Abstract
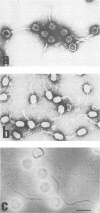
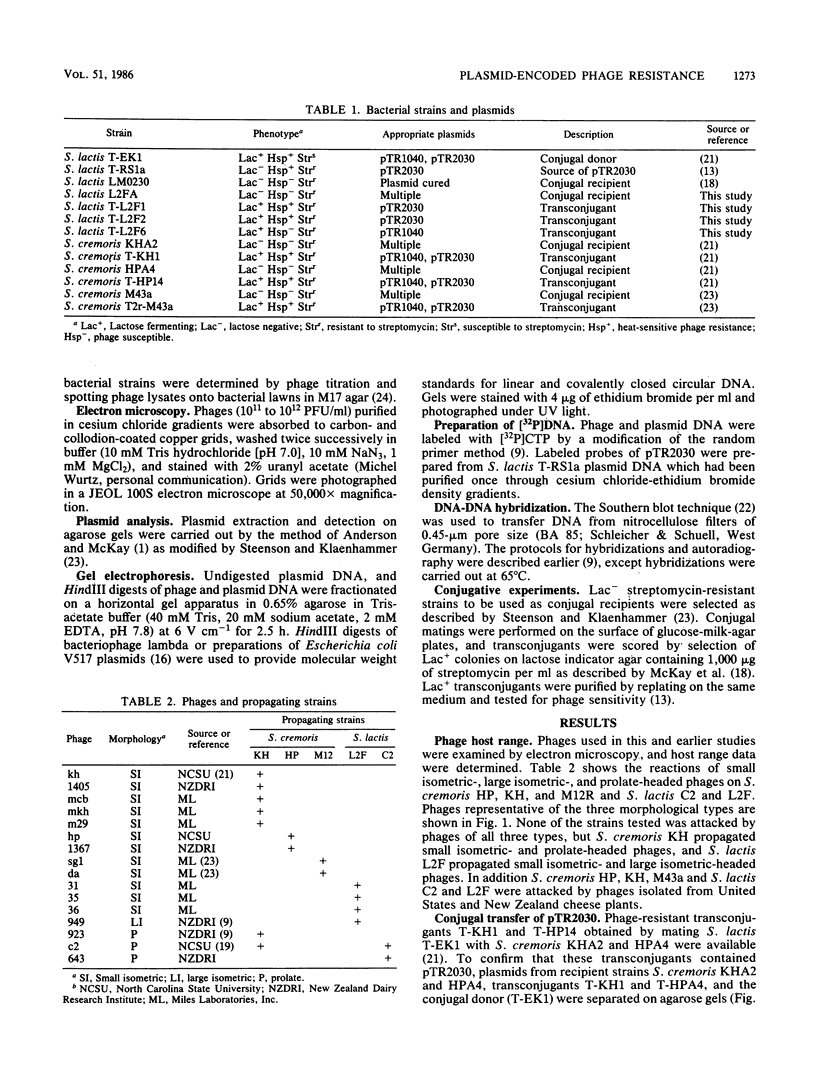
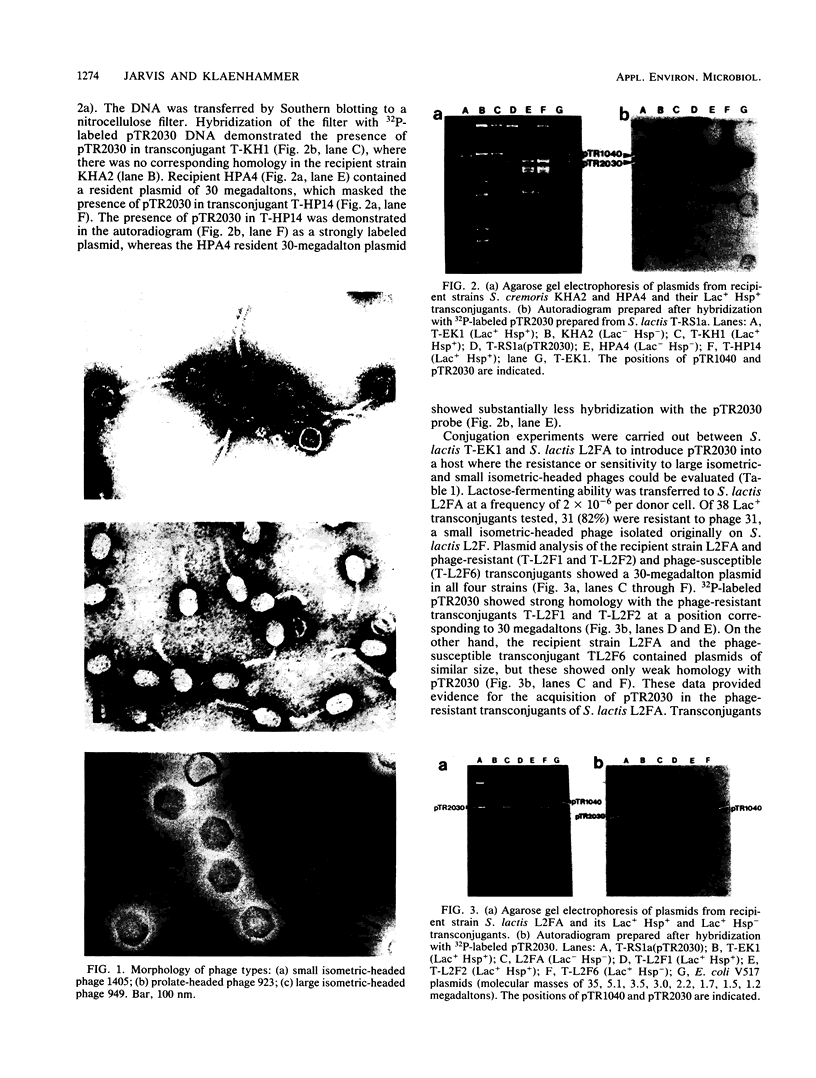
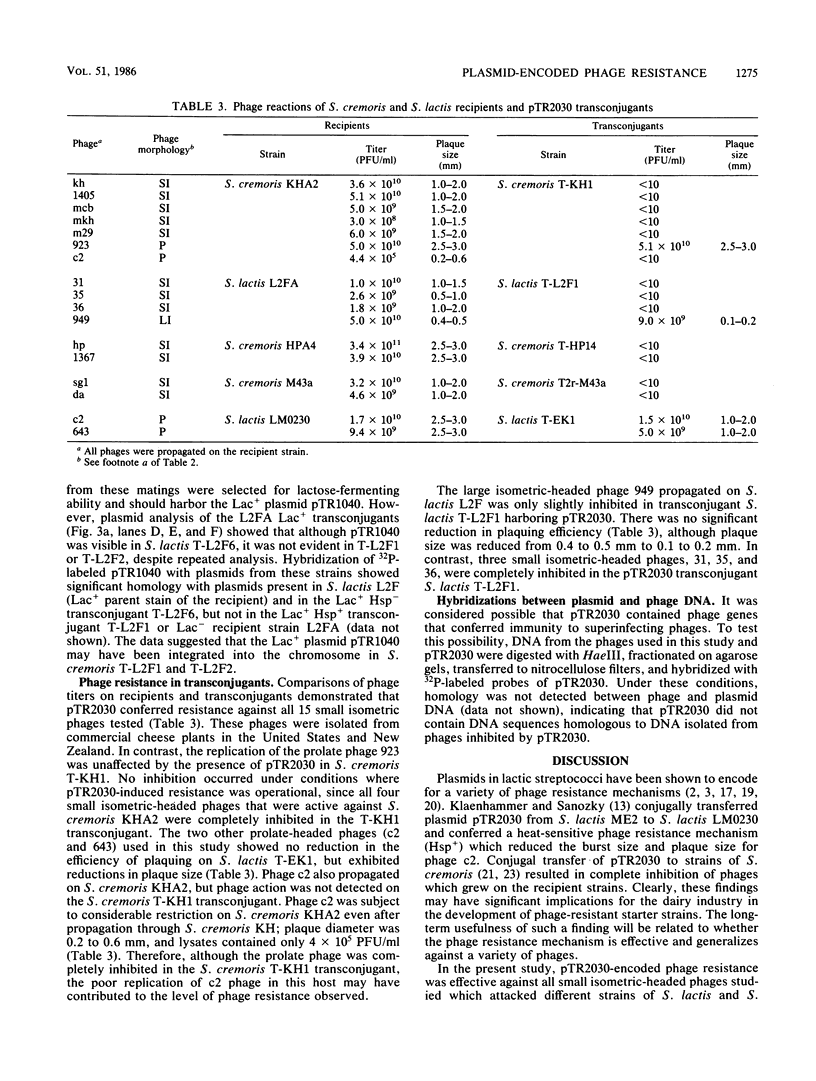
A series of reactions between phages, sensitive hosts, and transconjugants where the sensitivity of small isometric-, large isometric-, and prolate-headed phages to pTR2030-induced phage resistance was evaluated in Streptococcus lactis and Streptococcus cremoris strains. Phage-resistant transconjugants were constructed in the desired host by conjugal transfer of lactose-fermenting ability (Lac+, pTR1040) and phage resistance (Hsp+, pTR2030) from S. lactis TEK1. S. lactis and S. cremoris transconjugants harboring pTR2030 were resistant to all small isometric-headed phages examined. In contrast, prolate- and large isometric-headed phages were either not inhibited in the pTR2030 transconjugants or exhibited a reduction in plaque size without a reduction in the efficiency of plaquing. Small isometric-headed phages subject to pTR2030 induced inhibition shared no significant DNA homology with pTR2030, suggesting that phage immunity genes are not harbored on the plasmid or responsible for resistance. The general effectiveness of pTR2030 against small isometric-headed phages was highly significant since these are the phages which have been isolated most commonly from dairy fermentation plants.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson D. G., McKay L. L. Simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl Environ Microbiol. 1983 Sep;46(3):549–552. doi: 10.1128/aem.46.3.549-552.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopin A., Chopin M. C., Moillo-Batt A., Langella P. Two plasmid-determined restriction and modification systems in Streptococcus lactis. Plasmid. 1984 May;11(3):260–263. doi: 10.1016/0147-619x(84)90033-7. [DOI] [PubMed] [Google Scholar]
- Jarvis A. W. Differentiation of lactic streptococcal phages into phage species by DNA-DNA homology. Appl Environ Microbiol. 1984 Feb;47(2):343–349. doi: 10.1128/aem.47.2.343-349.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis A. W., Meyer J. Electron microscopic heteroduplex study and restriction endonuclease cleavage analysis of the DNA genomes of three lactic streptococcal bacteriophages. Appl Environ Microbiol. 1986 Mar;51(3):566–571. doi: 10.1128/aem.51.3.566-571.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keogh B. P., Shimmin P. D. Morphology of the bacteriophages of lactic streptococci. Appl Microbiol. 1974 Feb;27(2):411–415. doi: 10.1128/am.27.2.411-415.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaenhammer T. R., Sanozky R. B. Conjugal transfer from Streptococcus lactis ME2 of plasmids encoding phage resistance, nisin resistance and lactose-fermenting ability: evidence for a high-frequency conjugative plasmid responsible for abortive infection of virulent bacteriophage. J Gen Microbiol. 1985 Jun;131(6):1531–1541. doi: 10.1099/00221287-131-6-1531. [DOI] [PubMed] [Google Scholar]
- Macrina F. L., Kopecko D. J., Jones K. R., Ayers D. J., McCowen S. M. A multiple plasmid-containing Escherichia coli strain: convenient source of size reference plasmid molecules. Plasmid. 1978 Jun;1(3):417–420. doi: 10.1016/0147-619x(78)90056-2. [DOI] [PubMed] [Google Scholar]
- McKay L. L., Baldwin K. A. Conjugative 40-megadalton plasmid in Streptococcus lactis subsp. diacetylactis DRC3 is associated with resistance to nisin and bacteriophage. Appl Environ Microbiol. 1984 Jan;47(1):68–74. doi: 10.1128/aem.47.1.68-74.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay L. L., Baldwin K. A., Walsh P. M. Conjugal transfer of genetic information in group N streptococci. Appl Environ Microbiol. 1980 Jul;40(1):84–89. doi: 10.1128/aem.40.1.84-91.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders M. E., Klaenhammer T. R. Characterization of Phage-Sensitive Mutants from a Phage-Insensitive Strain of Streptococcus lactis: Evidence for a Plasmid Determinant that Prevents Phage Adsorption. Appl Environ Microbiol. 1983 Nov;46(5):1125–1133. doi: 10.1128/aem.46.5.1125-1133.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders M. E., Klaenhammer T. R. Evidence for Plasmid Linkage of Restriction and Modification in Streptococcus cremoris KH. Appl Environ Microbiol. 1981 Dec;42(6):944–950. doi: 10.1128/aem.42.6.944-950.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sing W. D., Klaenhammer T. R. Conjugal Transfer of Bacteriophage Resistance Determinants on pTR2030 into Streptococcus cremoris Strains. Appl Environ Microbiol. 1986 Jun;51(6):1264–1271. doi: 10.1128/aem.51.6.1264-1271.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Steenson L. R., Klaenhammer T. R. Streptococcus cremoris M12R transconjugants carrying the conjugal plasmid pTR2030 are insensitive to attack by lytic bacteriophages. Appl Environ Microbiol. 1985 Oct;50(4):851–858. doi: 10.1128/aem.50.4.851-858.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzaghi B. E., Sandine W. E. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975 Jun;29(6):807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsaneva K. P. Electron microscopy of virulent phages for Streptococcus lactis. Appl Environ Microbiol. 1976 Apr;31(4):590–601. doi: 10.1128/aem.31.4.590-601.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]