We observed that Rad52-dependent DNA annealing is repressed by free Rad51 ∼16-fold and by Rad51 nucleoprotein filaments more than 60-fold. Rad59 mitigated by ∼2-fold this inhibitory effect. It is known that Rad59 interacts with Rad52 in vivo (32) and that it enhances the DNA annealing function of Rad52 in vitro (37). The ability of Rad59 to partially rescue DNA annealing could be a consequence of its capacity to interact with Rad52 and, in so doing, make Rad52 less susceptible to inhibition by Rad51. Another possibility, which is not mutually exclusive, is that Rad59 could act by binding to Rad51 and thereby block Rad51 from interacting with Rad52. Consistent with the latter idea, we demonstrated a weak but direct interaction between Rad59 and Rad51 in vitro. Direct interaction between Rad51 and Rad59 was not observed in vivo in the absence of Rad52 (31), perhaps because Rad52 is required for Rad59 nuclear localization (39). Rad59 shares sequence homology with the N terminus of Rad52 but lacks the C-terminal Rad51-interaction domain. Hence, the conserved DNA annealing domain of Rad52 and Rad59 may contain an additional site for Rad51 interaction. These two mechanisms for stimulation by Rad59 do not preclude one another and both may function in vivo. However, because 1) the interaction between Rad52 and Rad59 is stronger than that between Rad51 and Rad59, 2) increasing the Rad59 concentration does not further alleviate the inhibition by Rad51, and 3) mitigation by Rad59 saturates at a 1:1 molar ratio with Rad52, it is more likely that Rad59 largely functions through its interaction with Rad52. Thus, by showing that Rad59 counteracts the impediment imposed by Rad51 on DNA annealing, our observations provide a biochemical basis for the enhancement of recombination by RAD59 via RAD52-dependent annealing in vivo.
Controlling Rad52-mediated DNA Annealing by Rad51 Provides a Surveillance Mechanism That Favors High Fidelity Repair of DNA Damage—Compared with RAD52-dependent SSA, RAD51-dependent recombination repairs DNA with higher fidelity, in part, because it requires longer sequence homology (6, 17), reducing the chance of recombination between imperfect homeologous sequences (40). Therefore, it would be more advantageous for cells to utilize the RAD51-dependent pathway for DSB repair and use SSA only when repair by the RAD51-dependent pathway is not possible. Consistent with this view, our results suggest that the pathway utilized for DSB repair is under the control of Rad51 protein itself; formation of Rad51 filament not only favors repair by the pathway using Rad51-mediated DNA strand invasion, but it also reduces the effectiveness of repair by the pathway using ssDNA annealing by inhibiting Rad52-mediated DNA annealing.
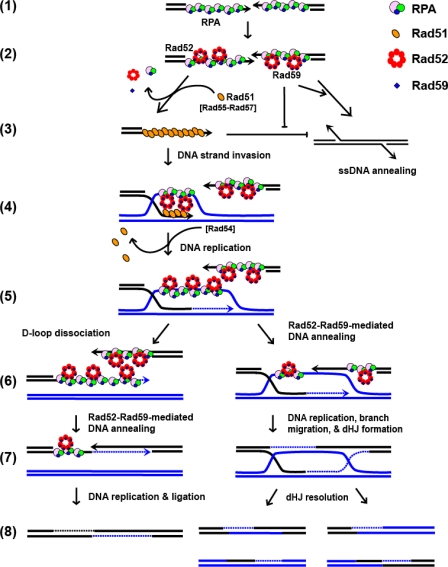
All homologous recombination pathways share a common initiation step: resection of dsDNA to generate 3′-terminated ssDNA, which is subsequently bound by RPA (Fig. 7, step 1). This step is followed by the binding of Rad52, and we would add Rad59 at this step (step 2). The Rad52-Rad59-RPA-ssDNA complex can participate in either Rad51-ssDNA filament formation or Rad52-mediated DNA annealing (step 3). These two choices are in competition with one another, as is another subsequent choice (step 6).
FIGURE 7.
A model for selection among the homologous recombination pathways of DSB repair. The RAD51-dependent and -independent recombination pathways are represented by two biochemical reactions: Rad51-mediated DNA strand exchange and Rad52-mediated DNA annealing. Both pathways share the common step of DSB resection and RPA binding to the 3′-ssDNA tails (step 1). The species-specific interaction between RPA and Rad52 protein recruits Rad52 and Rad59 to the RPA-ssDNA complex (step 2). In the RAD51-independent pathway (step 3, right arrow), Rad52 promotes annealing of RPA-ssDNA with a complementary sequence from the other end of the processed DSB. Rad59 plays an important role at this step by enhancing DNA annealing activity of Rad52 and counteracting the inhibitory effect of Rad51 protein. In the RAD51-dependent pathway (step 3, left arrow), with the help by Rad52 (and Rad55-Rad57), Rad51 displaces RPA and Rad52 from ssDNA to form the presynaptic complex; formation of the Rad51 nucleoprotein filament strongly inhibits Rad52-mediated DNA annealing. DNA strand invasion and exchange with homologous DNA duplex follow (step 4). Next, Rad51 protein is stripped off DNA by Rad54 protein, and DNA replication initiates from the invading strand (step 5). After the newly synthesized DNA is unwound from the displacement-loop (D-loop) intermediate (step 6, left), it anneals to the second processed end of the DSB in a Rad52-dependent manner, aided by Rad59 (step 7, left). Further DNA synthesis, branch migration, cleavage, and ligation complete repair of the DSB (step 8, left). Alternatively, the second end of the DSB can be directly annealed to the D-loop by Rad52-Rad59 proteins (step 6, right). After DNA replication, branch migration, and DNA ligation, the double Holliday junction (dHJ) structure is formed (step 7, right). Resolution of the double Holliday junction structure completes repair of the DSB (step 8, right).
The pathway choice is also likely moderated by other cellular proteins. For example, disassembly of the Rad51-ssDNA filament is controlled by the Srs2 helicase (41, 42), and Rad52-mediated annealing between divergent homologous sequences is subjected to heteroduplex rejection by the Sgs1 helicase (43, 44). The interconnections between these two processes can also explain why deletion of RAD54, which stabilizes Rad51-ssDNA filament formation (45) and promotes DNA strand invasion (36, 46), also increases SSA efficiency (8). Therefore, the competition is a dynamic and reversible process, and the pathway chosen for DSB repair is likely determined by DNA target availability, as well as the availability of the required multi-protein complexes. Based on this model, we propose that RAD59 modulates the biochemical pathway utilized; Rad59 enhances DNA annealing by Rad52, thereby increasing the chance of repair via SSA.
Rad52-59 May Play an Important Role at Post-synaptic Steps of the RAD51-dependent Recombination—Many recombination proteins have been reported to promote ssDNA annealing in vitro, including RecA (47), Rad51 (48), Rad52 (22, 23, 49), Rad59 (50), and Mre11 (51). However, Rad52 is the only protein from S. cerevisiae that can promote DNA annealing in the presence of RPA (22, 23, 37). Importantly, neither of the DNA strand exchange proteins, E. coli RecA (52) and S. cerevisiae Rad51 (this study), can anneal ssDNA that is complexed with an ssDNA-binding protein. Nevertheless, DNA annealing is an important step in the RAD51-dependent pathways. After DNA strand invasion (Fig. 7, step 4) and DNA replication (step 5), the second processed DSB needs to engage to complete DNA repair. In the SDSA model, the newly synthesized DNA is unwound from the D-loop intermediate (step 6, left), and capture of the second end of DSB is proposed to be via a DNA annealing-dependent mechanism (step 7, left). Alternatively, in the DSBR model, the second end of the DSB can be directly annealed to the D-loop intermediate to form a double Holliday junction structure (53, 54) (step 6, right). The strand that is displaced from the D-loop is bound by RPA (55), and the second end of the processed DSB will be coated with either RPA or Rad51. Given that Rad52 is the only known protein to anneal RPA-bound ssDNA, we believe that an essential function of Rad52 is to capture the second end by DNA annealing (54).
It is at step 4 that control by the Rad51 nucleoprotein filament can be exerted locally; until Rad51 is removed from the DNA heteroduplex by Rad54 (56) to permit DNA replication (57), second end capture by DNA annealing is not possible. It is during this time that the ssDNA displaced from the single-ended invasion joint molecule could be prevented from participating in Rad52-mediated annealing with ssDNA that is only partly complementary. Given that DNA annealing has lower sequence stringency than joint molecule formation, this repression might exist as a fidelity control mechanism to limit spurious annealing of the joint molecule intermediate with partially homologous ssDNA sequences during the time before DNA synthesis from the joint molecule. Removal of Rad51 and the subsequent DNA synthesis from the joint molecule would permit annealing over a potentially longer displaced ssDNA target (steps 5 and 6, right). In SDSA, we would imagine that DNA annealing is enabled by removal of Rad51 as part of the D-loop dissociation step (steps 5 and 6, left). The remaining alternative, not illustrated, is DNA strand invasion by the other processed DNA end in a second Rad51-mediated event that would require neither DNA replication nor Rad52-mediated DNA annealing as a prerequisite.
Because Rad59 alleviates the inhibitory effect of Rad51 and facilitates Rad52-promoted DNA annealing, we believe that RAD59 also plays a role at the post-synaptic step of the RAD51-dependent recombination. By counteracting the inhibitory effect of Rad51, Rad59 helps promote a more efficient second end capture. Genetically, deletion of RAD59 has little effect on RAD51-dependent recombination. It is possible that the absence of Rad59 only delays but does not affect the outcome of DSB repair events, as was shown previously for mating type switching (16). Alternatively, the cell might employ overlapping mechanisms to overcome the negative effect of Rad51. For example, the Rad51 nucleoprotein filament might be disrupted by the Srs2 helicase (41, 42) so that second end capture could be promoted by Rad52-dependent DNA annealing without the interference from Rad51. Deletion of SRS2 selectively inhibits the noncross-over DSB repair pathway, indicating that a later step, but not the initial strand invasion step, is affected in the srs2Δ mutant (58). The rad59 srs2 double mutant shows a synergistic reduction in DSB repair via RAD51-dependent recombination (6, 17), showing that when avenues of Rad51 control and Rad52 augmentation are eliminated, the RAD51-dependent pathway is severely impaired.
The presynaptic role of Rad52 in facilitating Rad51-mediated DNA strand invasion is well documented in the literature. In contrast, little is known about the post-synaptic role of Rad52. Because deletion of RAD52 abolishes mitotic recombination at the strand invasion step (59, 60), it is difficult to examine its role at a later step using a genetic approach. However, considerable evidence suggests that the function of Rad52 in the RAD51-dependent recombination pathway is not limited to the presynaptic stage. In contrast to the rad52 null mutant, truncation of the C-terminal Rad51-interacting domain of Rad52 (61) or simply removal of the essential four amino acid residues in the rad52Δ409-412 mutation (62) partially compromises homologous recombination. This defect is only partially suppressed by RAD51 overexpression (63), implying that deletion of RAD52 affects an essential function that is downstream of presynaptic filament formation, e.g. second end capture by annealing or repair by synthesis-dependent strand annealing. Also, in cytological studies, Rad52 foci were found to persist at DSB sites after Rad51 foci disassembled (64, 65), suggesting that Rad52 was also functioning after the presumptive DNA strand invasion step. Finally, recent biochemical analysis showed that Rad52 can anneal a “second” ssDNA to the ssDNA displaced from a joint molecule, which was formed by Rad51-mediated DNA strand exchange in the presence of RPA (54). These observations support our view that the DNA annealing function of the Rad52-Rad59 is important at the post-synaptic stage of DSBR, as well as in SSA and SDSA. Previously, we established that Rad52 is the functional counterpart of the E. coli RecO protein and suggested that the annealing of ssDNA complexed with the cognate ssDNA-binding protein is a universally conserved step of recombinational DNA repair (53). Here we extend this idea to show that DNA annealing is controlled by interaction with Rad51. In further support of our views, we have also observed that RecA inhibits RecO-promoted DNA annealing.5 These collected observations further reinforce our hypothesis that DNA annealing is a regulated step of recombinational DNA repair that is coordinated with the action of a DNA strand exchange protein, suggesting a mechanism that is conserved throughout evolution.
Acknowledgments
We thank Drs. Wolf-Dietrich Heyer and Neil Hunter for fruitful discussions and comments on the manuscript, and we are also grateful to Andrei Alexeev, Ichiro Amitani, Aura Carreira, Clarke Conant, Cynthia Haseltine, Jovencio Hilario, Amitabh Nimonkar, Behzad Rad, Edgar Valencia-Morales, Jason Wong, and Liang Yang for critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant AI-18987 (to S. C. K.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: DSB, double-stranded break; ssDNA, single-stranded DNA; SSA, single-stranded annealing; SDSA, synthesis-dependent strand annealing; MES, 4-morpholineethanesulfonic acid; Ni-NTA, nickel-nitrilotriacetic acid; dsDNA, double-stranded DNA; ATPγS, adenosine 5′-O-(thiotriphosphate).
Y. Wu, N. Kantake, T. Sugiyama, and S. C. Kowalczykowski, unpublished observations.
E. Valencia-Morales and S. C. Kowalczykowski, unpublished observations.
References
- 1.Pâques, F., and Haber, J. E. (1999) Microbiol. Mol. Biol. Rev. 63 349-404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Symington, L. S. (2002) Microbiol. Mol. Biol. Rev. 66 630-670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bianco, P. R., Tracy, R. B., and Kowalczykowski, S. C. (1998) Front Biosci. 3 D570-D603 [DOI] [PubMed] [Google Scholar]
- 4.Sugawara, N., and Haber, J. E. (1992) Mol. Cell. Biol. 12 563-575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bai, Y., and Symington, L. S. (1996) Genes Dev. 10 2025-2037 [DOI] [PubMed] [Google Scholar]
- 6.Sugawara, N., Ira, G., and Haber, J. E. (2000) Mol. Cell. Biol. 20 5300-5309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jablonovich, Z., Liefshitz, B., Steinlauf, R., and Kupiec, M. (1999) Curr. Genet 36 13-20 [DOI] [PubMed] [Google Scholar]
- 8.Ivanov, E. L., Sugawara, N., Fishman-Lobell, J., and Haber, J. E. (1996) Genetics 142 693-704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sugawara, N., Paques, F., Colaiacovo, M., and Haber, J. E. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 9214-9219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ivanov, E. L., and Haber, J. E. (1995) Mol. Cell. Biol. 15 2245-2251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aguilera, A. (1995) Curr. Genet. 27 298-305 [DOI] [PubMed] [Google Scholar]
- 12.Wu, X., Wu, C., and Haber, J. E. (1997) Genetics 147 399-407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rattray, A. J., and Symington, L. S. (1995) Genetics 139 45-56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kang, L. E., and Symington, L. S. (2000) Mol. Cell. Biol. 20 9162-9172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cortes-Ledesma, F., Malagon, F., and Aguilera, A. (2004) Genetics 168 553-557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bai, Y., Davis, A. P., and Symington, L. S. (1999) Genetics 153 1117-1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ira, G., and Haber, J. E. (2002) Mol. Cell. Biol. 22 6384-6392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsukamoto, M., Yamashita, K., Miyazaki, T., Shinohara, M., and Shinohara, A. (2003) Genetics 165 1703-1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.New, J. H., Sugiyama, T., Zaitseva, E., and Kowalczykowski, S. C. (1998) Nature 391 407-410 [DOI] [PubMed] [Google Scholar]
- 20.Shinohara, A., and Ogawa, T. (1998) Nature 391 404-407 [DOI] [PubMed] [Google Scholar]
- 21.Sung, P. (1997) J. Biol. Chem. 272 28194-28197 [DOI] [PubMed] [Google Scholar]
- 22.Shinohara, A., Shinohara, M., Ohta, T., Matsuda, S., and Ogawa, T. (1998) Genes Cells 3 145-156 [DOI] [PubMed] [Google Scholar]
- 23.Sugiyama, T., New, J. H., and Kowalczykowski, S. C. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 6049-6054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sugiyama, T., and Kowalczykowski, S. C. (2002) J. Biol. Chem. 277 31663-31672 [DOI] [PubMed] [Google Scholar]
- 25.Davis, A. P., and Symington, L. S. (2004) Mol. Cell. Biol. 24 2344-2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nassif, N., Penney, J., Pal, S., Engels, W. R., and Gloor, G. B. (1994) Mol. Cell. Biol. 14 1613-1625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pâques, F., and Haber, J. E. (1997) Mol. Cell. Biol. 17 6765-6771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Debrauwere, H., Loeillet, S., Lin, W., Lopes, J., and Nicolas, A. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 8263-8269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Symington, L. S. (1998) Nucleic Acids Res. 26 5589-5595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loeillet, S., Palancade, B., Cartron, M., Thierry, A., Richard, G. F., Dujon, B., Doye, V., and Nicolas, A. (2005) DNA Repair 4 459-468 [DOI] [PubMed] [Google Scholar]
- 31.Davis, A. P., and Symington, L. S. (2003) DNA Repair 2 1127-1134 [DOI] [PubMed] [Google Scholar]
- 32.Davis, A. P., and Symington, L. S. (2001) Genetics 159 515-525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zaitseva, E. M., Zaitsev, E. N., and Kowalczykowski, S. C. (1999) J. Biol. Chem. 274 2907-2915 [DOI] [PubMed] [Google Scholar]
- 34.Spies, M., and Kowalczykowski, S. C. (2006) Mol. Cell 21 573-580 [DOI] [PubMed] [Google Scholar]
- 35.Namsaraev, E. A., and Berg, P. (1998) Biochemistry 37 11932-11939 [DOI] [PubMed] [Google Scholar]
- 36.Mazin, A. V., Bornarth, C. J., Solinger, J. A., Heyer, W. D., and Kowalczykowski, S. C. (2000) Mol. Cell 6 583-592 [DOI] [PubMed] [Google Scholar]
- 37.Wu, Y., Sugiyama, T., and Kowalczykowski, S. C. (2006) J. Biol. Chem. 281 15441-15449 [DOI] [PubMed] [Google Scholar]
- 38.Kagawa, W., Kurumizaka, H., Ikawa, S., Yokoyama, S., and Shibata, T. (2001) J. Biol. Chem. 276 35201-35208 [DOI] [PubMed] [Google Scholar]
- 39.Lisby, M., Mortensen, U. H., and Rothstein, R. (2003) Nat. Cell Biol. 5 572-577 [DOI] [PubMed] [Google Scholar]
- 40.Spell, R. M., and Jinks-Robertson, S. (2003) Genetics 165 1733-1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krejci, L., Van Komen, S., Li, Y., Villemain, J., Reddy, M. S., Klein, H., Ellenberger, T., and Sung, P. (2003) Nature 423 305-309 [DOI] [PubMed] [Google Scholar]
- 42.Veaute, X., Jeusset, J., Soustelle, C., Kowalczykowski, S. C., Le Cam, E., and Fabre, F. (2003) Nature 423 309-312 [DOI] [PubMed] [Google Scholar]
- 43.Sugawara, N., Goldfarb, T., Studamire, B., Alani, E., and Haber, J. E. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 9315-9320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spell, R. M., and Jinks-Robertson, S. (2004) Genetics 168 1855-1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mazin, A. V., Alexeev, A. A., and Kowalczykowski, S. C. (2003) J. Biol. Chem. 278 14029-14036 [DOI] [PubMed] [Google Scholar]
- 46.Petukhova, G., Stratton, S., and Sung, P. (1998) Nature 393 91-94 [DOI] [PubMed] [Google Scholar]
- 47.Weinstock, G. M., McEntee, K., and Lehman, I. R. (1979) Proc. Natl. Acad. Sci. U. S. A. 76 126-130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim, H. K., Morimatsu, K., Norden, B., Ardhammar, M., and Takahashi, M. (2002) Genes Cells 7 1125-1134 [DOI] [PubMed] [Google Scholar]
- 49.Mortensen, U. H., Bendixen, C., Sunjevaric, I., and Rothstein, R. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 10729-10734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Petukhova, G., Stratton, S. A., and Sung, P. (1999) J. Biol. Chem. 274 33839-33842 [DOI] [PubMed] [Google Scholar]
- 51.de Jager, M., Dronkert, M. L., Modesti, M., Beerens, C. E., Kanaar, R., and van Gent, D. C. (2001) Nucleic Acids Res. 29 1317-1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McEntee, K. (1985) Biochemistry 24 4345-4351 [DOI] [PubMed] [Google Scholar]
- 53.Kantake, N., Madiraju, M. V., Sugiyama, T., and Kowalczykowski, S. C. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 15327-15332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sugiyama, T., Kantake, N., Wu, Y., and Kowalczykowski, S. C. (2006) EMBO J. 25 5539-5548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eggler, A. L., Inman, R. B., and Cox, M. M. (2002) J. Biol. Chem. 277 39280-39288 [DOI] [PubMed] [Google Scholar]
- 56.Solinger, J. A., Kiianitsa, K., and Heyer, W. D. (2002) Mol. Cell 10 1175-1188 [DOI] [PubMed] [Google Scholar]
- 57.McIlwraith, M. J., Vaisman, A., Liu, Y., Fanning, E., Woodgate, R., and West, S. C. (2005) Mol. Cell 20 783-792 [DOI] [PubMed] [Google Scholar]
- 58.Ira, G., Malkova, A., Liberi, G., Foiani, M., and Haber, J. E. (2003) Cell 115 401-411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Aylon, Y., Liefshitz, B., Bitan-Banin, G., and Kupiec, M. (2003) Mol. Cell. Biol. 23 1403-1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sugawara, N., Wang, X., and Haber, J. E. (2003) Mol. Cell 12 209-219 [DOI] [PubMed] [Google Scholar]
- 61.Boundy-Mills, K. L., and Livingston, D. M. (1993) Genetics 133 39-49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Krejci, L., Song, B., Bussen, W., Rothstein, R., Mortensen, U. H., and Sung, P. (2002) J. Biol. Chem. 277 40132-40141 [DOI] [PubMed] [Google Scholar]
- 63.Milne, G. T., and Weaver, D. T. (1993) Genes Dev. 7 1755-1765 [DOI] [PubMed] [Google Scholar]
- 64.Miyazaki, T., Bressan, D. A., Shinohara, M., Haber, J. E., and Shinohara, A. (2004) EMBO J. 23 939-949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lisby, M., Barlow, J. H., Burgess, R. C., and Rothstein, R. (2004) Cell 118 699-713 [DOI] [PubMed] [Google Scholar]
- 66.Kantake, N., Sugiyama, T., Kolodner, R. D., and Kowalczykowski, S. C. (2003) J. Biol. Chem. 278 23410-23417 [DOI] [PubMed] [Google Scholar]
- 67.Mirshad, J. K., and Kowalczykowski, S. C. (2003) Biochemistry 42 5937-5944 [DOI] [PubMed] [Google Scholar]
- 68.LeBowitz, J. (1985) Biochemical Mechanism of Strand Initiation in Bacteriophage Lambda DNA Replication. Ph. D. thesis, Johns Hopkins University, Baltimore, MD
- 69.Bugreev, D. V., and Mazin, A. V. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 9988-9993 [DOI] [PMC free article] [PubMed] [Google Scholar]