Abstract
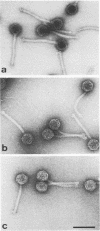
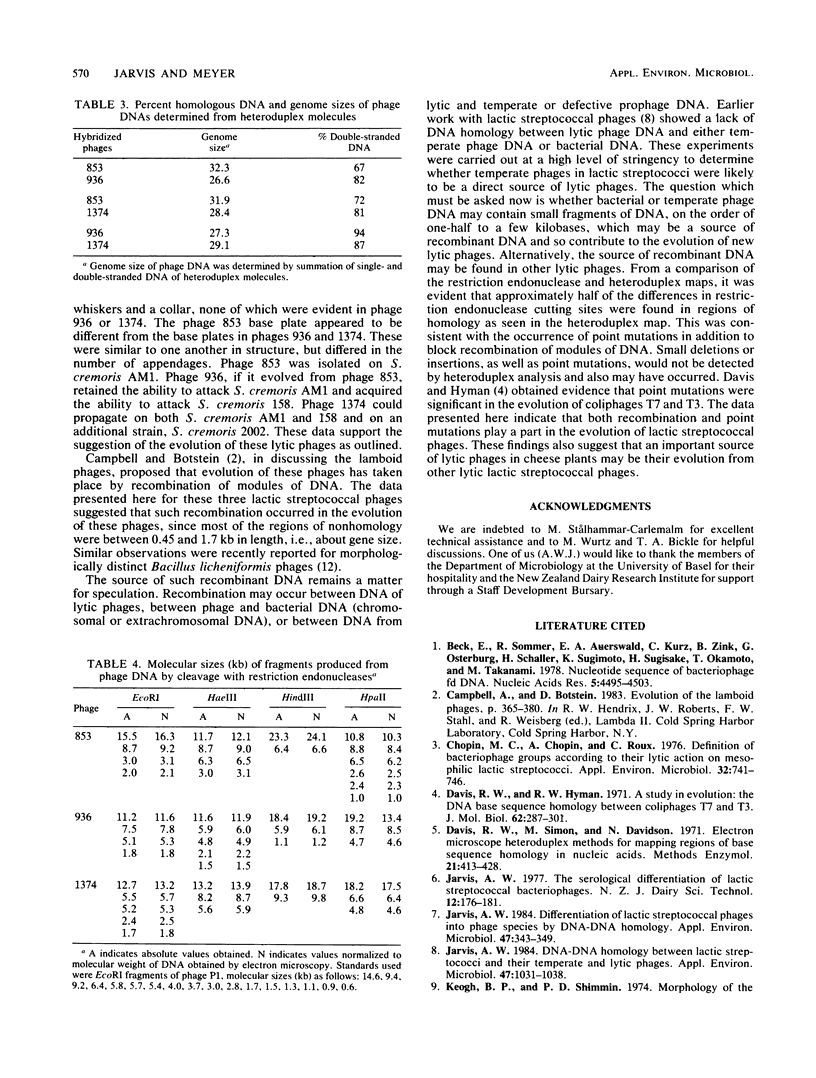
Three lactic streptococcal bacteriophages were compared with one another by electron microscopic analysis of heteroduplex DNA molecules. The phages were almost identical in morphology and had been isolated over a period of 10 years on different strains of Streptococcus cremoris from cheese plants situated in different parts of New Zealand. There was a high degree of homology between the DNAs, in agreement with Southern blot hybridization data reported earlier. There were, however, distinct regions of nonhomology, mostly between 0.45 and 1.71 kilobases in length, suggestive of the occurrence of block recombination events. A deletion of 2.23 kilobases in the two more recently isolated phages, or an insertion in the first isolate, was found. All three phage DNAs showed differences in restriction endonuclease cleavage sites. Alignment of the restriction endonuclease maps with the heteroduplex maps showed that differences in cleavage sites occurred most frequently in regions of nonhomology. However, differences in cleavage sites in regions of apparent homology were also detected, indicating that point mutations may have occurred in addition to block recombination events.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beck E., Sommer R., Auerswald E. A., Kurz C., Zink B., Osterburg G., Schaller H., Sugimoto K., Sugisaki H., Okamoto T. Nucleotide sequence of bacteriophage fd DNA. Nucleic Acids Res. 1978 Dec;5(12):4495–4503. doi: 10.1093/nar/5.12.4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopin M. C., Chopin A., Roux C. Definition of bacteriophage groups according to their lytic action on mesophilic lactic streptococci. Appl Environ Microbiol. 1976 Dec;32(6):741–746. doi: 10.1128/aem.32.6.741-746.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R. W., Hyman R. W. A study in evolution: the DNA base sequence homology between coliphages T7 and T3. J Mol Biol. 1971 Dec 14;62(2):287–301. doi: 10.1016/0022-2836(71)90428-1. [DOI] [PubMed] [Google Scholar]
- Jarvis A. W. DNA-DNA Homology Between Lactic Streptococci and Their Temperate and Lytic Phages. Appl Environ Microbiol. 1984 May;47(5):1031–1038. doi: 10.1128/aem.47.5.1031-1038.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis A. W. Differentiation of lactic streptococcal phages into phage species by DNA-DNA homology. Appl Environ Microbiol. 1984 Feb;47(2):343–349. doi: 10.1128/aem.47.2.343-349.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler C., Neumaier P. S., Wolf W. Recognition sequences of restriction endonucleases and methylases--a review. Gene. 1985;33(1):1–102. doi: 10.1016/0378-1119(85)90119-2. [DOI] [PubMed] [Google Scholar]
- Storchová H., Meyer J., Doskocil J. An electron microscopic heteroduplex study of the sequence relations between the bacteriophages LP52 and theta. Mol Gen Genet. 1985;199(3):476–480. doi: 10.1007/BF00330761. [DOI] [PubMed] [Google Scholar]