Summary
The bacterial flagellum and the highly related injectisome (or needle complex) are among the most complicated multi-protein structures found in Gram-negative microorganisms. The assembly of both structures is dependent upon a type III secretion system. An interesting regulatory feature unique to these systems is the coordination of gene expression with type III secretory activity. This means of regulation ensures that secretion substrates are expressed only when required during the assembly process or upon completion of the fully functional structure. Prominent within the regulatory scheme are secreted proteins and type III secretion chaperones that exert effects on gene expression at the transcriptional and post-transcriptional levels. Although the major structural components of the flagellum and injectisome systems are highly conserved, recent studies reveal diversity in the mechanisms used by secretion substrates and chaperones to control gene expression.
Keywords: type III secretion, flagella, gene regulation, type III chaperone
Introduction
Gram-negative bacteria utilize at least six distinct secretion pathways to transport proteins across the inner and/or outer membranes of the cell envelope [1,2]. One of those pathways, the type III secretion system (T3SS), can be divided into two major classes, flagellar and non-flagellar. The flagellar T3SS is associated with the MS ring of the basal body and is responsible for secreting the extracytoplasmic components of the flagellum [3]. The non-flagellar T3SS is associated with the bacterial injectisome, which translocates effector proteins into the cytoplasm of eukaryotic host cells to promote the pathogenic/symbiotic lifestyle of the microorganism [4,5]. Both systems use secretion competency as a signal to coordinate gene expression [6]. In the case of the flagellum, coordinating gene expression with secretory activity ensures that structural components of the flagellum are expressed only when required during the different stages of flagellar assembly. In contrast, coupling gene expression to secretory activity in the injectisome systems provides a mechanism for sensing environmental stimuli such as contact of the bacterium with a eukaryotic target cell. The basic strategy involves a secretable protein and its cognate T3SS-specific chaperone. Secretion competency is detected by cells in one of two ways; (i) sensing a reduction in the cytoplasmic concentration of the secretable protein, (ii) sensing the presence of the newly released chaperone. The absence or presence of the secretable protein and/or chaperone influences gene expression at the transcriptional and post-transcriptional levels. In this review we discuss the variety of ways in which some secretion substrates and type III secretion chaperones couple gene expression to secretory activity.
Coupling secretory activity to flagellar assembly
The bacterial flagellum is assembled in a sequential manner involving more than 30 gene products and has been studied most extensively in Salmonella enterica serovar Typhimurium [3,7-9]. Flagellar genes are generally divided into early, middle, and late (expressed from class I, II, and III promoters, respectively) based upon their temporal expression patterns. The early genes (flhDC) encode the transcriptional activator for class II promoters. The middle genes encode the MS-, C-, P-, and L-rings, T3SS, rod, and hook proteins that together form the hook-basal body (HBB) complex spanning the inner membrane. Following assembly of the basal body, the rod and hook proteins are secreted by the T3SS and assemble at the distal end of the growing structure. Completion of the HBB complex triggers two related events. First, there is a switch in the substrate specificity of the T3SS from rod/hook proteins to the late secretion substrates (i.e., flagellin monomer, capping protein) which are required for the final stages of flagellar assembly [8]. The second event, a shift from middle to late gene expression, is mediated by the type III secretion chaperones FliT, FliA, and FlgN, and is intimately linked to secretion of their cognate secretion substrates.
Down-regulation of middle gene expression
FliT is the chaperone for the filament capping protein FliD [10]. Upon completion of the HBB complex FliD is secreted to the tip of the hook where it facilitates polymerizaton of the flagellar filament [11]. The depletion of FliD from the cytoplasm permits FliT to bind to FlhC, thereby inhibiting transcription of the middle genes whose products are no longer required for the assembly process (Fig. 1A-B) [12].
Figure 1.
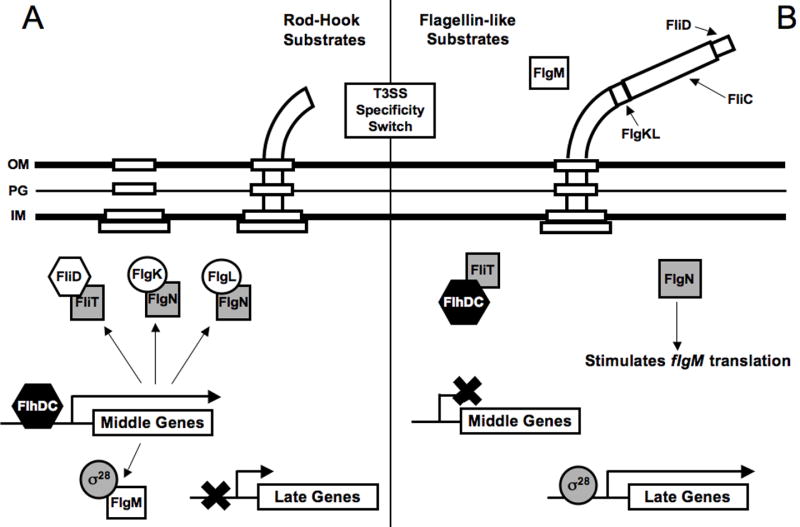
Schematic representation of the steps in flagellar gene regulation that are coupled to secretory activity in Salmonella enterica. Type III secretion chaperones are represented by shaded squares. (A) Hook-basal-body (HBB) assembly. During HBB assembly the hook-associated proteins (FlgK and FlgL), filament capping protein (FliD) and the FlgM anti-σ28 factor are expressed in an FlhDC dependant manner. Prior to switch in the substrate specificity of the T3SS, however, each of these proteins accumulates in the cytoplasm in association with their respective chaperones (FliT, FlgN, FlgN, and FliA [σ28]). The binding of FlgM to σ28 prevents transcription of the late genes. (B) Upon completion of the HBB the specificity of the T3SS switches to late substrates and FliD, FlgK, FlgL, and FlgM are secreted thereby releasing their respective chaperones. FliT binds to and sequesters FlhC resulting in the inhibtion of middle gene expression. σ28 directs transcription of the lates genes including the flagellin monomer (fliC). FlgN positively regulates FlgM translation from its class III promoter to allow for rapid down-regulation of late gene expression upon cessation of secretion.
Up-regulation of late gene expression
Two additional middle genes are fliA, encoding the flagella-specific sigma factor (σ28) required for activation of late gene transcription, and flgM, encoding the anti-σ28 factor FlgM, which is also expressed at a later stage from a class III promoter. In addition to its role in transcription, σ28 also functions as a chaperone that is required for FlgM secretion [13]. Prior to completion of the HBB complex, FlgM expressed from its class II promoter is retained in the cytoplasm where it directly binds to σ28 and inhibits expression of the late genes (Fig. 1A). Upon completion of the HBB complex, secretion of FlgM results in σ28-dependent transcription of the late genes, allowing for final assembly of the flagellum (Fig. 1B). In addition, the reduction in cytoplasmic FlgM levels limits the duration of late gene expression by decreasing the half-life of free σ28 in the cytoplasm where it is degraded by proteases [14].
Fine tuning late gene expression levels
FlgK and FlgL are among the first proteins to be secreted following the switch to late secretion substrates. The chaperone for FlgK and FlgL is FlgN [10]. Following secretion of FlgK and FlgL, FlgN enhances translation of FlgM expressed from its class III promoter through a poorly understood mechanism (Fig. 1B) [15,16]. At this stage during assembly FlgM is thought to be unavailable to interact with σ28 until completion of the flagellar filament terminates further secretion and FlgM once again accumulates in the cytoplasm. It has been suggested that the FlgN-dependent enhancement of FlgM translation may allow cells to rapidly down-regulate σ28- dependent genes following completion of the flagellar filament [16]. In addition to its role as an anti-sigma factor, FlgM may also regulate FliC (flagellin) expression at the post-transcriptional level [17].
Secretory activity as an inducing signal in the injectisome systems
The bacterial injectisome (or needle complex) is structurally similar to the flagellum and consists of a basal body-like structure, an associated T3SS, and a hollow needle-like filament that protrudes from the cell surface and serves as the conduit through which proteins are secreted [18]. Much like the flagellum, assembly of the injectisome is thought to proceed in a sequential manner starting with formation of a basal structure and ending with T3SS-dependent export of the needle protein to the distal tip [4]. The secretory activity of the fully assembled injectisome is tightly regulated and dependent upon specific activating signals, the most relevant being contact of the bacterium with eukaryotic targets cells. In most of the injectisome systems characterized from animal pathogens thus far, activation of the injectisome by host cell contact triggers three events: (i) secretion of the translocator proteins which form a pore in the target cell plasma membrane, (ii) translocation of the pre-formed effector proteins into the target cell, and (iii) increased expression of injectisome-related genes. In many of those pathogens the increase in gene expression is intimately coupled to secretion competency. It should be noted, however, that the secretion-dependent regulatory mechanisms described below are often times subject to additional levels of regulatory control that are beyond the scope of this review; for further information on the role of temperature, metabolic stress, and environmental signals we refer the reader to previous reviews of these topics [19-21].
Shigella flexneri
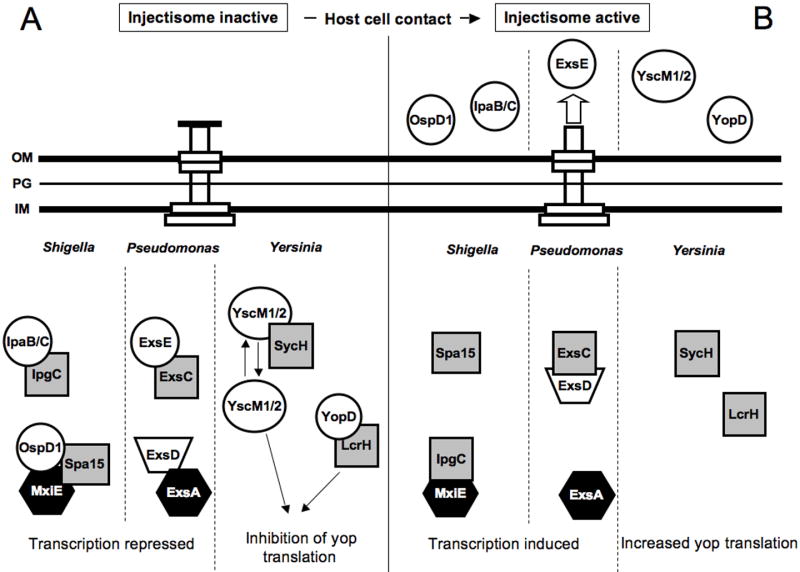
The injectisome of Shigella flexneri is expressed and assembled upon growth of the organism at 37°C. Contact of Shigella with host cells activates secretion by the injectisome and triggers transcription of ∼15 effector genes [22]. Expression of the effector genes is controlled by MxiE, an AraC-like transcriptional activator. The transcriptional activity of MxiE is dependent upon binding its co-activator (IpgC) [23]. Furthermore, transcription from MxiE-dependent promoters in E. coli is dependent upon co-expression of IpgC and MxiE [24], highly suggestive of a direct interaction. IpgC also functions as a type III secretion chaperone for the translocator proteins IpaB and IpaC [25]. Prior to activation of the injectisome, MxiE-dependent transcription is inhibited through sequestration of IpgC by IpaB/IpaC and by a co-complex of the Spa15 co-anti-activator and the OspD1 anti-activator, which directly binds MxiE (Fig. 2A) [26]. Spa15 also serves as a secretion chaperone for OspD1, a secretable substrate. Upon activation of the injectisome, OspD1, IpaB, and IpaC are secreted from the cell thereby releasing MxiE and IpgC which are then thought to form the functional activator of transcription (Fig. 2B).
Figure 2.
Schematic representation of injectisome regulatory systems that couple gene expression to secretory activity in Shigella flexneri, Pseudomonas aeruginosa, Yersinia sp. Type III secretion chaperones are represented by shaded squares, black hexagons represent transcriptional activators, and white circles represent secretion substrates. (A) Prior to activation of the injectisome regulatory proteins accumulate in the cytoplasm and inhibit gene expression. In S. flexneri the complex of OspD1/Spa15 binds to and inhibits MxiE-dependent transcripton. In P. aeruginosa ExsD inhibits ExsA-dependent transcription. In the Yersiniae YopD, LcrH, and YscM1/M2 cooperate to inhibit yop translation. (B) Activation of the injectisome following contact of the pathogen withhost cells triggers secretion and an increase in gene expression. In S. flexneri secretion of OspD1 disrupts the negative regulatory activity of the OspD1/Spa15 complex. In addition, secretion of IpaB/C releases IpgC, which functions as a co-activator for MxiE-dependent transcription of the type III effector genes. Secretion of ExsE in P. aeruginosa allows ExsC to sequester ExsD thereby freeing ExsA to activate transcription. In Yersinia sp. secretion of YopD and YscM1/2 relieves the block on yop translation.
The injectisome regulon of S. enterica is highly homologous to that of S. flexneri. As mentioned above S. enterica has homologs of both MxiE (InvF) and IpgC (SicA) that fulfill the same regulatory role as described for S. flexneri [27]. An OspD1-like factor, however, appears to be absent from S. enterica.
Pseudomonas aeruginosa
In the absence of inducing conditions the T3SS regulon of Pseudomonas aeruginosa is expressed at a basal level possibly allowing for 1-2 injectisomes to be assembled per cell [28]. Unlike the S. flexneri and S. enterica systems, where secretory activity results in induction of only the effector genes, activation of the P. aeruginosa injectisome triggers increased expression of the entire system including genes encoding the effectors, translocators, regulators, chaperones, and injectisome structural proteins [21]. Each of these genes is under the transcriptional control of ExsA, an AraC-like family member. ExsA-dependent transcription is coupled to the secretory activity of the injectisome by a cascade of three interacting proteins (ExsC, ExsD, and ExsE). ExsD is an anti-activator that binds to and inhibits ExsA-dependent transcription (Fig. 2A) [29]. ExsC functions as an anti-anti-activator by binding to and inhibiting the negative regulatory activity of ExsD [30]. ExsC also serves as a type III secretion chaperone for ExsE [31] [32]. Activation of the injectisome by host cell contact triggers translocation of ExsE resulting in a corresponding decrease in the cytoplasmic concentration of ExsE [33]. The reduced level of ExsE are though to liberate the ExsC chaperone, which then binds to and sequesters ExsD, thereby freeing ExsA to activate transcription of the entire system (Fig. 2B).
As is true for all injectisome systems the requirement for increased levels of the effector protein expression likely relates to the amount required to intoxicate host cells. The necessity for increased expression of the P. aeruginosa injectisome structural components is less obvious. One potential explanation might stem from the observation that P. aeruginosa likely uses the injectisome to protect itself in the environment from predators such as amoeba [34]. Low basal expression of the energetically expensive injectisome might be necessary to initiate intoxication but may not be sufficient for full intoxication unless additional injectisomes are assembled on the cell surface.
Yersinia sp.
The first observation of gene expression being coupled to secretory activity in the injectisome systems was made in Yersinia pseudotuberculosis [35]. Each of the three pathogenic species of Yersinia (Y. entereocolitica, Y. pestis, Y. pseudotuberculosis) utilize the injectisome to translocate Yop effector proteins into host cells. Growth of the organisms at 37°C induces transcription of the injectisome-related genes. Prior to activation of injectisome-dependent secretion by host cell contact, however, a complex consisting of LcrH (also called SycD) and YopD binds to the 5′ untranslated region of yop mRNAs and represses translation (Fig. 2A) [36,37]. LcrH also functions as a type III chaperone for YopB and YopD (secreted translocator proteins) [38,39]. For reasons that remain obscure, the negative regulatory activity of the LcrH-YopD complex is also dependent upon YscM1 (also called LcrQ) and YscM2 [36]. Neither protein is thought to directly interact with the LcrH-YopD complex. In the most simplistic model, activation of the injectisome by host cell contact triggers secretion of YopD and translocation of YscM1 and YscM2 into host cells (Fig. 2B). The corresponding decrease in the intracellular levels of these proteins then relieves the block on yop translation. In reality, however, the regulatory system is much more complicated with the potential for extensive cross-talk between type III secretion chaperones and secretion substrates (YscY, the chaperone for YscX which is essential for secretory activity, also interacts with LcrH; SycH, the chaperone for YscM1/M2 also interacts with YopH; SycO, the chaperone for YopO, also interacts with YscM1; and SycE, the chaperone for YopE also interacts with YscM1/M2) [40-42].
Contact of Yersiniae with host cells also results in increased yop transcription through an undefined mechanism [35]. Transcription in the Yersinia injectisome system is controlled by an AraC-like protein (LcrF/VirF). Unlike the Shigella, Salmonella, and Pseudomonas systems, however, where the transcriptional activity of the AraC homologs (MxiE and ExsA) is controlled by type III chaperones or chaperone-associated proteins that function as co-activators or anti-activators, there is no evidence that LcrF/VirF is controlled in such a manner.
Conclusions
The coupling of gene expression to secretion is a clever means of coordinating gene expression to coincide with stages in the assembly or completion of a complex secretory apparatus. In the flagellar systems this mechanism prevents expression of the late genes prior to completion of the HBB, serves as a way to conserve energetic resources, and prevents the late flagellin-like substrates from competing with rod/hook-like substrates for the secretion channel. In the injectisome systems, however, the coupling of gene expression to secretion serves as a mechanism to up-regulate gene expression in response to environmental signals that stimulate the secretory activity of the injectisome. It is not yet clear in the inectisome systems whether there is hierarchical regulation whereby secretion of an early substrate might release a chaperone which either inhibits expression of specific early substrates or allows expression of secreted substrates required during a secondary phase in the infection process (or both).
One of the important findings in the injectisome systems is the diversity by which these regulatory systems function. This might be best illustrated by comparing the roles of the homologous chaperones (IpgC/SicA in S. flexneri and S. enterica, respectively; LcrH in Yersinia sp., and PcrH in P. aeruginosa) for the translocator proteins. Whereas IpgC/SicA function as co-activators for transcription, LcrH functions as the post-transcriptional level to repress yop translation. In contrast, the LcrH homolog from P. aeruginosa (PcrH) lacks detectable regulatory activity [43,44]. This diversity likely reflects environmental pressures that have allowed each pathogen to fine tune gene expression patterns to meet the demands of specific niches.
Acknowledgments
Work on the Pseudomonas aeruginosa type III secretion system in the Yahr laboratory is supported by the National Institutes of Health (RO1-AI055042).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended readings
* of special interest
** of outstanding interest
- 1.Gerlach RG, Hensel M. Protein secretion systems and adhesins: the molecular armory of Gram-negative pathogens. Int J Med Microbiol. 2007;297:401–415. doi: 10.1016/j.ijmm.2007.03.017. [DOI] [PubMed] [Google Scholar]
- 2.Saier MH., Jr Protein secretion and membrane insertion systems in gram-negative bacteria. J Membr Biol. 2006;214:75–90. doi: 10.1007/s00232-006-0049-7. [DOI] [PubMed] [Google Scholar]
- 3.Apel D, Surette MG. Bringing order to a complex molecular machine: The assembly of the bacterial flagella. Biochim Biophys Acta. 2007 doi: 10.1016/j.bbamem.2007.07.005. [DOI] [PubMed] [Google Scholar]; * A comprehensive review of injectisome structure/function.
- 4.Cornelis GR. The type III secretion injectisome. Nat Rev Microbiol. 2006;4:811–825. doi: 10.1038/nrmicro1526. [DOI] [PubMed] [Google Scholar]
- 5.Galan JE, Wolf-Watz H. Protein delivery into eukaryotic cells by type III secretion machines. Nature. 2006;444:567–573. doi: 10.1038/nature05272. [DOI] [PubMed] [Google Scholar]; ** This review covers the T3SSs from of a number of animal pathogens. Topics covered include assembly, secretion/specificity signals, sensing cell contact, effector translocation, and effector molecular mimicry.
- 6.Miller VL. Connections between transcriptional regulation and type III secretion? Curr Opin Microbiol. 2002;5:211–215. doi: 10.1016/s1369-5274(02)00303-x. [DOI] [PubMed] [Google Scholar]
- 7.Aldridge P, Hughes KT. Regulation of flagellar assembly. Curr Opin Microbiol. 2002;5:160–165. doi: 10.1016/s1369-5274(02)00302-8. [DOI] [PubMed] [Google Scholar]
- 8.Ferris HU, Minamino T. Flipping the switch: bringing order to flagellar assembly. Trends Microbiol. 2006;14:519–526. doi: 10.1016/j.tim.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 9.McCarter LL. Regulation of flagella. Curr Opin Microbiol. 2006;9:180–186. doi: 10.1016/j.mib.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 10.Bennett JC, Thomas J, Fraser GM, Hughes C. Substrate complexes and domain organization of the Salmonella flagellar export chaperones FlgN and FliT. Mol Microbiol. 2001;39:781–791. doi: 10.1046/j.1365-2958.2001.02268.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ikeda T, Oosawa K, Hotani H. Self-assembly of the filament capping protein, FliD, of bacterial flagella into an annular structure. J Mol Biol. 1996;259:679–686. doi: 10.1006/jmbi.1996.0349. [DOI] [PubMed] [Google Scholar]
- 12.Yamamoto S, Kutsukake K. FliT acts as an anti-FlhD2C2 factor in the transcriptional control of the flagellar regulon in Salmonella enterica serovar typhimurium. J Bacteriol. 2006;188:6703–6708. doi: 10.1128/JB.00799-06. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** This study shows that FliT, in addition its role as a type III chaperone, regulates class II promoters of the flagellar hierarchy by binding to and inhibiting the FlhC. FliT serves as a sensor of late substrate secretion as it is only free to bind FlhC after secretion of its cognate substrate.
- 13.Aldridge PD, Karlinsey JE, Aldridge C, Birchall C, Thompson D, Yagasaki J, Hughes KT. The flagellar-specific transcription factor, sigma28, is the Type III secretion chaperone for the flagellar-specific anti-sigma28 factor FlgM. Genes Dev. 2006;20:2315–2326. doi: 10.1101/gad.380406. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** The authors describe a novel role for a σ70-family transcription factor as a type III chaperone. FliA(σ28) serves as the chaperone for FlgM, facilitating its secretion, and this activity can be genetically separated from transcriptional activation.
- 14.Barembruch C, Hengge R. Cellular levels and activity of the flagellar sigma factor FliA of Escherichia coli are controlled by FlgM-modulated proteolysis. Mol Microbiol. 2007;65:76–89. doi: 10.1111/j.1365-2958.2007.05770.x. [DOI] [PubMed] [Google Scholar]
- 15.Karlinsey JE, Lonner J, Brown KL, Hughes KT. Translation/secretion coupling by type III secretion systems. Cell. 2000;102:487–497. doi: 10.1016/s0092-8674(00)00053-2. [DOI] [PubMed] [Google Scholar]
- 16.Aldridge P, Karlinsey J, Hughes KT. The type III secretion chaperone FlgN regulates flagellar assembly via a negative feedback loop containing its chaperone substrates FlgK and FlgL. Mol Microbiol. 2003;49:1333–1345. doi: 10.1046/j.1365-2958.2003.03637.x. [DOI] [PubMed] [Google Scholar]; * In conjunction with earlier work (Karlinsey et al., 2000) this paper demonstrates that FlgN serves as both a type III chaperone as well as a translational regulator of FlgM. FlgN, normally sequestered in the cytoplasm by its chaperone substrates FlgK and FlgL, is released upon the initiation of secretion and serves to maintain a sufficient level of FlgM.
- 17.Rosu V, Chevance FF, Karlinsey JE, Hirano T, Hughes KT. Translation inhibition of the Salmonella fliC gene by the fliC 5′ untranslated region, fliC coding sequences, and FlgM. J Bacteriol. 2006;188:4497–4507. doi: 10.1128/JB.01552-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blocker A, Komoriya K, Aizawa S. Type III secretion systems and bacterial flagella: insights into their function from structural similarities. Proc Natl Acad Sci U S A. 2003;100:3027–3030. doi: 10.1073/pnas.0535335100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Francis MS, Wolf-Watz H, Forsberg A. Regulation of type III secretion systems. Curr Opin Microbiol. 2002;5:166–172. doi: 10.1016/s1369-5274(02)00301-6. [DOI] [PubMed] [Google Scholar]
- 20.Ellermeier JR, Slauch JM. Adaptation to the host environment: regulation of the SPI1 type III secretion system in Salmonella enterica serovar Typhimurium. Curr Opin Microbiol. 2007;10:24–29. doi: 10.1016/j.mib.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 21.Yahr TL, Wolfgang MC. Transcriptional regulation of the Pseudomonas aeruginosa type III secretion system. Mol Microbiol. 2006;62:631–640. doi: 10.1111/j.1365-2958.2006.05412.x. [DOI] [PubMed] [Google Scholar]
- 22.Le Gall T, Mavris M, Martino MC, Bernardini ML, Denamur E, Parsot C. Analysis of virulence plasmid gene expression defines three classes of effectors in the type III secretion system of Shigella flexneri. Microbiology. 2005;151:951–962. doi: 10.1099/mic.0.27639-0. [DOI] [PubMed] [Google Scholar]
- 23.Pilonieta MC, Munson GP. The Chaperone IpgC Copurifies With The Virulence Regulator MxiE. J Bacteriol. 2008 doi: 10.1128/JB.01824-07. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mavris M, Page AL, Tournebize R, Demers B, Sansonetti P, Parsot C. Regulation of transcription by the activity of the Shigella flexneri type III secretion apparatus. Mol Microbiol. 2002;43:1543–1553. doi: 10.1046/j.1365-2958.2002.02836.x. [DOI] [PubMed] [Google Scholar]
- 25.Menard R, Sansonetti P, Parsot C, Vasselon T. Extracellular association and cytoplasmic partitioning of the IpaB and IpaC invasins of S. flexneri. Cell. 1994;79:515–525. doi: 10.1016/0092-8674(94)90260-7. [DOI] [PubMed] [Google Scholar]
- 26.Parsot C, Ageron E, Penno C, Mavris M, Jamoussi K, d'Hauteville H, Sansonetti P, Demers B. A secreted anti-activator, OspD1, and its chaperone, Spa15, are involved in the control of transcription by the type III secretion apparatus activity in Shigella flexneri. Mol Microbiol. 2005;56:1627–1635. doi: 10.1111/j.1365-2958.2005.04645.x. [DOI] [PubMed] [Google Scholar]; ** The authors demonstrate that the activity of MxiE, the transcriptional activator of ∼15 effectors in Shigella flexneri, is antagonized by the anti-activator OspD1 and co-anti-activator Spa15. OspD1 is shown to be secreted further linking MxiE function to secretory activity.
- 27.Darwin KH, Miller VL. Type III secretion chaperone-dependent regulation: activation of virulence genes by SicA and InvF in Salmonella typhimurium. Embo J. 2001;20:1850–1862. doi: 10.1093/emboj/20.8.1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rietsch A, Mekalanos JJ. Metabolic regulation of type III secretion gene expression in Pseudomonas aeruginosa. Mol Microbiol. 2006;59:807–820. doi: 10.1111/j.1365-2958.2005.04990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCaw ML, Lykken GL, Singh PK, Yahr TL. ExsD is a negative regulator of the Pseudomonas aeruginosa type III secretion regulon. Mol Microbiol. 2002;46:1123–1133. doi: 10.1046/j.1365-2958.2002.03228.x. [DOI] [PubMed] [Google Scholar]
- 30.Dasgupta N, Lykken GL, Wolfgang MC, Yahr TL. A novel anti-anti-activator mechanism regulates expression of the Pseudomonas aeruginosa type III secretion system. Mol Microbiol. 2004;53:297–308. doi: 10.1111/j.1365-2958.2004.04128.x. [DOI] [PubMed] [Google Scholar]
- 31.Rietsch A, Vallet-Gely I, Dove SL, Mekalanos JJ. ExsE, a secreted regulator of type III secretion genes in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2005;102:8006–8011. doi: 10.1073/pnas.0503005102. [DOI] [PMC free article] [PubMed] [Google Scholar]; ** Along with the reference below (Urbanowski et al., 2005) the authors demonstrate that ExsE serves as a sensor for secretion competence of the Pseudomonas aeruginosa T3SS. Through a series of intermediate proteins secretion of ExsE results in derepression of ExsA-dependent transcription.
- 32.Urbanowski ML, Lykken GL, Yahr TL. A secreted regulatory protein couples transcription to the secretory activity of the Pseudomonas aeruginosa type III secretion system. Proc Natl Acad Sci U S A. 2005;102:9930–9935. doi: 10.1073/pnas.0504405102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Urbanowski ML, Brutinel ED, Yahr TL. Translocation of ExsE into Chinese hamster ovary cells is required for transcriptional induction of the Pseudomonas aeruginosa type III secretion system. Infect Immun. 2007;75:4432–4439. doi: 10.1128/IAI.00664-07. [DOI] [PMC free article] [PubMed] [Google Scholar]; * The authors demonstrate that ExsE, whose secretion is required for transcriptional activation of T3SS genes, is also translocated into host cells by Pseudomonas aeruginosa and that its translocation is required for upregulation of T3SS genes in response to host cell contact.
- 34.Pukatzki S, Kessin RH, Mekalanos JJ. The human pathogen Pseudomonas aeruginosa utilizes conserved virulence pathways to infect the social amoeba Dictyostelium discoideum. Proc Natl Acad Sci U S A. 2002;99:3159–3164. doi: 10.1073/pnas.052704399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pettersson J, Nordfelth R, Dubinina E, Bergman T, Gustafsson M, Magnusson KE, Wolf-Watz H. Modulation of virulence factor expression by pathogen target cell contact. Science. 1996;273:1231–1233. doi: 10.1126/science.273.5279.1231. [DOI] [PubMed] [Google Scholar]
- 36.Cambronne ED, Schneewind O. Yersinia enterocolitica type III secretion: yscM1 and yscM2 regulate yop gene expression by a posttranscriptional mechanism that targets the 5′ untranslated region of yop mRNA. J Bacteriol. 2002;184:5880–5893. doi: 10.1128/JB.184.21.5880-5893.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anderson DM, Ramamurthi KS, Tam C, Schneewind O. YopD and LcrH regulate expression of Yersinia enterocolitica YopQ by a posttranscriptional mechanism and bind to yopQ RNA. J Bacteriol. 2002;184:1287–1295. doi: 10.1128/JB.184.5.1287-1295.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neyt C, Cornelis GR. Role of SycD, the chaperone of the Yersinia Yop translocators YopB and YopD. Mol Microbiol. 1999;31:143–156. doi: 10.1046/j.1365-2958.1999.01154.x. [DOI] [PubMed] [Google Scholar]
- 39.Olsson J, Edqvist PJ, Broms JE, Forsberg A, Wolf-Watz H, Francis MS. The YopD translocator of Yersinia pseudotuberculosis is a multifunctional protein comprised of discrete domains. J Bacteriol. 2004;186:4110–4123. doi: 10.1128/JB.186.13.4110-4123.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]; * The authors use systematic mutagenesis to identify discrete functional domains within YopD in Yersinia pseudotuberculosis demonstrating that YopD is important for a number of individual steps during Yop secretion and regulation.
- 40.Dittmann S, Schmid A, Richter S, Trulzsch K, Heesemann J, Wilharm G. The Yersinia enterocolitica type three secretion chaperone SycO is integrated into the Yop regulatory network and binds to the Yop secretion protein YscM1. BMC Microbiol. 2007;7:67. doi: 10.1186/1471-2180-7-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Day JB, Plano GV. The Yersinia pestis YscY protein directly binds YscX, a secreted component of the type III secretion machinery. J Bacteriol. 2000;182:1834–1843. doi: 10.1128/jb.182.7.1834-1843.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Swietnicki W, O'Brien S, Holman K, Cherry S, Brueggemann E, Tropea JE, Hines HB, Waugh DS, Ulrich RG. Novel protein-protein interactions of the Yersinia pestis type III secretion system elucidated with a matrix analysis by surface plasmon resonance and mass spectrometry. J Biol Chem. 2004;279:38693–38700. doi: 10.1074/jbc.M405217200. [DOI] [PubMed] [Google Scholar]
- 43.Broms JE, Edqvist PJ, Carlsson KE, Forsberg A, Francis MS. Mapping of a YscY binding domain within the LcrH chaperone that is required for regulation of Yersinia type III secretion. J Bacteriol. 2005;187:7738–7752. doi: 10.1128/JB.187.22.7738-7752.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Broms JE, Forslund AL, Forsberg A, Francis MS. PcrH of Pseudomonas aeruginosa is essential for secretion and assembly of the type III translocon. J Infect Dis. 2003;188:1909–1921. doi: 10.1086/379898. [DOI] [PubMed] [Google Scholar]