Abstract
Specialized cells are the essence of complex multicellular life. Fossils allow us to study the modification of specialized, multicellular features such as jaws, scales, and muscular appendages. But it is still unclear what organismal properties contributed to the transition from undifferentiated organisms, which contain only a single cell type, to multicellular organisms with specialized cells. Using digital organisms I study this transition. My simulations show that the transition to specialized cells happens faster in organism composed of many cells than in organisms composed of few cells. Large organisms suffer less from temporarily unsuccessful evolutionary experiments with individual cells, allowing them to evolve specialized cells via evolutionary trajectories that are unavailable to smaller organisms. This demonstrates that the evolution of simple multicellular organisms which are composed of many functionally identical cells accelerates the evolution of more complex organisms with specialized cells.
Keywords: division of labor, somatic cells, differentiated multicellularity, early life
Introduction
In multicellular organisms cells differentiate and specialize to form tissues which cooperate to form organs such as brains, kidneys, hearts, stomachs, and lungs. Without specialized cells multicellular organisms would be nothing more than a homogeneous lump of cells. It is a widely accepted consequence of evolutionary theory that differentiated organisms with specialized cells evolved from undifferentiated ancestors (Darwin, 1859; Buss, 1988; Knoll, 2003; King, 2004).
It is believed that the preexistence of undifferentiated multicellularity conveys advantages for the evolution of specialized cells (Buss, 1988; Maynard-Smith, 1989). One argument regards the alleviation of reproductive competition in organisms that develop from a single cell. In such organisms cells are genetically identical and genes that encode for the development of specialized cells would not curtail their propagation by creating non-reproductive cells (Buss, 1988; Maynard-Smith, 1989; Dawkins, 1999; Maynard-Smith & Szathmary, 1997; Michod & Roze, 2001).
In this work I demonstrate that undifferentiated multicellularity conveys an additional advantage for the evolution of specialized cells. I find that the size of a multicellular organism affects its fitness landscape. Mutations that differentiate individual cells are less detrimental in organisms composed of many cells than in organisms composed of few cells. This changes the evolutionary landscape and accelerates the evolution of specialized cells. The insight that the size of an organism affects its ability to evolve new, specialized cells is vital for our understanding of how complex multicellular life evolved.
To study the evolution of a complex feature like differentiated multicellularity it is desirable to use an experimental system in which the evolutionary path from one stage to another is not preset but discovered by evolution itself. Digital organisms provide such a framework. Digital organisms are entities that are able to replicate and perform specific tasks. They compete for a common resource and are exposed to mutations. The combination of replication, competition, and mutation results in an evolutionary process, which can be used to address biological questions (Adami et al., 2000; Wilke et al., 2001; Yedid & Bell, 2002; Lenski et al., 2003; Chow et al., 2004; Kim et al., 2007).
So far, however, digital organisms have not been equipped with the ability to evolve multicellularity. To close this gap I developed and implemented digital organisms that are able to evolve multicellularity of varying complexity. The resulting digital self-replicating cellular organisms (DISCOs) are similar to the digital organisms used by the Avida software platform (Ofria & Wilke, 2004). The supporting online material (SOM) contains details about how a single DISCO cell works and how multicellularity is implemented. Besides providing insight into the evolution of specialized cells, this paper demonstrates how readily existing artificial life systems can be extended to study the evolution of complex multicellular features.
For the following it is sufficient to know that a DISCO has a genome that can encode logic functions as well as the development of a multicellular organism. The fitness of a DISCO is determined by its merit and its speed of replication. The merit is determined by the logic functions that the DISCO can execute, which are encoded in its genome. Logic functions differ in their complexity. The higher the complexity of the logic function, the larger the merit increase (see SOM and Lenski et al. (2003)).
The speed of replication is mainly determined by the number of cells a DISCO is composed of. Several types of cells exist. The default (D) cell is the replicative cell. Every DISCO has exactly one D cell and it is the only cell type in a unicellular organism. D cells can, if instructed by the genome, produce somatic X and Y cells. Somatic cells are always associated with a cost since a multicellular organism spends time and energy growing them while a unicellular organism can use these resources to produce offspring. On the other hand, by computing logic functions somatic cells can increase a DISCOs merit yielding a benefit that outweighs these costs. Some functions, however, can only be utilized by specific, specialized cells.
Specialized cells are common in biology. The model structure studied in this work is motivated by the heterocysts of cyanobacteria. Heterocysts are cells specialized on the fixation of nitrogen. They provide an oxygen-free environment for the nitrogen-fixing enzyme. To accomplish this, they develop thick cell walls that shut out oxygen. They also degrade photosystem II, which produces oxygen. These features allow heterocysts to fix nitrogen but prevent them from carrying out functions of non-specialized cells, such as cell division and photosynthesis via photosystem II.
Y cells are specialized cells in DISCOs and analogous to heterocysts in cyanobacteria. Y cells are different from normal (D and X) cells. The differences allow Y cells to utilize the three most complex logic functions which cannot be utilized by the non-specialized D and X cells. This specialization, however, makes it impossible for Y cells to utilize the six logic functions that non-specialized D and X cells can utilize. Thus, similar to heterocysts, Y cells are specialized for certain tasks (see first column in Fig. 11b and SOM).
It is important to emphasize that a multicellular DISCO can only benefit from a given logic function if (a) the function is encoded in the genome and (b) cell types that are able to utilize the function are present. This is analogous to heterocystous cyanobacteria that can only benefit from nitrogen fixation if (a) the nitrogen-fixing enzyme is correctly encoded in the genome and (b) cells with degenerated photosynthesis II and thick, oxygen-impermeable cell walls exist.
Simulations and Results
In this work I am interested in the transition from simple multicellularity to a more complex multicellularity with specialized cells. In particular, I would like to know if the pre-existence of undifferentiated multicellular organisms has an effect on the evolution of specialized cells. To study this, I will compare “-X” and “+X” simulations. In +X simulations X cells are able to increase the merit of a DISCO. A DISCO composed of one D cell and n X cells has n + 1 times the merit of a unicellular DISCO. In +X simulations the evolution of X cells, that is, undifferentiated multicellularity, is encouraged. This is not the case in -X simulations in which X cells are not able to increase the merit of the organism and are therefore disadvantageous. In -X simulations a transition to differentiated multicellularity with specialized cells has to occur directly from an unicellular ancestor.
To study the transition to differentiated organisms I evolved, as a first step, undifferentiated DISCOs. To ensure that DISCOs do not evolve specialized cells, I suspended the ability of Y cells to increase the merit of the organism for this initial set of simulations (see SOM Table 1 and Figure 1b). For each set of simulations (-X and +X), I conducted 500 independent runs that differ only with respect to the seed for the random number generator. Each simulation was initiated with a genome that encodes only replication. In other words, the ancestral DISCO was unicellular and could not compute any logic function. For computational reasons, I used an effective population size of 200 organisms and stopped a simulation after 10 000 generations (see SOM for more details). Following the logic of Lenski et al. (2003), at the end of each simulation I determined the most recent common ancestor of the population and its line of descent. Similar to a paleontologist, I use this (digital) fossil record to determine when each trait appeared. But in contrast to a paleontologist, I know the fitness of each fossil and study 500 independent instances of one evolutionary process. This gives me the opportunity to discover general properties of the process at hand.
Figure 1.
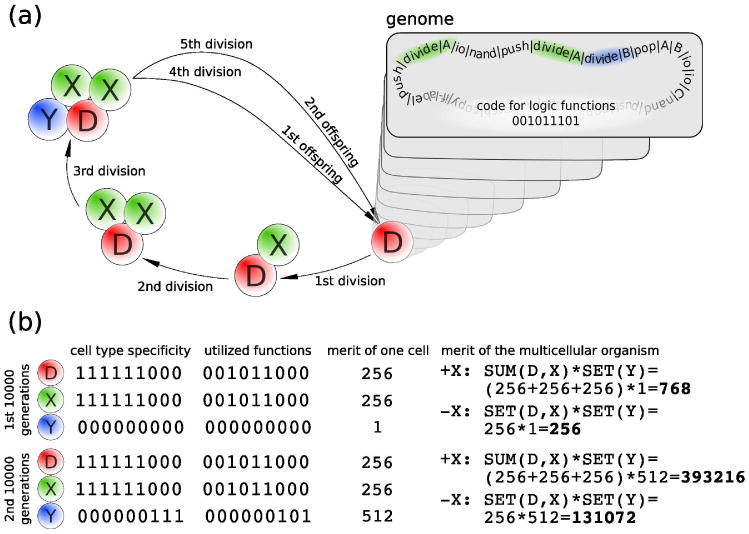
Multicellularity in DISCOs. (a) The first five cell division of a DISCO with a genome that encodes for two X cells (green shaded regions) and one Y cell (blue shaded region). The first three cell divisions produce the somatic X and Y cells. Every further division produces offspring which is released into the environment. (b) The merit of this multicellular organism during the first and second 10 000 generations of the -X and +X simulations. The genome encodes for five (out of nine) logic functions as indicated by the nine-digit binary sequence. D, X, and Y cells can utilize these functions (second column) only according to their cell type specificity (first column) and receive a corresponding merit (third column) which is used to calculate the merit of the organism (fourth column). Note that Y cells are not able to increase the merit of the organism during the first 10 000 generations and that X cells are not able to increase the merit of the organism during the -X simulations. Cells that do not increase merit are disadvantageous, since they increase the number of cell divisions that are required to reach maturity. A more detailed description is available in the SOM.
As expected, none of the -X simulations evolved multicellularity during these first 10 000 generations. All DISCOs remained unicellular. On the other hand, 491 of the 500 +X simulations evolved undifferentiated multicellularity. Multicellular DISCOs are very diverse with respect to their size. They have body sizes ranging from two to thirteen cells, with size five as the most frequent. The organisms were also very successful in evolving logic functions. Most simulations evolved DISCOs that can compute all six available functions; few evolved “just” five functions.
To study the evolution of specialized cells, I used each of these MRCAs as a starting point for another 2 × 500 simulations. This time, Y cells were able to utilize functions that had not been available so far. They can increase the fitness of a DISCO substantially (see SOM and Figure 1b). In such a situation, one expects the evolution of DISCOs with Y cells and Y cell specific functions, which was indeed the case. As expected multicellular DISCOs in the -X simulations were exclusively bicellular, composed of one D and one Y cell. Specialized cells were discovered in 197 of the 500 -X and in 308 of the 500 +X simulations. This difference is highly significant (2-sample test for equality of proportions: χ2 = 48.40, df=1, p = 3.5 · 10−12). Apparently, the pre-existence of undifferentiated multicellular organisms promotes the evolution of more complex multicellular organisms with specialized cells.
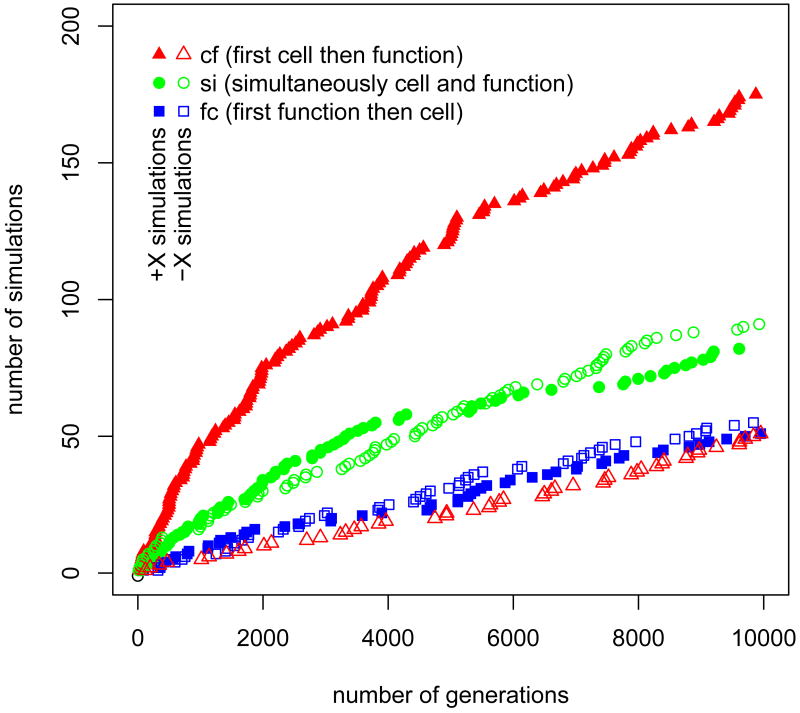
To study why undifferentiated multicellularity promotes the evolution of Y cells, I examined the evolutionary paths that lead to DISCOs with specialized cells. Considering the order of events we have three possibilities. Mutations can result in the simultaneous (si) appearance of Y cells and Y cell specific functions, or the two traits may appear in succession, either first the cell and then the function (cf) or first the function and then the cell (fc). The digital fossil record allows us to determine via which path and at which time Y cells were discovered (see Fig. 2). Two features are conspicuous. First, for the -X simulations si is the most frequently traveled path; about 46% of the specialized cells are discovered simultaneously with the cell function. Second, the -X and the +X simulations differ noticeably only with respect to cf (red triangles). Apparently, evolving first the cell and then the function is much easier for undifferentiated multicellular organisms than it is for unicellular ones and accounts for the significant difference in the number of simulations that discovered Y cells between the +X and the -X simulations.
Figure 2.
Number of simulations that evolved specialized cells as a function of time. Simulations are grouped according to the evolutionary paths cf, si, and fc (see Figure legend and main text) that led to the evolution of new cell types that utilize new functions. Noticeable differences between simulations with unicellular (-X) and undifferentiated multicellular (+X) ancestors exists only with respect to evolutionary path cf along which specialized cell (Y cells) appear before the genome encodes for the specialized functions.
To explain these observations we have to consider the fitness of organisms along the three possible paths. Especially the intermediates for cf and fc are of interest. Let Dc and Df denote DISCOs along the evolutionary paths cf and fc. That is, Dc is a DISCO that encodes Y cells but not (yet) Y cell specific logic functions, and Df is a DISCO that has acquired Y cell specific functions but not (yet) Y cells. If Dc and Df have a low fitness, then they are not maintained for long in the population and there is less opportunity for a second mutation to give rise to the missing Y cell function or the missing Y cell. In such a case evolution along the corresponding paths is impaired and one would expect most specialized cells to evolve directly via si (Iwasa et al., 2003, 2004).
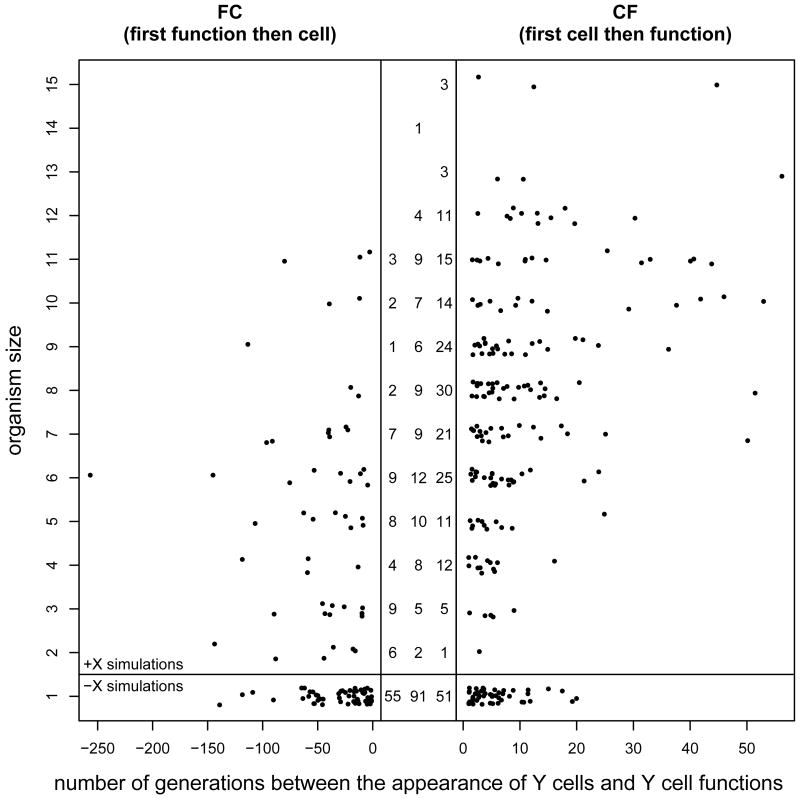
The digital fossil record provides information about the time, t, that Dc and Df are maintained in the population, as well as the organism size at that point (see Fig. 3). Let us first discuss the data for organisms of size one, that is, the -X simulations. None of the 51 simulations that evolved Y cells via cf maintained Dc for more than 20 generations in the population. This suggests that mutations that provide a DISCO with Y cells are deleterious. The digital fossil record shows that the fitness decrease is not a result of a merit decrease due to a loss of logic functions. Rather, the loss in fitness is caused by developmental costs. DISCOs with Y cells but without Y cell functions grow one additional, unused cell. This constitutes a fitness burden. In the SOM I show that mutations that transform a unicellular DISCO into a bicellular decrease the relative fitness from 1 to about 0.62. Thus, the fitness of Dc is indeed low.
Figure 3.
Time, t, in number of generations between the appearance of specialized cells (Y cells) and the appearance of specialized functions. The data is grouped according to the size of the DISCO immediately before the appearance of Y cells. The plot contains data from the +X (organism size greater than one) and the -X (organism size equals one) simulations. The pannel in the middle shows the number of simulations that evolved Y cells via fc, si, and cf, respectively. The correlation between t and the size of the organism for evolutionary path cf is evident (Kendall's tau statistic: z = 5.15, p = 2.64 · 10−7). It shows how organism size affects the evolution of specialized cells by reducing the detrimental effect of temporarily unsuccessful evolutionary experiments with individual cells. To increase expressiveness I added small random noise to the organism size and used different plot regions for fc and cf.
What about the fitness of Df? The data in Fig. 3 shows that Df is easier to maintain in the population than Dc and suggests that mutations along path fc are less deleterious than mutations along cf. But for the following reasons we can actually expect most mutations that generate Y cell functions in DISCOs without Y cells (mutations along path fc) to be very deleterious. The digital fossil record shows that most mutations (> 95%) that generate Y cell functions in DISCOs with Y cells (along evolutionary path cf) destroy at least one of the previously evolved logic functions. This is not detrimental for DISCOs with Y cells because they trade a Y cell specific function for a non-specific function. However, DISCOs without Y cells cannot utilize the newly discovered logic function and experience “just” a loss of already evolved logic functions. Hence, most mutations that lead to Df are actually deleterious and evolution via fc can only use a small (< 5%) subset of neutral mutations. All things considered, Df and Dc have on average a low fitness and we should not be surprised that many specialized cells evolve via si (Iwasa et al., 2003, 2004).
But why and how does the situation change for undifferentiated, multicellular organisms? Why is the rate of evolution via cf higher in +X than in -X simulations (see Fig. 2)? We can answer this question by considering the developmental cost of Y cells for organisms of different sizes. As one would expect, the burden of developing one additional unused cell is more substantial for small organisms than it is for large ones. For example, a size increase by one decreases the relative fitness of a unicellular DISCO to 0.62, whereas the relative fitness of a ten-celled DISCO is decreased to only 0.93 (see SOM). Hence, the intermediates along cf are less deleterious for large organisms and are therefore maintained longer in the population. This is evidenced by a significant (Kendall's tau statistic: z = 5.15, p = 2.64 · 10−7) correlation between t and the size of the organism for cf (see Fig. 3). Consequently, the rate of evolution along cf increases with organism size and large organisms are significantly more likely to evolve Y cells via cf (Pearson's χ2 test: χ2 = 37.20, df=13, p = 3.9 · 10−4, see numbers in Fig. 3). By lowering the barriers along one of the evolutionary paths, organism size promotes the evolution of specialized cells and, therefore, of more complex multicellular organisms.
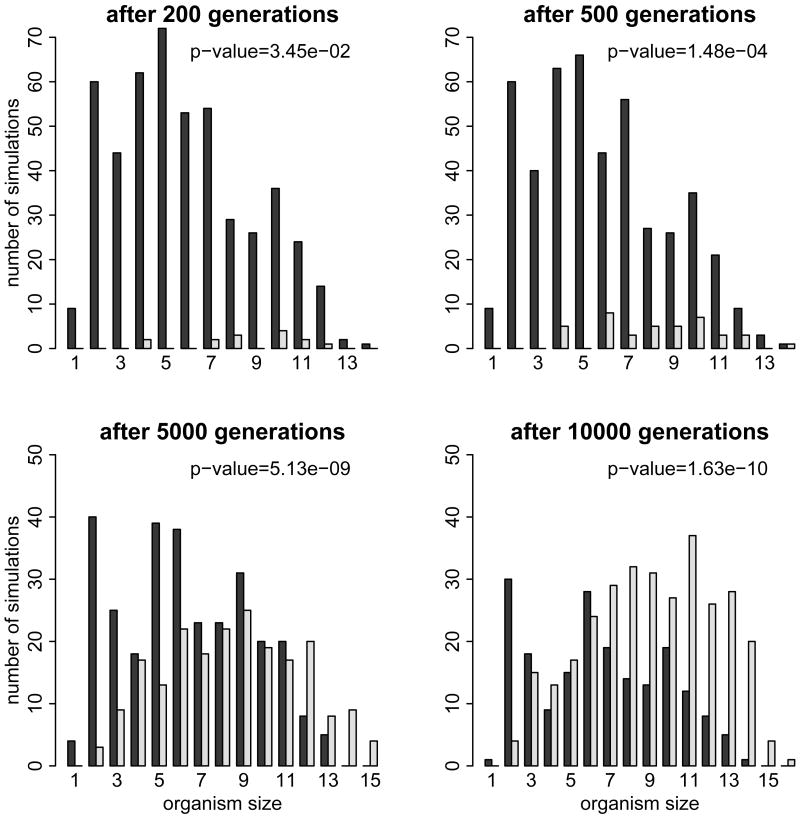
If this is the case, then we should also find evidence for this phenomenon within the +X simulations. In particular, organisms composed of many cells should evolve specialized cells earlier than organisms composed of few cells. Figure 4 shows the size distribution of DISCOs with and without Y cells at different time points of the +X simulations. For example, after 200 generations 14 simulations discovered Y cells. Only two of those (< 15%) were discovered in organisms smaller than seven cells, even though, most simulations (> 60%) contained organisms smaller than seven cells. This bias towards specialized cells in larger organisms is even more pronounced in later stages of the simulations and remains significant (see p-values in Fig. 4). Even within the +X simulations we observe that an increase in organism size eases the evolution of specialized cells.
Figure 4.
Number of simulations that evolved organisms with (white bars) and without (black bars) specialized cells after 200, 500, 5000, and 10000 generations, grouped according to the size of the organism. Large organisms show a significant bias (see p-values of a Pearson's chi-squared test) towards discovering specialized cells earlier than small organisms.
Discussion
For multicellular organisms (Bonner, 1965; Bell & Mooers, 1997; Bonner, 2004), as well as insect (Wilson, 1971) or human (Blau, 1974) societies it is known that the degree of specialization increases with the size of the system. Large systems seem to be able to benefit more from specialized units. Consequently, the lack of specialization in very small multicellular organisms (Bell & Mooers, 1997) might be explained by the existence of a minimum threshold size at which specialization becomes advantageous. It is important to emphasize that this is not the case in this paper. Specialized cells can increase the fitness of small and large DISCOs substantially. Nonetheless, there is a significant correlation between organism size and the presence of specialized cells (see Fig. 4). this correlation is based on evolutionary constraints in small organisms and not on a minimum threshold size at which specialization becomes advantageous.
This paper demonstrates how artificial life simulations can be used to study the evolution of complex multicellularity. Currently, the implementation of DISCOs allows only for a sudden change of cell types which results in an equally sudden change of the logic functions that the cell can utilize. For biological systems, one might favor models in which new cell types evolve more gradually with intermediate, “chimeric” types that can perform new and old functions, but both just sub-optimally due to an inherent trade off. Even for such models similar results can be expected since allowing for chimeric cell types does not change the fact that individual cells affect the fitness of the whole organisms less in large organisms (organisms composed of many cells) than in small organisms (organisms composed of few cells).
In general, loss of function mutations are more frequent than gain of function mutations. It is therefore likely that many evolutionary trajectories exists that lead to specialized cells but involve intermediate fitness losses due to loss of function mutations. As I have demonstrated in this paper, such mutations are less harmful to organisms composed of many cells. In such organisms mutations that create chimeric cells with (temporarily) mediocre functionality can be maintained in the population until additional mutations accumulate that allow the new cell type to execute the new function at its full potential. Since loss of function mutations are so frequent, an increase in organism size can be expected to substantially increase the arsenal of evolutionary trajectories that are available for the evolution of specialized cells and is therefore an important step towards the evolution of complex multicellularity.
The insight that the size of a biological system affects its evolutionary landscape can also be applied to other aspects of biology. Take, for example, gene duplication. It is commonly accepted that gene duplication accelerates the discovery of new gene functions. After gene duplication one copy of the gene can execute the old function whereas the other copy can accumulate mutations which might eventually lead to a new function (Lynch & Conery, 2000). The results from this paper suggest that the genome size of the organism has a crucial impact on the organism's ability to discover new gene functions by means of gene duplication. For very small genomes (e.g., the genome of a virus) a duplicated gene constitutes a substantial fitness burden and might not be maintained in the population long enough to adopt a new function. For big genomes (e.g., eukaryotic genomes) a single gene duplication constitutes in-significant additional baggage and can be maintained long enough to discover a new role. What has been learned about organism size and the evolution of complex multicellularity also applies to genome sizes and the evolution of complex genomes.
Organism size has always been considered an important factor for the evolution of multicellularity. In fact, benefits of increased size are thought to have promoted the transition from unicellular to undifferentiated multicellular life (Bonner, 1965; Kirk, 1997; Bonner, 2001; Kirk, 2003; King, 2004). Advantages of increased size include predator evasion (Boraas et al., 1998), increased motility (Kirk, 2003), or increased capacity to store nutrients (Koufopanou & Bell, 1993; Kerszberg & Wolpert, 1998). In this work I observe another, less obvious benefit of organism size. This benefit —the ability to discover new specialized cells via trajectories that are inaccessible to small organisms— does not concern the fitness of the organism itself but its ability to evolve more complex multicellular forms.
Supplementary Material
Acknowledgments
I am grateful to Erick Matsen for mesmerizing and amazing discussions, Matthew Hegreness and Reinhard Bürger for help with the manuscript, and Martin Nowak for invaluable input. I am supported by a Merck-Wiley fellowship. Support from the nsf/nih joint program in mathematical biology (nih grant r01gm078986) is gratefully acknowledged. The Program for Evolutionary Dynamics at Harvard University is sponsored by J. Epstein.
References and Notes
- Adami C, Ofria C, Collier TC. Evolution of biological complexity. Proc Natl Acad Sci U S A. 2000;97:4463–8. doi: 10.1073/pnas.97.9.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell G, Mooers A. Size and complexity among multicellular organisms. Biol J Linnean Soc. 1997;60:345–363. [Google Scholar]
- Blau P. On the nature of organizations. Wiley; New York: 1974. [Google Scholar]
- Bonner JT. Perspective: the size-complexity rule. Evolution Int J Org Evolution. 2004;58:1883–90. doi: 10.1111/j.0014-3820.2004.tb00476.x. [DOI] [PubMed] [Google Scholar]
- Bonner JT. Size and Cycle. Princeton University Press; 1965. [Google Scholar]
- Bonner JT. First Signals: The Evolution of Multicellular Development. Princeton University Press; 2001. [Google Scholar]
- Boraas M, Seale D, Boxhorn J. Phagotrophy by a flagellate selects for colonial prey: A possible origin of multicellularity. Evol Ecol. 1998;12:153–164. [Google Scholar]
- Buss LW. The Evolution of Individuality. Princeton University Press; 1988. [Google Scholar]
- Chow SS, Wilke CO, Ofria C, Lenski RE, Adami C. Adaptive radiation from resource competition in digital organisms. Science. 2004;305:84–6. doi: 10.1126/science.1096307. [DOI] [PubMed] [Google Scholar]
- Darwin C. On the Origin of Species: By Means of Natural Selection. Murray; London: 1859. [Google Scholar]
- Dawkins R. The Extended Phenotype. revised Oxford University Press; 1999. [Google Scholar]
- Iwasa Y, Michor F, Nowak MA. Evolutionary dynamics of escape from biomedical intervention. Proc Biol Sci. 2003;270:2573–8. doi: 10.1098/rspb.2003.2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasa Y, Michor F, Nowak MA. Stochastic tunnels in evolutionary dynamics. Genetics. 2004;166:1571–9. doi: 10.1534/genetics.166.3.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerszberg, Wolpert The origin of metazoa and the egg: a role for cell death. J Theor Biol. 1998;193:535–537. doi: 10.1006/jtbi.1998.0714. [DOI] [PubMed] [Google Scholar]
- Kim I, Cha S, Kim M, Lee Y, Lee K, Choi Y, Hwang J, Jin B, Han Y. Polymorphism and genomic structure of the a+t-rich region of mitochondrial dna in the oriental mole cricket, gryllotalpa orientalis (orthoptera: Gryllotalpidae) Biochem Genet. 2007 doi: 10.1007/s10528-007-9099-5. [DOI] [PubMed] [Google Scholar]
- King N. The unicellular ancestry of animal development. Dev Cell. 2004;7:313–25. doi: 10.1016/j.devcel.2004.08.010. [DOI] [PubMed] [Google Scholar]
- Kirk DL. Volvox : A Search for the Molecular and Genetic Origins of Multicellularity and Cellular Differentiation. Cambridge University Press; 1997. [Google Scholar]
- Kirk D. Seeking the ultimate and proximate causes of volvox multicellularity and cellular differentiation. Integr Comp Biol. 2003;43:247–253. doi: 10.1093/icb/43.2.247. [DOI] [PubMed] [Google Scholar]
- Knoll AH. Life on a Young Planet: The First Three Billion Years of Evolution on Earth. Princeton University Press; 2003. [Google Scholar]
- Koufopanou V, Bell G. Soma and germ - an experimental approach using volvox. Proc R Soc Lond Ser B-Biol Sci. 1993;254:107–113. [Google Scholar]
- Lenski RE, Ofria C, Pennock RT, Adami C. The evolutionary origin of complex features. Nature. 2003;423:139–44. doi: 10.1038/nature01568. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–5. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- Maynard-Smith J. Evolutionary Progress, chapter Evolutionary progress and levels of selection. University Of Chicago Press; 1989. pp. 219–230. [Google Scholar]
- Maynard-Smith J, Szathmary E. The Major Transitions in Evolution. reprint Oxford University Press; 1997. [Google Scholar]
- Michod RE, Roze D. Cooperation and conflict in the evolution of multicellularity. Heredity. 2001;86:1–7. doi: 10.1046/j.1365-2540.2001.00808.x. [DOI] [PubMed] [Google Scholar]
- Ofria C, Wilke CO. Avida: a software platform for research in computational evolutionary biology. Artif Life. 2004;10:191–229. doi: 10.1162/106454604773563612. [DOI] [PubMed] [Google Scholar]
- Wilke CO, Wang JL, Ofria C, Lenski RE, Adami C. Evolution of digital organisms at high mutation rates leads to survival of the flattest. Nature. 2001;412:331–3. doi: 10.1038/35085569. [DOI] [PubMed] [Google Scholar]
- Wilson EO. The insect societies. Harvard University Press; Cambridge: 1971. [Google Scholar]
- Yedid G, Bell G. Macroevolution simulated with autonomously replicating computer programs. Nature. 2002;420:810–2. doi: 10.1038/nature01151. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.