Abstract
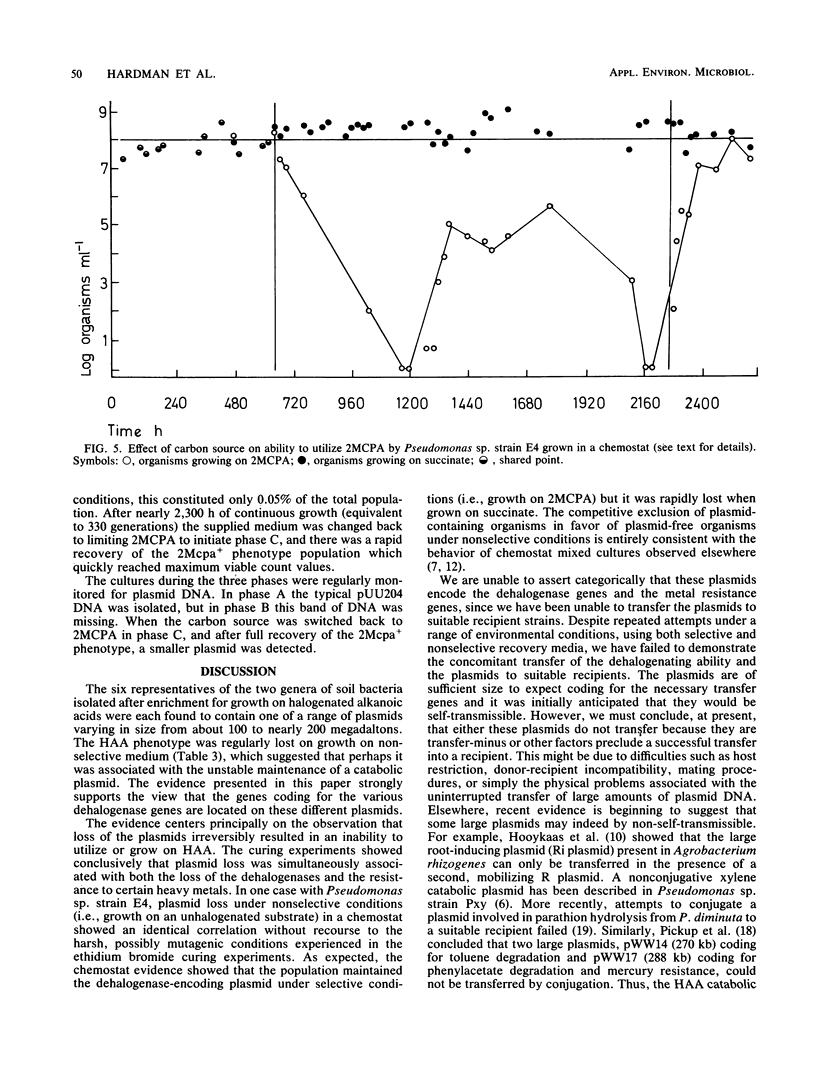
Four Pseudomonas species and two Alcaligenes species were isolated from soil with a capacity to grow on halogenated alkanoic acids. They were shown to contain one of five large plasmids. The plasmids had molecular weights ranging from 98,800 to 190,000. They were associated with the ability to utilize the halogenated substrates 2-monochloropropionic acid and 2-monochloroacetic acid and with resistance towards one or more of the heavy metals mercury, selenium, and tellurium. The largest plasmid, pUU204, was shown to be unstable in continuous-flow culture when the organism was supplied with succinate as the sole carbon source. The dehalogenase gene associated with pUU204 appeared to be readily transferred to an incP group plasmid, R68-45.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baxter-Gabbard K. L. A simple method for the large-scale preparation of sucrose gradients. FEBS Lett. 1972 Jan 15;20(1):117–119. doi: 10.1016/0014-5793(72)80031-0. [DOI] [PubMed] [Google Scholar]
- Eckhardt T. A rapid method for the identification of plasmid desoxyribonucleic acid in bacteria. Plasmid. 1978 Sep;1(4):584–588. doi: 10.1016/0147-619x(78)90016-1. [DOI] [PubMed] [Google Scholar]
- Fennewald M., Prevatt W., Meyer R., Shapiro J. Isolation of inc P-2 plasmid DNA from Pseudomonas aeruginosa. Plasmid. 1978 Feb;1(2):164–173. doi: 10.1016/0147-619x(78)90036-7. [DOI] [PubMed] [Google Scholar]
- Friello D. A., Mylroie J. R., Gibson D. T., Rogers J. E., Chakrabarty A. M. XYL, a nonconjugative xylene-degradative plasmid in Pseudomonas Pxy. J Bacteriol. 1976 Sep;127(3):1217–1224. doi: 10.1128/jb.127.3.1217-1224.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godwin D., Slater J. H. The influence of the growth environment on the stability of a drug resistance plasmid in Escherichia coli K12. J Gen Microbiol. 1979 Mar;111(1):201–210. doi: 10.1099/00221287-111-1-201. [DOI] [PubMed] [Google Scholar]
- Hooykaas P. J., den Dulk-Ras H., Schilperoort R. A. Method for the transfer of large cryptic, non-self-transmissible plasmids: ex planta transfer of the virulence plasmid of Agrobacterium rhizogenes. Plasmid. 1982 Jul;8(1):94–96. doi: 10.1016/0147-619x(82)90046-4. [DOI] [PubMed] [Google Scholar]
- Jones I. M., Primrose S. B., Robinson A., Ellwood D. C. Maintenance of some ColE1-type plasmids in chemostat culture. Mol Gen Genet. 1980;180(3):579–584. doi: 10.1007/BF00268063. [DOI] [PubMed] [Google Scholar]
- KING E. O., WARD M. K., RANEY D. E. Two simple media for the demonstration of pyocyanin and fluorescin. J Lab Clin Med. 1954 Aug;44(2):301–307. [PubMed] [Google Scholar]
- Philippsen P., Kramer R. A., Davis R. W. Cloning of the yeast ribosomal DNA repeat unit in SstI and HindIII lambda vectors using genetic and physical size selections. J Mol Biol. 1978 Aug 15;123(3):371–386. doi: 10.1016/0022-2836(78)90085-2. [DOI] [PubMed] [Google Scholar]
- Serdar C. M., Gibson D. T., Munnecke D. M., Lancaster J. H. Plasmid Involvement in Parathion Hydrolysis by Pseudomonas diminuta. Appl Environ Microbiol. 1982 Jul;44(1):246–249. doi: 10.1128/aem.44.1.246-249.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. Gel electrophoresis of restriction fragments. Methods Enzymol. 1979;68:152–176. doi: 10.1016/0076-6879(79)68011-4. [DOI] [PubMed] [Google Scholar]
- Wheatcroft R., Williams P. A. Rapid methods for the study of both stable and unstable plasmids in Pseudomonas. J Gen Microbiol. 1981 Jun;124(2):433–437. doi: 10.1099/00221287-124-2-433. [DOI] [PubMed] [Google Scholar]


