Abstract
The availability of the complete DNA sequence of the Chlamydomonas reinhardtii genome and advanced computational biology tools has allowed elucidation and study of the small ubiquitin-like modifier (SUMO) system in this unicellular photosynthetic alga and model eukaryotic cell system. SUMO is a member of a ubiquitin-like protein superfamily that is covalently attached to target proteins as a post-translational modification to alter the localization, stability, and/or function of the target protein in response to changes in the cellular environment. Three SUMO homologs (CrSUMO96, CrSUMO97, and CrSUMO148) and three novel SUMO-related proteins (CrSUMO-like89A, CrSUMO-like89B, and CrSUMO-like90) were found by diverse gene predictions, hidden Markov models, and database search tools inferring from Homo sapiens, Saccharomyces cerevisiae, and Arabidopsis thaliana SUMOs. Among them, CrSUMO96, which can be recognized by the A. thaliana anti-SUMO1 antibody, was studied in detail. Free CrSUMO96 was purified by immunoprecipitation and identified by mass spectrometry analysis. A SUMO-conjugating enzyme (SCE) (E2, Ubc9) in C. reinhardtii was shown to be functional in an Escherichia coli-based in vivo chimeric SUMOylation system. Antibodies to CrSUMO96 recognized free and conjugated forms of CrSUMO96 in Western blot analysis of whole-cell extracts and nuclear localized SUMOylated proteins with in situ immunofluorescence. Western blot analysis showed a marked increase in SUMO conjugated proteins when the cells were subjected to environmental stresses, such as heat shock and osmotic stress. Related analyses revealed multiple potential ubiquitin genes along with two Rub1 genes and one Ufm1 gene in the C. reinhardtii genome.
POST-TRANSLATIONAL modification can regulate protein function and cellular processes in a rapid and reversible manner. In addition to protein modification by small molecules such as phosphate and carbohydrates, peptides and small proteins also serve as modifiers. The three most studied small polypeptides that covalently modify other cellular proteins are ubiquitin, small ubiquitin-like modifier (SUMO), and neural precursor cell-expressed developmentally downregulated (Nedd)8 (Johnson 2004; Kerscher et al. 2006; Geiss-Friedlander and Melchior 2007; Palancade and Doye 2008). Ubiquitin amino acid sequence is highly conserved and the conjugation of ubiquitin to target proteins usually, but not always, results in their degradation by the 26S proteasome (Pickart 2000, 2001, 2004). Nedd8 shares high similarity with ubiquitin (60% identity and 80% similarity), and the primary substrates for Nedd8 in yeast and mammalian cells are Cullin proteins that play an important role in ubiquitin-mediated proteolysis (Kamitani et al. 1997; Yeh et al. 2000; Pan et al. 2004).
The three-dimensional (3-D) structure of human and yeast SUMO closely resembles that of ubiquitin (Melchior 2000; Hay 2001; Weissman 2001; Seeler and Dejean 2003; Johnson 2004). A prominent structural feature of SUMO is a long and highly flexible N terminus, which protrudes from the globular core of the protein. Despite the similarities in overall conformation, SUMO functions quite differently from ubiquitin. That is, SUMOylation often enables target proteins to participate in new and diverse cellular processes, including nuclear transportation, transcriptional regulation, maintenance of genome integrity, and signal transduction (Seeler and Dejean 2003; Colby et al. 2006).
In yeast and invertebrates, a single SUMO gene has been identified and has been shown to be essential for viability in Caenorhabditis elegans and Saccharomyces cerevisiae, while in Schizosacchromyces pombe, mutants lacking the single SUMO gene remain viable, but suffer severe defects in genome maintenance (Tanaka et al. 1999; Li and Hochstrasser 2003; Broday et al. 2004). Organisms have different numbers of SUMO isoforms and some SUMO isoforms appear to fulfill specialized functions. In humans, four major SUMO family members have been described, namely SUMO-1 to -4 (Melchior 2000; Hay 2001; Guo et al. 2004). Human SUMO-2 and -3 share 95% identity and their conjugation is strongly induced in response to various stresses (Holmstrom et al. 2003). In Arabidopsis thaliana, eight genes encoding SUMOs have been described (Kurepa et al. 2003). Similarity analysis clustered these SUMO proteins into five subfamilies: SUMO1/2, SUMO3, SUMO5, SUMO4/6, and SUMO7/8. As A. thaliana SUMO1 amino acid sequence is equally related to human SUMO-1, -2, and -3, it is difficult to group the A. thaliana SUMO proteins with animal and yeast homologs. As SUMOs from more plant and algal species are fully characterized, the relationship between SUMO sequence and function in plant biology likely will become clearer.
SUMOylation, the conjugation of SUMO peptide(s) to the target protein, results in an isopeptide bond between the C-terminal carboxyl group of a double-glycine (GG) motif in SUMO and the ɛ-amino group of a lysine residue in the target protein. A SUMO-specific protease generates a mature SUMO by cleaving C-terminal amino acids immediately following the double-glycine motif in precursor SUMO molecules (Bayer et al. 1998; Toshiaki et al. 1999; Nishida et al. 2001). The conjugating system is an ATP-dependent enzymatic cascade that takes place in three steps (E1, E2, and E3). In the first step, SUMO is activated to form a thiolester linkage with the cysteine residue of the SUMO-activating enzyme (SAE) (E1). After activation, SUMO is transferred to the active-site cysteine of the SUMO-conjugating enzyme (SCE), E2 (Ubc9), forming a SUMO-Ubc9 thiolester intermediate (Desterro et al. 1997; Johnson and Blobel 1997; Schwarz et al. 1999; Sampson et al. 2001). For some target proteins, such as Ran GTPase-activating protein 1 (RanGAP1), SUMO can be transferred directly from E2 to the substrate (Matunis et al. 1996). However, in most cases, a specific SUMO ligase (E3) is required for efficient and proper transfer of SUMO from E2 to a target protein (Hochstrasser 2001). In mammalian cells, RWD-containing SUMOylation enhancer (RSUME) has been shown to interact with Ubc9 and enhances SUMO-1, -2, and -3 conjugation (Carbia-Nagashima et al. 2007). For deconjugation, a specific protease/hydrolase/isopeptidase is required to cleave the isopeptide bond between SUMO and its substrate (Melchior et al. 2003). In yeast, ubiquitin-like protease 1 (Ulp1) catalyzes both SUMO maturation and SUMO deconjugation (Li and Hochstrasser 1999).
Numerous proteins have been identified as SUMO target proteins since the discovery of SUMO in 1996, including the important regulatory proteins c-Jun, p53, PCNA, histone, and histone deacetylase (Seeler and Dejean 2003; Kerscher et al. 2006). Target proteins generally contain a consensus motif, ψKXE, where ψ represents a large hydrophobic amino acid, X is any residue, and E (glutamine) can be substituted by D (aspartic acid) (Sternsdorf et al. 1999; Bernier-Villamor et al. 2002). This motif is sufficient for SUMOylation in vitro; however, for in vivo SUMOylation a nuclear localization signal is often required and interactions beyond those between the ψKXE motif and Ubc9, a SUMO conjugase, are likely to be critical for substrate selection (Kurtzman and Schechter 2001). In addition to ψKXE, several other sequence motifs have been found to be sites for SUMO attachment. These include TKXE, TKED, AKCP, VKYC, and VKFT (Johnson 2004). The requirement of both the nuclear localization sequence and SUMO consensus sequences can be used to search for putative target proteins in the Chlamydomonas reinhardtii genome. However, we have found that those short, unspecific motifs alone do not support in vivo SUMOylation. Fusion of putative target sequences to Ubc9 has been shown to aid in vivo detection of putative SUMOylation target proteins (Jakobs et al. 2007).
In plants, there is evidence that SUMOylation plays an important role in responding to stress and pathogens (Miura et al. 2007). The tomato SUMO homolog, LeSUMO, was shown in a yeast two-hybrid assay to interact with ethylene-inducing xylanase from the fungus Trichoderma viride, a strong elicitor of the rapid defense response in tomato. Moreover, the expression of LeSUMO in transgenic tobacco plants suppressed the induction of the defense response by ethylene-inducing xylanase, indicating that LeSUMO is likely to be a repressor in the plant defense pathway (Hanania et al. 1999). In A. thaliana, Western blot analysis of SUMO1/2 showed a significant increase of SUMO1/2 conjugates after exposure of seedlings to several stress conditions, such as heat shock, H2O2, ethanol, and the amino acid analog canavanine (Kurepa et al. 2003). In transgenic A. thaliana plants, overexpression of SUMO1/2 caused increased global SUMOylation levels, attenuated the abscisic acid-mediated growth inhibition, and induced the expression of abscisic acid and stress-responsive genes, such as RD29A (Lois et al. 2003). Dominant-negative A. thaliana mutants of the SUMO-conjugating enzyme ESD4 yield plants that are smaller and show delayed flowering (Reeves et al. 2002; Murtas et al. 2003). A mutation in a putative SUMOylation attachment site of long after far red light 1 (LAF1), a transcriptional activator for phytochrome A signaling, alters LAF1 accumulation in the nucleus (Ballesteros et al. 2001). A. thaliana SUMO E3 ligase (SIZ1) gene mutants show altered innate immunity (Lee et al. 2007) and an increased susceptibility to drought stress (Miura et al. 2007).
Although extensive proteomic analyses of SUMOylated proteins have been conducted in S. cerevisiae and mammalian cells (Denison et al. 2004; Rosas-Acosta et al. 2005), the sumoylation systems of other unicellular organisms, including C. reinhardtii, have not been investigated in depth. Indeed, even in plants where SUMOylation has been studied for some time (Kurepa et al. 2003), only three proteins have been verified as modified by SUMO, all of which are regulators of transcription (Miura et al. 2007). Thus, the identity, function, and regulation of plant SUMO target proteins and the cellular processes they regulate are largely unknown. In this work, we applied computational biology paradigms to gain insight into the SUMOylation system encoded in the genome of C. reinhardtii, a single-cell alga and model plant cell system. This includes the identification of three SUMO genes, three SUMO-like genes, and several putative Ubc9-like E2 conjugases and their characterization. Of the three SUMO genes, CrSUMO96 and CrSUMO97 are the most similar to other known SUMO proteins. CrSUMO148 represents a unique SUMO-like protein with tandem repeats of C-terminal domains each with a potential GG maturation cleavage site. CrSUMO-like89A, CrSUMO-like89B, and CrSUMO-like90 appear to encode proteins that may be fusions of SUMO sequences with the C termini of different proteins and, thus, represent a newly discovered class of SUMO-containing molecules that are akin to previously identified proteins containing ubiquitin-associated (UBA) domains (Spikes et al. 1994; Kerscher et al. 2006). Heat and osmotic pressure stress conditions increase the appearance of proteins recognized by CrSUMO96 antibodies, indicating that SUMOylation may be an important modification system for responses to stress conditions and in the regulation of gene expression in C. reinhardtii.
MATERIALS AND METHODS
Computational biology methods:
Releases 3.1 (R3.1) and 4.0 of the C. reinhardtii genome, a collection of 40,294 expressed sequence tags (ESTs), and R3.1 annotations were obtained from the Department of Energy Joint Genome Institute, Walnut Creek, California (http://genome.jgi-psf.org/Chlre3/Chlre3.home.html). We performed diverse searches (see below) on the genomes directly and on the annotated R3.1 proteome, as well as on our own gene prediction results. Because R4 has no publicly available gene or protein annotations, we also assembled the ESTs over the R4 genome as a reference by using the GMAP tool (Wu and Watanabe 2005). GMAP creates the optimal projection of the ESTs to the genome and identifies small exons, allowing mismatches and insertion/deletion events, using a minimal sampling strategy for genomic mapping, oligomer chaining for approximate alignment, and sandwich dynamic programming.
GMAP EST assemblies and de novo gene predictions by the augustus algorithm (Stanke and Waack 2003) were integrated via the program to assemble spliced alignments (PASA) pipeline (Haas et al. 2003) that generates maximal alignment assemblies, comprehensively incorporating all available transcript data.
SUMO genes in the C. reinhardtii genome were inferred from a reference set of known SUMO proteins. This reference set was seeded by all well-characterized SUMO proteins in A. thaliana, Homo sapiens, and S. cerevisiae (supplemental Table I). Potential homologs of the seed sequences were retrieved from the Nonredundant Protein Database at the National Center for Biotechnology Information using BLAST searches (Altschul et al. 1997). We retained database sequences annotated as SUMO proteins with significant similarity (e ≤ 10−20) to the query. The final reference set included SUMO proteins from tomato, S. cerevisiae, S. pombe, Schistosoma mansoni (Cabral et al. 2008), and other species.
Direct TBLASTN (Altschul et al. 1997) searches against genomes produced a high number, but only marginally significant, of hits due to missing certain short, primarily 5′-end exons. Accuracy was improved by selecting the 200,000-bp neighborhoods centered around each of the TBLASTN hits and performing genewiseDB searches (Birney et al. 2004) against these neighborhoods. GenewiseDB applies hidden Markov models (HMMs) for the exon/intron boundary predictions and for the assessment of the similarity to the query sequence. Narrowing the search space not only decreased the heavy computational load of the brute-force genewiseDB searches against the whole R4 genome but also resulted in more realistic gene predictions. In whole-genome searches, genewiseDB merged exons >0.5 million bp apart, an approach necessary for sizeable mammalian introns but not for the considerably shorter introns of the compact C. reinhardtii genome. We also performed estwiseDB searches (Birney et al. 2004) against the compact EST assemblies, where no such measures were necessary.
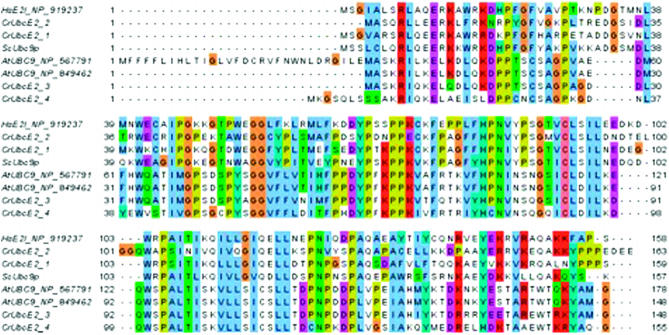
It is remarkable that the only apparent extensive modular feature of the SUMO architecture is moderately similar to the ubiquitin domain as indicated by HMMs from the PFAM (Finn et al. 2008) and the SMART (Schultz et al. 1998) protein domain databases. Unfortunately, the ubiquitin domain also is shared with a number of SUMO proteases, SUMO-activating enzymes, and other ubiquitin-related proteins that are not SUMOs. Also, the ubiquitin model missed some bona fide C. reinhardtii SUMOs. This may explain why searches querying either the ubiquitin domain HMM or individual reference sequences produced a considerable number of hits, many of them false positives. This motivated our quest for more selective and sensitive HMMs. We selected well-characterized SUMO proteins (Figure 1) from the reference set, created their multiple alignment, and trained and calibrated a HMM using the HMMER package (Eddy 1998). This specific HMM query produced highly significant hits to the C. reinhardtii SUMO proteins.
Figure 1.—
SUMO homologs in C. reinhardtii: amino acid sequence alignment of yeast, human, and Arabidopsis SUMOs against SUMO homologs of C. reinhardtii.
The close sequence similarity of SUMO and other, functionally divergent ubiquitin-related proteins mandated conservative analyses. Therefore we eliminated a number of ubiquitin-related but not SUMO proteins. Such cases may be identified when the most similar sequences to a candidate are not annotated as SUMOs. We also eliminated proteins with less significant similarity that lacked the canonical glycine–glycine/asparagine cleavage site motif in the vicinity of the C terminus. We also discarded a number of pseudogenes produced by gene duplication events. In these pseudogenes, the loss of function or the lack of transcription was indicated by in-frame stop codons, unusual codon usage, or overly divergent ubiquitin domains.
Scripts were written in the PERL programming language or MATLAB (MathWorks, Nantucket, MA). All computations were performed under the LINUX CentOS Operation System on Intel Xeon 64-bit processors. BLAST, genewiseDB, and estwiseDB searches, augustus predictions, and EST mappings were processed on a compute farm of 80 nodes under the Portable Batch System (PBS-PRO) job-scheduling software. Multiple alignments were created by the T-Coffee package (Notredame et al. 2000) and displayed by the Jalview Java Alignment Viewer (Clamp et al. 2004). Similarity trees were constructed by the first-order algorithm of the neighbor-joining method (Gascuel 1997). Nuclear and subnuclear localization was predicted by the method of Lei and Dai (2005). Three-dimensional structure predictions were performed by using the Swiss-Model automated comparative protein modeling server (http://www.expasy.org/swissmod/SWISS-MODEL.html).
Chlamydomonas strains, growth conditions, and stress treatments:
C. reinhardtii wall-less, wild-type strain CC-503 and walled, wild-type strain CC-125 were originally obtained from the Chlamydomonas Genetics Center at Duke University (Durham, NC). They were maintained on Tris-acetate phosphate (TAP) plates containing 1.2% agar (Harris 1989) at 25° under constant light (60 μE · m−2 · sec−1). For RNA or protein isolation, cells were inoculated into liquid TAP media (Harris 1989), unless indicated otherwise, and allowed to grow under continuous light at 25° on a rotary shaker at 135 rpm to a density of ∼0.5–1 × 107 cells/ml. For heat-shock experiments, midlog-phase cells were transferred to incubators prewarmed to 37° or 42° and grown for the indicated time. For osmotic stress treatments, sorbitol or sodium chloride was added to midlog cells to final concentrations of 200 or 100 mm, respectively.
Quantitative real-time RT–PCR reactions:
Total RNA was isolated from 25 ml of CC125 and CC503 cells grown at 25° and shifted to 42° for 1 hr by PureYield RNA isolation (Promega, Madison, WI) according to the manufacturer's instructions. Complementary DNA was first synthesized with oligo(dT) priming of 1 μg of total RNA in a Plexor Two-Step qRT–PCR system (Promega) reaction both with and without reverse transcriptase and subsequently diluted 1:20 in 1 mm MOPS, 0.1 mm EDTA. Quantitative PCR was carried out in a 25-μl reaction mixture that contained the vendor's master mix, 0.25 mm of each primer (Biosearch Technologies, Novato, CA), and 5 μl cDNA. The primer sets were separately tested for efficiency. The reaction conditions for the ABI 7500 were 95° for 2 min, followed by 40 cycles of 95° for 5 sec, 65° for 30 sec, with dissociation conditions of 95° for 15 sec, 60° for 1 min, and 95° for 15 sec. Primer sets used for real-time RT–PCR reactions were as follows: CrSUMO-like89A, 5′-GTTGAACACGAAGCGGTGGTT-3′ and 5′-AATCAACACGTATGGGCAGAGTC-3, 89% efficiency; CrSUMO-like90, 5′-CAGGCCCTTCTTGTTGCAGTAG-3′ and 5′-CAACATCATTATCAAAGGACAGGGTG-3′, 90% efficiency; CrSUMO96, 5′-AGTTCCATCCACAATTACCGACC-3′ and 5′-CTCACCGGTCATGGAGTGATTG-3′, 92% efficiency; CrSUMO97, 5′-GTACTGTCTGCTCTACCGACTGAA-3′ and 5′-TGAATGGTTTGGATTAGACGGTTGG-3′, 96% efficiency; CrSUMO148, 5′-GTCGCAACGCCCTTCTTCTG-3′ and 5′-GCGATCAACATCTCAATCAAGTCTACT-3′, 96% efficiency; and CIA5, 5′-GGGTCCCGTCAAACAACAACC-3′ and 5′-TCAGGTCAGGTCGGTGCATGA-3′, 92% efficiency. Quantitative PCR on minus reverse transcriptase reactions did not show signal, confirming the absence of detectable DNA in the input total RNA. The 2−ΔΔCt method (Livak and Schmittgen 2001) was used to compare relative transcript abundance in the 25° and 42° samples normalized to an endogenous reference gene, CIA5 (Xiang et al. 2001). Efficiency-corrected ΔCt values of the different CrSUMO mRNAs in each cell sample, normalized to CIA5 mRNA were compared to the lowest abundant transcript, CrSUMO89A mRNA, which was set at 1.0 for comparison purposes. Levels of CIA5 mRNA did not change under the conditions tested in this work.
Cloning, total RNA isolation, RT–PCR, and plasmid construction:
Total RNA was isolated as described before (Xiang et al. 2001), and cDNA coding regions for CrSUMO96, CrSUMO148, CrSUMO-like89A, and CrUBCE2_1 were reverse transcribed and amplified by using the following primer sets: CrSUMO96-forward, 5′-TCCGAATCCATGGCGGACGAGGAGGCTAAG-3′ (NcoI site underlined); CrSUMO96 reverse, 5′-CCTCGAGTGCGCAGCTGCAGCCGCCGACCT-3′ (XhoI site underlined); CrSUMO148-forward, 5′-ACCATGGCGGACGTTAAGGCTGAGGCGATCA-3′ (NcoI site underlined); CrSUMO148-reverse, 5′-CCTCGAGCCCTGTCCCACGGCGCAGCACAG-3′ (XhoI site underlined); CrSUMO-like89A-forward, 5′-CCCATGGAGGCGGCGGCGGAAGGGCAGCATGGTGAC-3′ (NcoI site underlined); CrSUMO-like89A-reverse, 5′-CTGCTCGAGGTTGCCAACTTGTGCAGGCACGCAATC-3′ (XhoI site underlined); CrUBCEB_1-forward, 5′-CCGAATTCATGTCTGGCGTCGCACGCTCAC-3′ (EcoRI site underlined); and CrUBCE2_1-reverse, 5′-GACCTCGAGTCACGAGGGTGGCGGGTAGT-3′ (XhoI site underlined). One microgram of total RNA was utilized for first-strand cDNA synthesis, and the reaction was performed at 42° for 1 hr by using the SuperScriptII reverse transcriptase (Invitrogen Life Technologies, Carlsbad, CA) and reverse primers for each gene. PCR was performed with Pfu DNA polymerase (Stratagene, La Jolla, CA) for 35 cycles annealed at 58° and extended for 1 min. RT–PCR products were recovered by using a GENECLEAN SPIN kit (Qbiogene, Carlsbad, CA), A-tailed with Taq DNA polymerase (Invitrogen Life Technologies), and ligated into pGEM-TEasy Vector (Promega). The NcoI/XhoI fragments from CrSUMO96, CrSUMO148, and CrSUMO-like89A were cloned into pET28b vector (Novagen; EMD Biosciences, San Diego). These plasmids were named pET28b-CrSUMO96-CterHis, pET28b-CrSUMO148-CterHis, and pET28b-CrSUMO-like89A-CterHis, respectively. The EcoRI/XhoI fragment from a CrUBCE2_1 cDNA clone was introduced into pGEX-4T-1 vector (Amersham Biosciences, Piscataway, NJ), and the resultant plasmid was named pGEX4T1-CrUbcE2-1.
Expression and purification of recombinant proteins:
Plasmids pET28b-CrSUMO96-CterHis, pET28b-CrSUMO148-CterHis, and pET28b-CrSUMO-like89A-CterHis were transformed into Escherichia coli BL21 (Novagen, EMD Biosciences), respectively. Following a 4-hr incubation at 37° with 0.5 mm isopropyl-1-thio-d-galactopyranoside (IPTG) Invitrogen Life Technologies), the BL21 cells were lysed with CelLytic bacterial cell lysis extraction reagent (Sigma, Saint Louis), and total soluble protein was applied to a HIS-Select nickel affinity gel (Sigma) column. His-tagged protein was eluted with 50 mm sodium phosphate, pH 8.0, containing 0.3 m sodium chloride and 250 mm imidazole and was dialyzed overnight against phosphate-buffered saline (PBS). Protein concentration was determined by a Bio-Rad (Hercules, CA) protein assay.
Antisera, immunoblots, and immunoprecipitation:
Polyclonal antibodies were raised against purified His-CrSUMO96, His-CrSUMO148, and His-CrSUMO-like89A (Cocalico Biologicals, Reamstown, PA). Antibody affinity purification was performed according to Ermolova et al. (2003). Arabidopsis anti-SUMO1 antibody was a gift from Richard Vierstra (University of Wisconsin, Madison, WI), and the anti-histidine tag antibody was purchased from Clontech Laboratories (Mountain View, CA).
For protein electrophoresis and immunoblots, exponentially growing cells were harvested and resuspended in a density of ∼0.5–1 × 108/ml with Tris-buffered saline (TBS). In experiments detecting SUMO-conjugated proteins, protease inhibitor cocktail (Sigma) and 2 mm N-ethylmaleimide (NEM) were added to TBS. Resuspended cells were mixed with an equal volume of 2× SDS sample buffer, boiled, and resolved by SDS–PAGE (Laemmli 1970), using 10% polyacrylamide (w/v) separation gels and 5% polyacrylamide (w/v) stacking gels. Proteins were transferred onto nitrocellulose membranes (Amersham Biosciences) with a semi-dry electrophoretic transfer cell (Bio-Rad Laboratories). The membranes were blocked in TBS containing 5% milk powder and probed with antibodies diluted in blocking buffer. Detection employed horseradish peroxidase (HRP)-labeled donkey anti-rabbit immunoglobulins (Amersham Biosciences) in conjunction with Super Signal chemiluminescence (Pierce, Rockford, IL) and X-Omat autoradiographic film (Eastman Kodak, Rochester, NY).
For two-dimensional PAGE analysis of proteins from control cells grown at 25° and from similar cell cultures exposed to a 42° heat-shock treatment for 1 hr, 20 ml of cells at ∼3 × 106 cells/ml were collected by centrifugation and extracted with acetone and phenol (Hajduch et al. 2005). A total of 400 μg of the resulting proteins from each sample were separated by isoelectric focusing using an 11-cm ReadyStrip IPG strip, pH 3–10 (Bio-Rad). Proteins separated by isoelectric focusing were submitted to electrophoresis on a 4–20% gradient SDS–polyacrylamide gel with a 4% stacking gel. After transfer of the proteins to a nitrocellulose membrane, the blot was incubated with anti-CrSUMO96 antibodies at a dilution of 1:500 overnight at 4° and then with secondary antibodies (ECL rabbit IgG, HRP-linked whole Ab, from donkey; GE Healthcare, Piscataway, NJ) at a 1:2500 dilution. SUMO and SUMOylated proteins on the blot were detected using a SuperSignal West Pico chemiluminescent substrate (Pierce).
For immunoprecipitation experiments, exponentially growing cells were harvested and washed with TBS and then resuspended to a density of ∼0.5–1 × 108/ml with lysis buffer [TBS, 1% (v/v) Tween-20, and protease inhibitor cocktail (Sigma)]. Cells were broken with a sonication pulse pattern (pulse on 1 sec, pulse off 1 sec, 1-min cycle repeated six times), using a tapered microtip and a VCX600 ultrasonic processor (Sonics & Materials, Danbury, CT). In some cases, sonicated cell lysate was heated to 90° for 30 min. Clarified cell lysate was mixed with 10 μg purified anti-CrSUMO96 polyclonal antibody, 20 μl 25% protein A agarose (Santa Cruz Biotechnology, Santa Cruz, CA), and protease inhibitor cocktail (Sigma), diluted with TBS to a final volume of 1 ml. The reaction was incubated at 4° for 2 hr, and the immunocomplexes were washed three times with lysis buffer and eluted with 2× SDS sample buffer. The resulting proteins were resolved by SDS–PAGE, detected by silver staining or transferred to nitrocellulose membranes, and detected with anti-CrSUMO96 antibody (Bio-Rad Laboratories).
Mass spectrometric peptide sequencing of CrSUMO96:
Proteins purified by immunoprecipitation were excised from a silver-stained gel. Each protein band was cut into ∼1-mm3 pieces and destained. After destaining, gel pieces were dehydrated with 100% acetonitrile and dried under vacuum for 15 min. The gel pieces were rehydrated in a solution of 40 mm NH4HCO3, 10% acetonitrile, and 20 μg/ml trypsin and incubated at 37° overnight. The peptides were subjected to HPLC/mass spectrometry (MS)/MS analysis performed by the mass spectrometry core facility in the Redox Biology Center of the University of Nebraska (Lincoln, NE).
Immunofluorescence localization assay:
For immunofluorescence experiments, cells at a density of ∼2–5 × 106 cells/ml were fixed by addition of 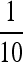 vol of formaldehyde (Fisher Scientific Chemicals, Fairlawn, NJ) and incubated at room temperature for 3 min. Cells were then collected by centrifugation (3 min at 3000 × g), washed with phosphate-buffered saline three times, resuspended in cold methanol, and kept overnight at −20°. An aliquot of 100 μl of cells at a density of ∼2–5 × 106 cells/ml was spotted onto a polylysine-treated coverslip (polyprep, Sigma) and allowed to stand for 10 min. Fixation and extraction of pigments were conducted by plunging the coverslip into a Coplin jar containing methanol at −20° and incubating the slide in the cold methanol for 5 min. This procedure was then repeated. The CrSUMO96 primary antibody and the cyanine-5-conjugated goat anti-rabbit secondary antibody (Jackson ImmunoResearch Laboratories, West Grove, PA) were diluted 1:100 and 1:60, respectively. Sytox green (Molecular Probes, Eugene, OR) in PBS was used for DNA staining at a 0.25-μm final concentration. Images were photographed using an Olympus Fluoroview FV500 confocal laser scanning system with an Olympus BX60 fluorescence microscope (Olympus America, Melville, NY). This immunofluorescence procedure was based on a protocol provided by Susan Dutcher (Washington University, St. Louis).
vol of formaldehyde (Fisher Scientific Chemicals, Fairlawn, NJ) and incubated at room temperature for 3 min. Cells were then collected by centrifugation (3 min at 3000 × g), washed with phosphate-buffered saline three times, resuspended in cold methanol, and kept overnight at −20°. An aliquot of 100 μl of cells at a density of ∼2–5 × 106 cells/ml was spotted onto a polylysine-treated coverslip (polyprep, Sigma) and allowed to stand for 10 min. Fixation and extraction of pigments were conducted by plunging the coverslip into a Coplin jar containing methanol at −20° and incubating the slide in the cold methanol for 5 min. This procedure was then repeated. The CrSUMO96 primary antibody and the cyanine-5-conjugated goat anti-rabbit secondary antibody (Jackson ImmunoResearch Laboratories, West Grove, PA) were diluted 1:100 and 1:60, respectively. Sytox green (Molecular Probes, Eugene, OR) in PBS was used for DNA staining at a 0.25-μm final concentration. Images were photographed using an Olympus Fluoroview FV500 confocal laser scanning system with an Olympus BX60 fluorescence microscope (Olympus America, Melville, NY). This immunofluorescence procedure was based on a protocol provided by Susan Dutcher (Washington University, St. Louis).
In vivo E2 ligase assay:
Plasmids pTE1E2S1 and pET28-RanGAP1-C2 were obtained from Hisato Saitoh (Kumamoto University, Japan). UBC9 (E2) in the original plasmid pT-E1E2S1 was eliminated by removing the small NdeI fragment and circularizing the plasmid, designated as pT-E1E2S1ΔE2. Plasmid cotransformation was conducted as follows: 1 μg of each of the pT-E1E2S1ΔE2 and pET28-RanGAP1-C2, with or without 1 μg pGEX4T1-CrUbcE2B-1, was mixed with 100 μl BL21 competent cells. After incubation for 30 min on ice followed by a heat-shock treatment, 1 ml of Luria–Bertani (LB) media was added. The bacteria were incubated at 37° for 2 hr and plated on LB plates containing 50 μg/ml chloramphenicol and 25 μg/ml kanamycin, with or without 100 μg/ml ampicillin. Colonies were picked randomly and inoculated in LB media containing appropriate antibiotics. After IPTG induction at 37° for 4 hr, total soluble protein was separated by SDS–PAGE. The expression of RanGAP1-C2 was detected by immunoblot analysis with HRP-conjugated anti-6× His antibody (Clontech Laboratories), and the expression of CrUbcE2_1 was detected with anti-GST antibody (Santa Cruz Biotechnology).
RESULTS
SUMO and SUMO-like proteins are encoded by at least six genes in C. reinhardtii:
C. reinhardtii SUMO genes and proteins in V3.1 of the genome determined by the Department of Energy Joint Genome Institute (JGI) were complemented by our gene/protein models for V4 of the genome. We performed the optimal mapping of the ESTs we assembled over the V4 genome by the GMAP tool (Wu and Watanabe 2005) and predicted genes using the augustus algorithm (Stanke and Waack 2003). Databases were queried by protein and translated BLAST, position-specific iterative BLAST (Altschul et al. 1997), HMMER (Eddy 1998), estwise, and TBLASTN-directed genewise (see Computational biology methods) (Birney et al. 2004) searches. Sequence reference sets and SUMO HMMs were developed as described in Computational biology methods. Initially, four SUMO homologs were identified and named CrSUMO96, CrSUMO148, and CrSUMO-like89A and CrSUMO-like89B, respectively, on the basis of the length of their predicted amino acid sequences (Table 1). They share 68, 54, 56, and 61% amino acid sequence similarity with the closest human SUMO homologs and 64, 34, 56, and 56% similarity with A. thaliana SUMO1, respectively (Figure 1, Table 1). In version 4 of the JGI database of the Chlamydomonas genome, a fifth SUMO homolog was discovered and named CrSUMO97. It shares 73% identity and 80% similarity to CrSUMO96. The close similarity of CrSUMO96 and CrSUMO97 and their separation by only 161 bp on chromosome 1 (Table 2) suggest a gene duplication in the recent past. In a subsequent database examination, an additional CrSUMO-like protein, CrSUMO-like90, was discovered. The amino acid sequences of the three CrSUMO proteins and the C-terminal amino acid sequences of the three CrSUMO-like proteins are presented in Figure 1 along with amino acid sequences of SUMOs from S. cerevisiae, H. sapiens, and A. thaliana. A high degree of similarity between the various SUMO sequences is observed (Table 1 and Figure 1).
TABLE 1.
SUMO proteins in C. reinhardtii and the extent of their similarity to their closest A. thaliana, S. cerevisiae, and human homologs, as well as scores for our SUMO-domain hidden Markov model
| C. reinhardtii SUMO name | GenBank protein ID | % similar residuesa to the most similar protein in
|
||||
|---|---|---|---|---|---|---|
| C. reinhardtii | A. thaliana | S. cerevisiae | H. sapiens | HMM score | ||
| CrSUMO89A | XP_001700385 | SUMO90, 69 | SUMO2b, 56 | Smt3pc, 64 | EAW52726d, 56 | 11.9 |
| CrSUMO89B | EU553548 | SUMO90, 78 | SUMO2b, 56 | Smt3pc, 55 | AAH65723e, 61 | 24.3 |
| CrSUMO90 | XP_001700386 | SUMO89A, 77 | SUMO2b, 54 | Smt3pc, 56 | AAH65723e, 64 | 35.0 |
| CrSUMO96 | XP_001695783 | SUMO97, 77 | SUMO2b, 64 | Smt3pc, 61 | SUMO-1f, 68 | 107.8 |
| CrSUMO97 | XP_001695782 | SUMO96, 80 | SUMO2b, 70 | Smt3pc, 61 | SUMO-3g, 59 | 112.7 |
| CrSUMO148 | XP_001697951 | SUMO96, 59 | SUMO2b, 34 | Smt3pc, 34 | SUMO-3g, 54 | 48.5 |
The number of identical plus other positively scoring amino acid residue positions divided by the total number of residues in the query protein. Residues downstream of the canonical GG/GN cleavage site motif were disregarded.
At5g55160 small ubiquitin-like modifier 2, A. thaliana.
NP_010798 ubiquitin-like protein of the SUMO family, S. cerevisiae.
hCG1766780.
LOC391257 protein.
Human small ubiquitin-related modifier 1 precursor (SUMO-1) (sentrin), NP_003343 SMT3 suppressor of mif two 3 homolog 1 of S. cerevisiae.
Human small ubiquitin-related modifier 3 precursor (SUMO-3) (ubiquitin-like protein SMT3A), NP_008867 small ubiquitin-like modifier protein 3.
TABLE 2.
Cleavage and binding domains of the C. reinhardtii SUMO proteins, their chromosomal localization, and their relative abundance in RT–PCR experiments (see also Figure 3)
| C. reinhardtii SUMO name | Patterns
|
N-term. ext.b | Nuclear localization | Chr. no. | Relative transcript abundancec | ||||
|---|---|---|---|---|---|---|---|---|---|
| GG | GN | ΨKXEa | Start | End | |||||
| CrSUMO89A | IGNd | 6 | Nuclear lamina | 17 | 2,511,141 | 2,510,908 | + | ||
| CrSUMO89B | VGNd | 19 | Nuclear lamina | 17 | 2,507,181 | 2,506,915 | ND | ||
| CrSUMO90 | VGNd | 19 | Nucleoplasm | 17 | 2,503,656 | 2,503,420 | ++ | ||
| CrSUMO96 | QVGG | VKTE | 20 | Nucleoplasm | 1 | 9,064,900 | 9,063,630 | +++ | |
| CrSUMO97 | QVGGG | VKSE | 23 | Nucleoplasm | 1 | 9,068,532 | 9,065,061 | — | |
| CrSUMO148 | QEGG (4) SRGG | VKAE | 14 | Nuclear lamina | 16 | 1,920,521 | 1,917,872 | ++ | |
N-term. ext., N-terminal extension; Chr. no., chromosome number.
Only the canonical ΨKXE motif [a large aliphatic (isoleucine, valine, or leucine), a lysine, any residue, followed by a glutamic or aspartic acid residue] was found. These proteins lack the alternative TKXE, TKED, AKCP, VKYC, and VKFT motifs.
The number of residues amino terminal to the ubiquitin domain.
Data are provided in Figure 3.
Does not contain double glycine motif.
The C-terminal segment, CSCALE, in CrSUMO96 and the GVSA segment in CrSUMO97 precursor proteins are cleaved to bring the canonical GG motif into the C-terminal position. This double-G motif functions as the attachment site for the SUMO-activating enzyme, E1, and, ultimately, it reacts with an ɛ-amino group of a lysine residue within a protein targeted for SUMOylation. CrSUMO148 presents a unique SUMO structure. It possesses five separate double GG potential cleavage sites within its long C-terminal domain. Three of the double-G motifs are part of a perfect IDAFVQEGG repeat (Figure 1).
The three CrSUMO-like proteins are distinctly different from the three CrSUMO proteins with regard to a number of features. First, they are distinguished by the lack of a double-G motif at the C termini. Instead, they possess nonprocessed glycine–asparagine C termini (i.e., the mRNAs for these proteins contain glycine and asparagine codons immediately upstream of a termination codon, producing “GN” C termini). Second, these proteins possess unusual proline inserts/substitutions near the C termini of the proteins (positions 92 and 104 for CrSUMO-like90, position 92 for CrSUMO-like89A, and position 104 for CrSUMO-like89B in Figure 1). Very close proximity of the three closely related CrSUMO-like genes on chromosome 17 (Table 2) suggests they may have arisen from two gene duplication events. Finally, the presence of long upstream reading frames for the genes encoding the three CrSUMO-like proteins (http://genome.jgi-psf.org/Chlre3/Chlre3.home.html) is consistent with the potential existence of these proteins as fusions of the SUMO-like proteins to the C termini of three separate and possibly unrelated proteins.
Protein identification numbers and other characteristics of each of the SUMO and SUMO-like proteins are provided in Tables 1 and 2. A similarity tree representing the amino acid sequence distance among SUMOs in different species was constructed, using the first-order algorithm of the neighbor-joining method (Gascuel 1997) (supplemental Figure 1).
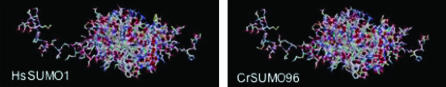
CrSUMO96 is encoded by a gene containing five exons. A ψKXE consensus motif, VKTE, was found at the N terminus of the polypeptide, indicating the possibility of poly-SUMOylation. Cleavage by an Ulp hydrolase/isopeptidase after the tandem glycine residues would produce a mature SUMO of 90 amino acids in length. The 3-D structure of CrSUMO96 was predicted using the Swiss-Model automated comparative protein-modeling server. The predicted 3-D structure is very similar to that of the human SUMO-1 except for a slight difference at the N terminus (Figure 2). The highly similar CrSUMO97 is encoded by a gene with eight exons. As with CrSUMO96, CrSUMO97 has a C-terminal GG motif followed by 4 amino acids in the immature precursor and contains an N-terminal ψKXE SUMOylation site motif.
Figure 2.—
Predicted 3-D structure of human SUMO1 and CrSUMO96.
The CrSUMO148 gene contains six exons with a predicted reading frame of 148 amino acids. The CrSUMO148 protein also contains a ψKXE consensus motif, VKAE, at the N terminus. Cleavage after the first double-glycine repeat would produce a mature protein of 83 amino acids in length. Cleavage after the second, third, fourth, and fifth double-glycine repeats would produce mature proteins of 93, 103, 113, and 135 amino acids, respectively.
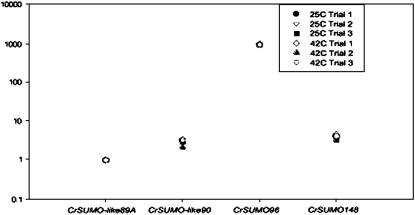
Real-time RT–PCR detected mRNAs of CrSUMO-like89A, CrSUMO-like90, CrSUMO96, and CrSUMO148 (Figure 3). CrSUMO97 mRNA was not detected in these experiments, even with input RNA of 1 μg. CrSUMO-like89B mRNA was not investigated. There was no detectable difference in the respective CrSUMO mRNA abundance between samples from cells that were shifted from 25° to 42° for 1 hr, and 2−ΔΔCt values for those conditions resulted in values close to 1.0. However, some CrSUMO transcripts are more highly expressed than others. CrSUMO96 mRNA was dramatically more abundant when compared to CrSUMO-like89A expression levels, and CrSUMO-like90 and CrSUMO148 mRNAs consistently showed slightly more abundance than CrSUMO89A mRNA (Figure 3). Samples from walled CC-125 cells and wall-less CC-503 cells revealed similar expression patterns (data not shown).
Figure 3.—
Relative abundance of C. reinhardtii CrSUMO transcripts. Expression of the candidate SUMO genes was analyzed by quantitative real-time reverse transcription–PCR for expression in CC-503 cells at 25° and 42°. Each 25-μl reaction mixture contained cDNA equivalent to 50 ng of total input mRNA. Relative abundance was calculated with efficiency-corrected ΔCt values. Each data point is the average of an experimental triplicate and represents an individual trial.
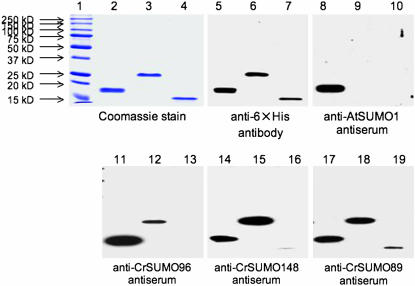
The cDNA coding regions for CrSUMO96 and CrSUMO148 were amplified by RT–PCR and that for CrSUMO-like89A by PCR. All three were cloned into the pET28 expression vector for protein expression in E. coli. Attachment of six histidines (6× His tag) to each protein allowed purification of C. reinhardtii SUMO proteins (Figure 4, lanes 2–4). These polypeptides were recognized by the anti-6× His antibody (Figure 4, lanes 5–7). Polyclonal antibodies were generated in rabbits against bacterially expressed and purified antigens. In immunoblot analysis, the anti-CrSUMO96 antibody recognized recombinant (r)CrSUMO96 and showed a slight cross-reaction with rCrSUMO148 (Figure 4, lanes 11–13). Similarly, the anti-CrSUMO148 antibody recognized rCrSUMO148 and showed cross-reaction with rCrSUMO96 (Figure 4, lanes 14–16). Because of the high degree of similarity between CrSUMO96 and CrSUMO97, we assume, but have not demonstrated, that the polyclonal antibodies raised against rCrSUMO96 likely will also recognize CrSUMO97—if, indeed, CrSUMO97 is produced (Figure 3). Interestingly, the anti-CrSUMO-like89A antibody detected rCrSUMO-like89A in an immunoblot but showed much weaker affinity for rCrSUMO-like89A than for rCrSUMO96 and rCrSUMO148 (Figure 4, lanes 17–19). Arabidopsis anti-SUMO1 antibody detected only rCrSUMO96 on immunoblots (Figure 4, lanes 8–10).
Figure 4.—
Immunodetection of C. reinhardtii CrSUMO96, CrSUMO148, and CrSUMO-like89A with anti-CrSUMOs and anti-AtSUMO-1 antibodies. Overexpression and purification of 6× His-tagged CrSUMOs from E. coli, antisera production, and immunoblot detection are shown. CrSUMO96 overexpressed in E. coli and affinity purified on a 6× His column (lanes 2, 5, 8, 11, 14, and 17), overexpressed and purified CrSUMO148 (lanes 3, 6, 9, 12, 15, and 18), and overexpressed and purified CrSUMO-like89A (lanes 4, 7, 10, 13, 16 and 19) were separated on 12% SDS–PAGE and stained with Coomassie blue (lanes 1–4) or detected after immunobloting using anti-6× His antibody (lanes 5–7), anti-AtSUMO1 antiserum (lanes 8–10), anti-CrSUMO96 antiserum (lanes 11–13), anti-CrSUMO148 antiserum (lanes 14–16), and anti-CrSUMO-like89A antiserum (lanes 17–19).
Purification and mass spectrometry analysis of CrSUMO96 in C. reinhardtii:
The decision to focus on CrSUMO96 for further study was made because of prior knowledge that the double-glycine motif was critical for the isopeptide bond formation when the activated SUMO conjugates to a target protein (Johnson 2004; Kerscher et al. 2006). The CrSUMO148 predicted coding region contains five double-glycine motifs, while the CrSUMO-like proteins contain none. Moreover, of the SUMO cDNAs we have cloned and expressed in E. coli, rCrSUMO96 was also the only C. reinhardtii SUMO homolog to be recognized by Arabidopsis anti-SUMO1 antibody.
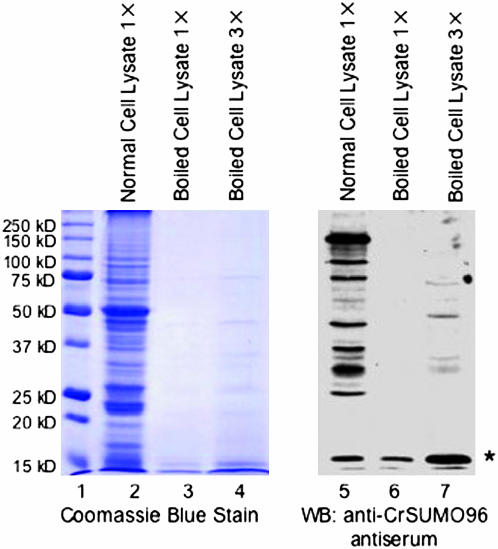
Because A. thaliana free SUMO1 and SUMO3 were reported to be resistant to treatment at 90° for 30 min (Kurepa et al. 2003), we tested to determine if this feature of SUMO could be used for enrichment of endogenous free SUMO from C. reinhardtii. After a C. reinhardtii cell extract was heated to 90° for 30 min and clarified by centrifugation, the endogenous free SUMO corresponding to the size of CrSUMO96 remained soluble and was detected by immunoblot analysis with the anti-CrSUMO96 antibody (Figure 5). These experiments also showed that approximately three times the amount of free SUMO was detected when the amount of heated cell extract was tripled (Figure 5, lanes 4 and 7).
Figure 5.—
Purification and identification of endogenous free CrSUMO96 from C. reinhardtii after boiling of cell extracts. Proteins in unboiled C. reinhardtii cell lysates (lanes 2 and 5) and boiled lysates at 1× (lanes 3 and 6) and 3× (lanes 4 and 7) loading concentrations were detected by Coomassie blue staining (lanes 1–4) or with anti-CrSUMO96 antiserum on immunoblots of the SDS–PAGE (lanes 5–7). The asterisk (*) indicates the endogenous free CrSUMO96. Molecular size markers are shown at the left (lane 1).
To confirm the identity of CrSUMO96, immunoprecipitation experiments were performed using anti-CrSUMO96 antibody. The immunoprecipitated fraction was shown to be enriched with CrSUMO96 by immunoblot (with anti-CrSUMO96 antibody; supplemental Figure 2, left) and by silver stain (supplemental Figure 2, right). A similar pattern was seen in that the free CrSUMO96 increased respective to the amount of heated cell extract added to the reaction. The endogenous free SUMO purified by immunoprecipitation was recovered from the silver-stained gel and analyzed by mass spectrometry. The presence of two peptide fragments containing amino acid sequences identical to predicted fragments from CrSUMO96 protease digestion confirmed the identity of the protein (supplemental Figure 3).
Detection of SUMO-conjugated proteins by immunoblot analysis:
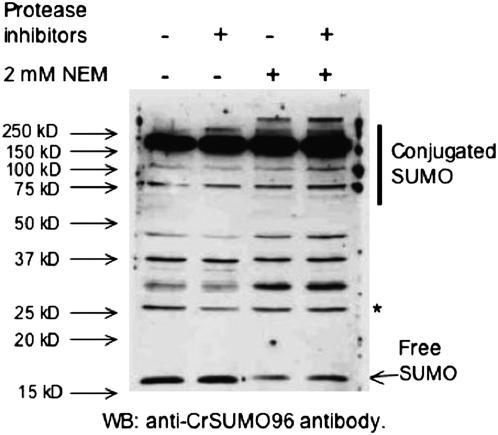
To detect SUMO-conjugated proteins in C. reinhardtii, immunoblot analysis was performed using anti-CrSUMO96 antibodies (Figure 4). In addition to free CrSUMO96 migrating at ∼15 kDa, a large number of SUMO-conjugated proteins were detected, suggesting that all the enzymes needed in the SUMO-conjugating system are present and functional in C. reinhardtii. NEM, an inhibitor of SUMO-specific isopeptidases (Li and Hochstrasser 1999), was added to protect SUMO-conjugated proteins from desumoylation. When NEM was added to the extraction buffer, more SUMO-conjugated proteins and less free SUMO were detected by the antibody (Figure 6, lanes 3 and 4). However, a few proteins still exist, such as the 26-kDa protein (* in Figure 6), that did not change intensity after NEM was added, indicating that antibodies to these proteins may also be present in our anti-CrSUMO96 antisera.
Figure 6.—
Detection of endogenous SUMO-conjugated proteins from C. reinhardtii. Cell extracts were prepared in the presence or the absence of protease inhibitor cocktail or the isopeptidase inhibitor, NEM. Anti-CrSUMO96 antibody was utilized to detect the endogenous SUMO-conjugated proteins. The 26-kDa band indicated with an asterisk (*) is one of the nonspecific reacting proteins that can serve as a loading control.
Subcellular localization of CrSUMO96 and its conjugated proteins:
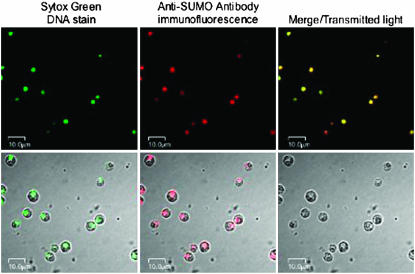
The subcellular localization of CrSUMO96 and its conjugated proteins was detected by immunofluorescence (Figure 7). Anti-CrSUMO96 antibody was recognized by a secondary cyanine-5-conjugated goat anti-rabbit antibody that emits a far-red fluorescence signal under illumination with light of 630 nm. Sytox green was employed to stain DNA and allow detection of the nucleus during confocal microscopy. As shown in Figure 7, CrSUMO96 and its protein conjugates are localized primarily, if not exclusively, to the nucleus of the cell. This observation is in agreement with previous observations that most of the SUMO conjugates in mammalian cells are found in the nucleus (Johnson 2004; Kerscher et al. 2006).
Figure 7.—
In situ localization of CrSUMO96 and its conjugated proteins by immunofluorescence. Wild-type C. reinhardtii cells grown in TAP media were stained with Sytox Green (top left) or detected with anti-CrSUMO96 antibody, which is recognized by the red fluorescent cyanine-5-conjugated goat anti-rabbit antibody (top middle). Images under transmitted light and confocal images are also shown in the top right and the bottom.
SUMO-conjugating enzyme (E2) from C. reinhardtii:
In contrast to most ubiquitin-conjugating systems, there is only one SUMO E2 enzyme in yeast, mammals, and A. thaliana. The number of CrUBCE2 genes present in the C. reinhardtii genome is uncertain. As many as 12 homologs can be discerned. Using HMM analysis and conservative selection parameters, four predicted CrUbcE2 molecules designated CrUbcE2_1, CrUbcE2_2, CrUbcE2_3, and CrUbcE2_4 were selected as most likely to function as SUMO-conjugating enzymes. All of the gene sequences predict an open reading frame that contains a conserved ubiquitin-conjugating enzyme catalytic (UBCC) domain (IPR000608). However, it is difficult to distinguish SUMO E2 from ubiquitin E2's by amino acid sequence alone. This is exemplified by the fact that the degree of sequence similarity between SUMO E2 and the various ubiquitin E2's in yeast is comparable with that between the various yeast ubiquitin E2's (Johnson et al. 1997).
It has been reported that SUMO E2 conjugases have a much more positive net charge at neutral pH than ubiquitin E2's (Johnson and Blobel 1997). The calculated isoelectric points (pI) for each of the four selected CrUbcE2 candidates are listed in supplemental Figure 5A. The potential C. reinhardtii E2 that ranked highest in similarity to authentic Ubc9-like E2 conjugases, CrUbcE2_1 (XP_001694849), was amplified by RT–PCR and used for additional studies.
The predicted amino acid sequence of CrUbcE2_1, as derived from the sequenced RT–PCR product, is shown in supplemental Figure 4. Alignment of CrUbcE2_1 and the amino acid sequence of other CrUbcE2's with Ubc E2's from other species (Figure 8) shows a marked similarity between the putative CrUbcE2's and the E2-conjugating enzymes from other eukaryotes. Calculation of the isoelectric point of CrUbc2EB_1 confirmed an alkaline pI of 8.81 for this molecule (supplemental Figure 5A). A similarity tree representing distances among E2 homologs is depicted in supplemental Figure 4B.
Figure 8.—
SUMO-conjugating enzyme E2 from C. reinhardtii: amino acid sequence alignment of human, yeast, A. thaliana, and C. reinhardtii SUMO-conjugating enzyme (E2).
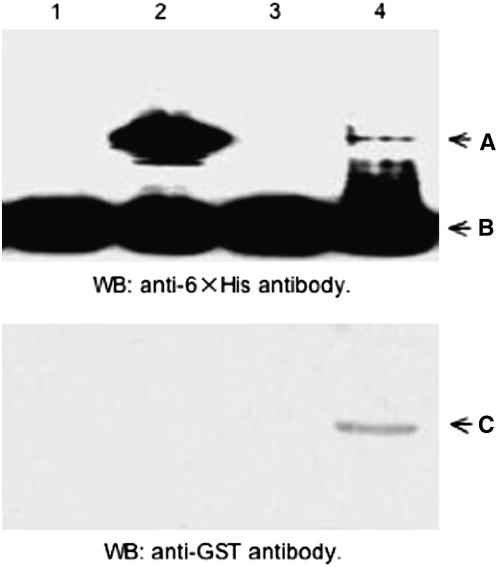
An in vivo SUMOylation experiment was conducted using an in vivo E. coli system established by the laboratory of Hisato Saitoh (Uchimura et al. 2004a,b) to determine if CrUbcE2_1, the C. reinhardtii SUMO E2 conjugase with the highest similarity to authentic human and yeast E2 conjugases as determined by HMM analysis (supplemental Figure 4A), was a potential SUMO conjugase. This system utilizes E. coli BL21 cells transformed with pT-E1E2S1, a plasmid that contains genetically engineered versions of genes encoding mouse E1-activating enzyme, Xenopus laevis E2-conjugating enzyme, and human SUMO1. Transformation with an additional plasmid, pRanGAP1-C2 leads to production of a histidine-tagged RanGAP1 C-terminal region (RanGAP1-C2), a well-known SUMO target protein. Incubation of BL21 cells carrying the pRanGAP1-C2 plasmid alone, as expected, did not produce SUMOylated RanGAP1-C2 because the SUMO-conjugating system is absent in E. coli (Figure 9, top, lane 1). However, when BL21 cells carry both plasmids (pT-E1E2S1 and pRanGAP1-C2), RanGAP-C2 is produced and a SUMOylated version of this target protein is synthesized in vivo (Figure 9, lane 2, band A). As expected, removal of the E2 conjugase gene from the pT-E1E2S1 plasmid to produce plasmid pT-E1E2S1ΔE2 eliminated the ability of BL21 cells to support in vivo SUMOylation of RanGAP1-C2 (Figure 9, lane 3). To determine if the putative C. reinhardtii E2 conjugase (from CrUbcE2_1 cDNA) could substitute for the X. laevis E2 conjugase, the cDNA coding region was inserted downstream of a bacterial promoter to produce the plasmid, pGEX4T1-CrUbcE2B-1. When this plasmid was cotransformed with pT-E1E2S1ΔE2 and pRanGAP1-C2, the resultant cells were capable of producing SUMOylated RanGAP1-C2 (Figure 9, lane 4, band A). Expression of the putative E2 CrUbcE2_1-conjugating enzyme in transformed BL21 cells was confirmed by immunoblot detection of the GST-tagged C. reinhardtii E2 conjugase (Figure 9, bottom, lane 4, band C). Compared with the vertebrate Ubc9, the heterologous C. reinhardtii SUMO-conjugating enzyme worked with a lower efficiency in this in vivo E. coli sumoylation system (Figure 9, lane 2 vs. lane 4, band A). This is not surprising, considering the multiple interactions required between the C. reinhardtii SUMO E2 and the three heterologous mammalian proteins (E1, SUMO, and the target protein RanGAP1-C2) for successful SUMOylation to take place. Because of its apparent successful function in the in vivo SUMO E2 conjugase assay, CrUbcE2_1 becomes a prime candidate as an authentic C. reinhardtii SUMO-conjugating enzyme.
Figure 9.—
Functional analysis of CrUbcE2_1. Total E. coli proteins isolated from the BL21 strain were transformed with pRanGAP1-C2 (lane 1), pT-E1E2S1 and pRanGAP1-C2 (lane 2), pT-E1E2S1ΔE2 and pRanGAP1-C2 (lane 3), and pT-E1E2S1ΔE2, pRanGAP1-C2, and pGEX4T1-CrUbcE2-1 (lane 4), respectively, and incubated under conditions allowing Ubc9 conjugation. Top: SUMOylated RanGAP1-C2 (A) and nonSUMOylated RanGAP1-C2 (B) detected on immunoblots with anti-6× His antibody. Bottom: GST-CrUbcE2_1 (C) detected with anti-GST antibody. WB, Western blot.
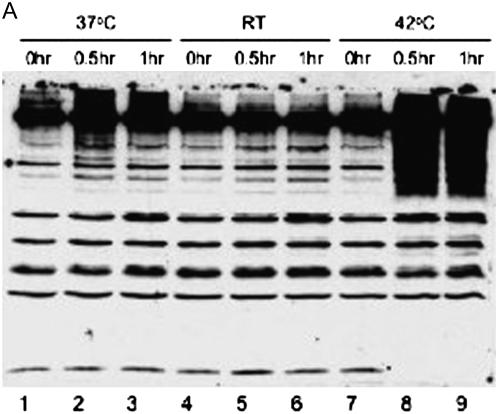
Stress-induced accumulation of SUMO-conjugated proteins:
Because it has been reported that SUMO conjugation is part of the stress response in animals and plants (Hong et al. 2001; Kurepa et al. 2003), immunoblot experiments were performed to detect SUMO-conjugated proteins isolated from C. reinhardtii cells grown in various stress and nonstress conditions (Figure 10). When C. reinhardtii cells were shifted from optimal growth temperature of 25° to 37°, an increase in SUMO-conjugated proteins was detected with anti-CrSUMO96 antibodies (Figure 10A, lanes 2 and 3). Moreover, when the cells were shifted from 25° to 42°, a marked increase in SUMO-conjugated proteins was detected in the molecular size range of ≥60 kDa. Interestingly, at this high temperature no free CrSUMO96 was detected, indicating that most free CrSUMO96 was incorporated into SUMO target proteins under this stress condition (Figure 10A, lanes 8 and 9). A more detailed examination of proteins from control and heat-shocked cells by 2-D PAGE analysis (supplemental Figure 6) demonstrates again that heat shock results in a marked decrease in the amount of nonconjugated CrSUMO's (thick-walled red boxes). Concomitantly, there is a pronounced increase in the degree of SUMOylation of several proteins (green boxes), including the de novo appearance of SUMOylated forms of a number of additional proteins. Reaction of identical protein blots with preimmune serum demonstrates the specificity of the CrSUMO96 antiserum for free CrSUMO96 and CrSUMO96 conjugated to larger proteins. Only weak detection of a few non-SUMO-related proteins was observed using the preimmune serum (supplemental Figure 6).
Figure 10.—
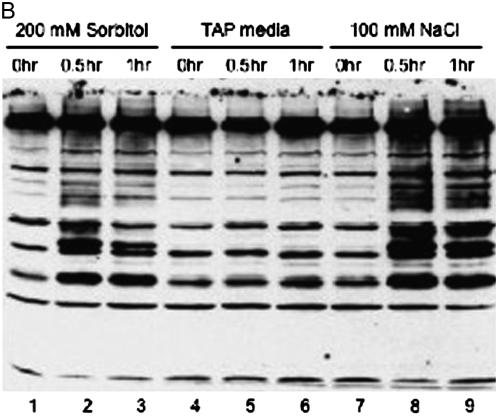
SUMO-conjugated proteins in different environmental conditions, including heat shock (A) and osmotic stress (B).
Osmotic stress also induced the accumulation of SUMO-conjugated proteins (Figure 10B). When 200 mm sorbitol or 100 mm NaCl was added to the cells, more SUMO-conjugated proteins were detected with the anti-CrSUMO96 antibody as compared with the cells grown in normal media (Figure 10B, lane 2, 3, 5, 6, 8, and 9). It appears that some SUMO-conjugated proteins induced by the osmotic stress are distinct from those induced by heat shock, because heat shock induced the accumulation of SUMO-conjugated proteins with a molecular size of >60 kDa. In contrast, osmotic stress induced SUMOylation of larger proteins, as well as proteins <60 kDa [Figure 10, A (lanes 2, 3, 8, and 9) and B (lanes 2, 3, 8, and 9)].
DISCUSSION
SUMO and other ubiquitin-like proteins in C.reinhardtii:
Our investigations have revealed six putative CrSUMO and CrSUMO-like proteins and several potential SUMO-conjugating enzymes (CrUbc9's) in the unicellular, photosynthetic alga, C. reinhardtii. The six C. reinhardtii SUMO homologs have been designated as CrSUMO96, CrSUMO97, and CrSUMO148 and CrSUMO-like89A, CrSUMO-like89B, and CrSUMO-like90. They share higher sequence identity with human SUMO-1, yeast SMT3, and A. thaliana SUMO1 than other ubiquitin-like proteins in the JGI C. reinhardtii database (Figure 1 and supplemental Figure 1). There are 15 proteins listed as ubiquitin-like proteins in the Cluster of EuKaryotic Orthologous Groups (KOG) Browser in the JGI C. reinhardtii database. Most contain a ubiquitin domain; however, only CrSUMO96, CrSUMO97, and CrSUMO148 and CrSUMO-like89A, CrSUMO-like89B, and CrSUMO-like90 have significant sequence similarity with known SUMOs in other species (Figure 1).
Of the six C. reinhardtii SUMO homologs, the closely related CrSUMO96 and CrSUMO97 contain a ψKXE consensus motif at their N termini. CrSUMO96 has a very similar predicted 3-D structure when compared with human SUMO1 (Figure 2). Moreover, CrSUMO96 is recognized by A. thaliana anti-SUMO1 antibody (Figure 4). Unlike any known SUMO proteins, CrSUMO148 contains five double-glycine motifs at the C terminus. The observations that all three CrSUMO-like proteins contain a GN dipeptide at their C termini instead of the canonical double-glycine motif (Figure 1) and the likely possibility that all three genes encoding the CrSUMO-like proteins contain potential large open reading frames upstream of the SUMO-like domain suggest the CrSUMO-like proteins may be related to a class of fusion proteins containing highly conserved ubiquitin sequences at either the N-terminal or the C-terminal ends (Kerscher et al. 2006; see Kramer et al. 1995 for an extensive list of fusion proteins with ubiquitin-like terminal domains). These fusion proteins, thus, can bind to ubiquitin-binding domains (UBDs) and carry along their fusion partner to perform a particular function. Two examples of proteins with ubiquitin sequences at their C termini, both involved in pre-mRNA splicing, are SF3a120 (Kramer et al. 1995) and CePRP21 (Spikes et al. 1994). Whether the three CrSUMO-like proteins perform functions analogous to ubiquitin-containing fusion proteins remains to be determined.
Other components of the SUMO-conjugating system in C. reinhardtii:
In addition to free CrSUMO96, proteins conjugated to CrSUMO96 also can be detected with anti-CrSUMO96 antibodies in immunoblot analyses of cell extracts (Figures 5, 6, and 10 and supplemental Figure 6). These observations strongly suggest that all enzymes needed for SUMO conjugation are present and functional in C. reinhardtii. We have demonstrated that XP_001694849 (CrUbcE2_1) is a prime candidate as an authentic SUMO-conjugating enzyme (SCE, E2) in C. reinhardtii (Figure 9). Predicted amino acid sequence of CrUbcE2_1 shares 43% identity and 56% similarity with yeast Ubc9p and 49% identity and 59% similarity with A. thaliana Ubc9 (Figure 6). Like SUMO E2's of other species, CrUbcE2_1 has a basic isoelectric point (i.e., pI = 8.81) that might be necessary for it to interact with free SUMO and other components in the SUMO-conjugating system (Johnson 2004). The most compelling argument suggesting that XP_001694849 (CrUbcE2_1) encodes a bona fide SUMO E2 conjugase is our demonstration of SUMO E2 conjugase activity in an E. coli in vivo SUMOylation system (Figure 9).
SAE1:
A putative C. reinhardtii SUMO-activating enzyme 1 (SAE1, XP_001690572) shares remarkable patterns with the yeast Aos1 (SAE1) (CAA97885) molecule. These patterns include an SGG and an RGG cleavage site at the C termini and a series of conserved charged residues as well as serines, threonines, and tyrosines both at the cores and at the C termini. It is intriguing whether the lack of similarity at the N terminus is a result of functional divergence. Overall, 40% of the residues are identical and 54% are similar between the two proteins.
SAE2:
Two putative C. reinhardtii SUMO-activating enzyme 2 (SAE2) proteins (XP_001691317 and XP_001690945) align with a similarity of 67 and 80%, respectively, to the A. thaliana SAE2 (NP_973506) at three extensive but somewhat disjunct domains. Clearly, these domains are not related to the ubiquitin or the SUMO domains. The two proteins have somewhat different variants (KFPLCTLAETP, LPPLCPAPASP, and KDPSCPCSAGVP) of the motif KXPV/GCTXXXP containing the cysteine in SAE2 that forms the thioester intermediate with SUMO in SAE2 molecules of other eukaryotes. We also note that XP_001690945, in spite of its original annotation, is more closely related to SAE2 than to SAE1 enzymes.
SUMO E3 ligase:
Currently it is difficult to distinguish SUMO E3 ligases from those responsible for adding ubiquitin or other ubiquitin-like peptides, such as NEDD8, to their target proteins. There are 26 “ubiquitin-protein ligases” and 34 “E3 ubiquitin ligases” predicted in the KOG Browser of the C. reinhardtii database.
Ubiquitin, UFM1, and RUB:
HMM and BLAST searches identified multiple potential ubiquitin genes and pseudogenes in C. reinhardtii. Nonetheless, a minimum of five ubiquitin proteins could be identified with a high level of confidence. They are ubiquitins XP_001694320 (242 residues), XP001702404 (244 residues), and XP_001147989 (77 residues, enigmatically 97% identical to a Pan troglodytes coiled-coil 99 protein), predicted protein 140045 (153 residues), and biubiquitin XP_001694608 (153 residues) (supplemental Figure 7). Each of the last two proteins is composed of two very similar internal duplicate segments. With the exception of XP_001694320, they all contain a predicted double-glycine motif near the C terminus of the peptide, and all have a PFAM ubiquitin domain in the predicted sequence. A protein sequence (XP_001696636) with 77% sequence similarity to human Ufm1 (Table 1) suggests that C. reinhardtii contains at least one representative of this rarer ubiquitin-related family of proteins. Each of C. reinhardtii proteins XP_001694608 (biubiquitin) and 1794 (both containing 153 residues) is >95% identical to other ubiquitin-related proteins in Populus trichocarpa, Tetrahymena thermophyla SB210, and other organisms. These two proteins are the closest homologs of the A. thaliana related to ubiquitin-1 protein (RUB1, NP_564379) and the XP_001694608 sequence is 94% identical to RUB1. Protein 1794 is 98% identical to A. thaliana UBQ10 (NP_974516) and 97% identical to a H. sapiens protein annotated as “similar to Ubiquitin-63E” (XP_001132949) and a P. troglodytes hypothetical protein (XP_001136911).
Additional components of the C. reinhardtii SUMOylation system are likely to emerge as studies of SUMOylation in other eukaryotes reveal new components. However, SUMOylation systems are often sufficiently unique that components in one organism may be missing in others. For example, the newly discovered RSUME protein found to enhance conjugation of SUMO-1, -2 and -3 to target proteins by binding to Ubc9 in mammalian cells (Carbia-Nagashima et al. 2007) has not been identified in our searches of the C. reinhardtii genome.
Stress—heat-shock, osmotic, nutrition, and photoautotrophic growth:
Some SUMO target proteins are conjugated under normal growth conditions, while some are conjugated preferentially upon stresses (Johnson 2004; Miura et al. 2007). The conjugation of SUMO to its target proteins may serve as a signal to guide regulation of expression of specific sets of genes under stress conditions (Kerscher 2007; Miura et al. 2007).
The accumulation of SUMO-conjugated proteins was detected when C. reinhardtii cells were shifted to 37° or 42° heat-shock conditions and to sorbitol- or NaCl-induced osmotic stresses (Figure 10, A and B, and supplemental Figure 6). The appearance of distinctly different sizes of SUMO-conjugated proteins produced under different stress conditions indicates that SUMOylation is involved in multiple cellular processes initiated by different environmental cues. Under each stress condition, 30 min of treatment induced more SUMO-conjugated proteins than 1 hr. This indicates that SUMOylation could be a rapid and transient reaction and that deSUMOylation may take place as soon as the signaling process is complete. Such a dynamic modification requires highly regulated SUMOylation and deSUMOylation enzymes.
Conclusions and future research:
We have demonstrated the presence of SUMO and SUMO-conjugating systems in C. reinhardtii, a unicellular photosynthetic alga and model plant cell system. Free and conjugated SUMOs were detected by immunoblot analysis and shown to be localized in the nucleus by immunofluorescence. Endogenous free CrSUMO96 was purified by immunoprecipitation and identified by LC/MS/MS analysis. SCE (E2) of C. reinhardtii was cloned and shown to be functional in an in vivo E. coli SUMOylation system. Accumulation of SUMO-conjugated proteins was detected when the cells were subjected to environmental stresses, such as heat-shock and osmotic stresses.
Investigations in regard to how SUMO affects biological processes are only in their early stages (Johnson 2004; Kerscher et al. 2006; Geiss-Friedlander and Melchior 2007; Miura et al. 2007; Palancade and Doye 2008). Identification of SUMO target proteins and an understanding of their biological functions in the SUMOylated and nonSUMOylated states lie ahead. The way forward will include challenges because (i) many SUMOylated proteins are present at a level below the normal detection limit (Li et al. 2004), (ii) for most SUMO target proteins, only a small fraction of the substrate is SUMOylated at any given time, and (iii) there are strong SUMO protease (isopeptidase) activities in native cell lysates.
C. reinhardtii is a valuable model system to study various cellular functions and biochemical pathways because of its small genome size, haploid nature, susceptibility to gene manipulations, and, most importantly, the ability to grow the organism in large quantities (Weeks 1992; Rochaix et al. 1998). The present bioinformatics studies of the completed C. reinhardtii genome sequence have revealed much about SUMO-associated protein families in this organism and suggest that C. reinhardtii may be an especially facile organism for defining the role of SUMOylation in controlling gene expression and cellular functions such as response to environmental changes, photosynthesis, and flagellar-based motility.
Acknowledgments
The authors thank Richard Vierstra (University of Wisconsin) for the generous gift of antibodies to A. thaliana SUMO-1, Tom Elthon (University of Nebraska Protein Core Facility) for assistance with 2-D PAGE analyses, Susan Dutcher for providing procedures for microscopic detection of proteins using immunofluorescence, and Ashraf Raza (University of Nebraska Redox Biology Center) for assistance in analyzing tryptic fragments of CrSUMO96 by mass spectrometry. Systems administration by J. J. M. Riethoven and F. Ma is gratefully acknowledged. This research was supported with funds from the U.S. National Science Foundation (IBN-0120802 to D.P.W.), the University of Nebraska Agricultural Research Division, and the National Science Foundation of China (no. 30500024 to Y.W.).
References
- Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang et al., 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballesteros, M. L., C. Bolie, L. M. Lois, J. M. Moore, J. P. Vielle-Calzada et al., 2001. LAF1, a MYB transcription activator for phytochrome A signaling. Genes Dev. 15 2613–2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer, P., A. Arndt, S. Metzger, R. Mahajan, F. Melchior et al., 1998. Structure determination of the ubiquitin-related modifier SUMO-1. J. Mol. Biol. 280 275–286. [DOI] [PubMed] [Google Scholar]
- Bernier-Villamor, V., D. A. Sampson, M. J. Matunis and C. D. Lima, 2002. Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell 108 345–356. [DOI] [PubMed] [Google Scholar]
- Birney, E., M. Clamp and R. Durbin, 2004. Genewise and genomewise. Genome Res. 14 988–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broday, L., I. Koloteuv, C. Didier, A. Bhoumik, B. P. Gupta et al., 2004. The small ubiquitin-like modifier (SUMO) is required for gonadal and uterine-vulval morphogenesis in Caenorhabditis elegans. Genes Dev. 18 2380–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabral, F. J., O. S. Pereira, Jr., C. S. Silva, R. Guerra-Sa and R. Vanderlei, 2008. Schistosoma mansoni encodes SMT3B and SMTC molecules responsible for post-translational modification of cellular proteins. Parasitol. Int. 57 172–178. [DOI] [PubMed] [Google Scholar]
- Carbia-Nagashima, A., J. Gerez, C. Perez-Castro, M. Paez-Pereda, S. Silberstein et al., 2007. RSUME, a small RWD-containing protein, enhances SUMO conjugation and stabilizes HIF-1α during hypoxia. Cell 131 309–323. [DOI] [PubMed] [Google Scholar]
- Clamp, M., J. Cuff, S. M. Searle and G. J. Barton, 2004. The Jalview Java alignment editor. Bioinformatics 20 426–427. [DOI] [PubMed] [Google Scholar]
- Colby, T., A. Matthai, A. Boeckelmann and H.-P. Stuible, 2006. SUMO-conjugating and SUMO-deconjugating enzymes from Arabidopsis. Plant Physiol. 142 318–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison, C., A. D. Rudner, S. A. Gerber, C. E. Bakalarski, D. Moazed et al., 2004. A proteomic strategy for gaining insights into protein sumolyation in yeast. Mol. Cell. Proteomics 4 246–254. [DOI] [PubMed] [Google Scholar]
- Desterro, J. M. P., J. Thomson and R. T. Hay, 1997. Ubc9 conjugates SUMO but not ubiquitin. FEBS Lett. 417 297–300. [DOI] [PubMed] [Google Scholar]
- Eddy, S. R., 1998. Profile hidden Markov models. Bioinformatics 14 755–763. [DOI] [PubMed] [Google Scholar]
- Ermolova, N. V., M. A. Cushman, T. Taybi, S. A. Condon, J. C. Cushman et al., 2003. Expression, purification, and initial characterization of a recombinant form of plant PEP-carboxylase kinase from CAM-induced Mesembryanthemum crystallinum with enhanced solubility in Escherichia coli. Protein Expr. Purif. 29 123–131. [DOI] [PubMed] [Google Scholar]
- Finn, R. D., J. Tate, J. Mistry, P. C. Coggill, S. J. Sammut et al., 2008. The Pfam protein families database. Nucleic Acids Res. 36 D281–D288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascuel, O., 1997. BIONJ: an improved version of the NJ algorithm based on a simple model of sequence data. Mol. Biol. Evol. 14 685–695. [DOI] [PubMed] [Google Scholar]
- Geiss-Friedlander, R., and F. Melchior, 2007. Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 8 947–956. [DOI] [PubMed] [Google Scholar]
- Guo, D., M. Li, Y. Zhang, P. Yang, S. Eckenrode et al., 2004. A functional variant of SUMO4, a new IκBα modifier, is associated with type 1 diabetes. Nat. Genet. 36 837–841. [DOI] [PubMed] [Google Scholar]
- Haas, B. J., A. L. Delcher, S. M. Mount, J. R. Wortman, R. K. Smith, Jr. et al., 2003. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 31 5654–5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajduch, M., A. Ganapathy, J. W. Stein and J. J. Thelen, 2005. A systematic proteomic study of seed filling in soybean. Establishment of high-resolution two-dimensional reference maps, expression profiles, and an interactive proteome database. Plant Physiol. 137 1397–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanania, U., N. Furman-Matarasso, M. Ron and A. Avni, 1999. Isolation of a novel SUMO protein from tomato that suppresses EIX-induced cell death. Plant J. 19 533–541. [DOI] [PubMed] [Google Scholar]
- Harris, E. H., 1989. The Chlamydomonas Sourcebook: A Comprehensive Guide to Biology and Laboratory Use. Academic Press, San Diego. [DOI] [PubMed]
- Hay, R. T., 2001. Protein modification by SUMO. Trends Biochem. Sci. 26 322–323. [DOI] [PubMed] [Google Scholar]
- Hochstrasser, M., 2001. SP-RING for SUMO: new functions bloom for a ubiquitin-like protein. Cell 107 5–8. [DOI] [PubMed] [Google Scholar]
- Holmstrom, S., M. E. Van Antwerp and J. A. Iniguez-Lluhi, 2003. Direct and distinguishable inhibitory roles for SUMO isoforms in the control of transcriptional synergy. Proc. Natl. Acad. Sci. USA 26 15758–15763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, Y., R. Rogers, M. J. Matunis, C. N. Mayhew, M. Goodson et al., 2001. Regulation of heat shock transcription factor 1 by stress-induced SUMO-1 modification. J. Biol. Chem. 276 40263–40267. [DOI] [PubMed] [Google Scholar]
- Jakobs, A., J. Koehnke, F. Himstead, M. Funk, B. Korn et al., 2007. Ubc9 fusion-directed SUMOylation (UFDS): a method to analyze function of protein SUMOylation. Nat. Methods 4 245–250. [DOI] [PubMed] [Google Scholar]
- Johnson, E. S., 2004. Protein modification by SUMO. Annu. Rev. Biochem. 73 355–382. [DOI] [PubMed] [Google Scholar]
- Johnson, E. S., and G. Blobel, 1997. Ubc9p is the conjugating enzyme for the ubiquitin-like protein Smt3p. J. Biol. Chem. 272 26799–26802. [DOI] [PubMed] [Google Scholar]
- Johnson, E. S., I. Schwienhorst, R. J. Dohmen and G. Blobel, 1997. The ubiquitin-like protein Smt3p is activated for conjugation to other proteins by an Aos1/Uba2p heterodimer. EMBO J. 16 5509–5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamitani, T., K. Kito, H. P. Nguyen and E. T. H. Yeh, 1997. Characterization of NEDD8, a developmentally down-regulated ubiquitin-like protein. J. Biol. Chem. 272 28557–28562. [DOI] [PubMed] [Google Scholar]
- Kerscher, O., 2007. SUMO junction—What's your function? New insights through SUMO-interacting motifs. EMBO J. 8 550–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerscher, O., R. Felberbaum and M. Hochstrasser, 2006. Modifications of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Biol. 22 159–180. [DOI] [PubMed] [Google Scholar]
- Kramer, A., F. Mulhauser, C. Wersig, K. Groning and G. Bible, 1995. Mammalian splicing factor SF3a120 represents a new member of the SURP family of proteins and is homologous to the essential splicing factor PR21p of Saccharomyces cerevisiae. RNA 1 260–272. [PMC free article] [PubMed] [Google Scholar]
- Kurepa, J., J. M. Walker, J. Smalle, M. M. Gosink, S. J. Davis et al., 2003. The small ubiquitin-like modifier (SUMO) protein modification in Arabidopsis. J. Biol. Chem. 278 6862–6872. [DOI] [PubMed] [Google Scholar]
- Kurtzman, A. L., and N. Schechter, 2001. Ubc9 interacts with a nuclear localization signal and mediates nuclear localization of the paired-like homeobox protein Vsx-1 independent of SUMO-1 modification. Proc. Natl. Acad. Sci. USA 98 5602–5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli, U. K., 1970. Cleavage of structural proteins during the assembly of the head bacteriophage T4. Nature 227 680–685. [DOI] [PubMed] [Google Scholar]
- Lee, J., J. Nam, H. C. Park, G. Na, K. Miura et al., 2007. Salicylic acid-mediated innate immunity in Arabidopsis is regulated by SIZ SUMO E3 ligase. Plant J. 49 79–90. [DOI] [PubMed] [Google Scholar]
- Lei, Z., and Y. Dai, 2005. An SVM-based system for predicting protein subnuclear localizations. BMC Bioinformatics 6 291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S.-L., and M. Hochstrasser, 1999. A new protease required for cell-cycle progression in yeast. Nature 398 246–251. [DOI] [PubMed] [Google Scholar]
- Li, S.-L., and M. Hochstrasser, 2003. The Ulp1 SUMO isopeptidase: distinct domains required for visibility, nuclear envelope localization and substrate specificity. J. Cell Biol. 160 1069–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, T., E. Evdokimov, R. F. Shen, C. C. Chao, E. Tekle et al., 2004. Sumoylation of heterogeneous nuclear ribonucleoproteins, zinc finger proteins, and nuclear pore complex proteins: a proteomic analysis. Proc. Natl. Acad. Sci. USA 101 8551–8556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak, K. J., and T. D. Schmittgen, 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 25 402–408. [DOI] [PubMed] [Google Scholar]
- Lois, L. M., C. D. Lima and N. H. Chua, 2003. Small ubiquitin-like modifier modulates abscisic acid signaling in Arabidopsis. Plant Cell 15 1347–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matunis, M. J., E. Coutavas and G. Blobel, 1996. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J. Cell Biol. 135 1457–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melchior, F., 2000. SUMO-nonclassical ubiquitin. Annu. Rev. Cell Biol. 16 591–626. [DOI] [PubMed] [Google Scholar]
- Melchior, F., M. Schergaut and A. Pichler, 2003. SUMO: ligases, isopeptidases and nuclear pores. Trends Biochem. Sci. 28 612–618. [DOI] [PubMed] [Google Scholar]
- Miura, K., J. B. Jin and P. M. Hasegawa, 2007. Sumolyation, a post-translational regulatory process in plants. Curr. Opin. Plant Biol. 10 495–502. [DOI] [PubMed] [Google Scholar]
- Murtas, G., P. H. Reeves, Y.-F. Fu, I. Bancroft, C. Dean et al., 2003. A nuclear protease required for flowering-time regulation in Arabidopsis reduces the abundance of small-ubiquitin-related-modifier conjugates. Plant Cell 15 2308–2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida, T., F. Kaneko, M. Kitagawa and H. Yasuda, 2001. Characterization of a novel mammalian SUMO-1/Smt3-specific isopeptidase, a homologue of rat axam, which is an axin-binding protein promoting β-catenin degradation. J. Biol. Chem. 276 39060–39066. [DOI] [PubMed] [Google Scholar]
- Notredame, C., D. G. Higgins and J. Heringa, 2000. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302 205–217. [DOI] [PubMed] [Google Scholar]
- Palancade, B., and V. Doye, 2008. Sumoylating and desumoylating enzymes at nuclear pores: Underpinning their expected duties? Trends Cell Biol. 18 174–183. [DOI] [PubMed] [Google Scholar]
- Pan, Z. Q., A. Kentsis, D. C. Dias, K. Yamoah and K. Wu, 2004. Nedd8 on cullin: building an expressway to protein destruction. Oncogene 15 1985–1997. [DOI] [PubMed] [Google Scholar]
- Pickart, C. M., 2000. Ubiquitin in chains. Trends Biochem. Sci. 25 544–548. [DOI] [PubMed] [Google Scholar]
- Pickart, C. M., 2001. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70 503–533. [DOI] [PubMed] [Google Scholar]
- Pickart, C. M., 2004. Back to the future with ubiquitin. Cell 116 181–190. [DOI] [PubMed] [Google Scholar]
- Reeves, P. H., G. Murtas, S. Dash and G. Coupland, 2002. Early in short days 4, a mutation in Arabidopsis that causes early flowering and reduces the mRNA abundance of the floral repressor FLC. Development 129 5349–5361. [DOI] [PubMed] [Google Scholar]
- Rochaix, J.-D., M. Goldschmidt-Clermont and S. Merchant, 1998. The Molecular Biology of Chloroplasts and Mitochondria of Chlamydomonas. Kluwer Academic/Plenum Publishers, New York.
- Rosas-Acosta, G., W. K. Russell, A. Seyrieux, D. H. Russell and V. G. Wilson, 2005. A universal strategy for proteomic studies of SUMO and other ubiquitin-like modifiers. Mol. Cell. Proteomics 4 56–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson, D. A., M. Wang and M. J. Matunis, 2001. The small ubiquitin-like modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. J. Biol. Chem. 276 21664–21669. [DOI] [PubMed] [Google Scholar]
- Schultz, J., F. Milpetz, P. Bork and C. P. Ponting, 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. USA 95 5857–5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz, S. E., K. Matuschewski, D. Liakopoulos and M. Scheffner, 1999. The ubiquitin-like proteins SMT3 and SUMO-1 are conjugated by the UBC9 E2 enzyme. Proc. Natl. Acad. Sci. USA 95 560–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeler, J. S., and A. Dejean, 2003. Nuclear and unclear functions of SUMO. Nat. Rev. Mol. Cell Biol. 4 690–699. [DOI] [PubMed] [Google Scholar]
- Spikes, D. A., J. Kramer, P. H. Bingham and K. Van Doren, 1994. SWAP pre-mRNA splicing regulators are a novel, ancient protein family sharing a highly conserved sequence motif with the prp21 family of constitutive splicing factors. Nucleic Acids Res. 22 4510–4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke, M., and S. Waack, 2003. Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics 19(Suppl. 2): ii215–ii225. [DOI] [PubMed] [Google Scholar]
- Sternsdorf, T., K. Jensen, B. Reich and H. Will, 1999. The nuclear dot protein Sp100, characterization of domains necessary for dimerization, subcellular localization, and modification by small ubiquitin-like modifiers. J. Biol. Chem. 274 12555–12566. [DOI] [PubMed] [Google Scholar]
- Tanaka, K., J. Nishide, K. Okazaki, H. Kato, H. Niwa et al., 1999. Characterization of a fission yeast SUMO-1 homologue, Pmtp3, required for multiple nuclear events, including the control of telomere length and chromosome segregation. Mol. Cell. Biol. 19 8660–8672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toshiaki, S., A. Ichiyama, H. Saitoh, T. Kawakami, M. Omata et al., 1999. A new 30-k-Da Ubiquitin-related SUMO-1 hydrolase from bovine brain. J. Biol. Chem. 44 31131–31134. [DOI] [PubMed] [Google Scholar]
- Uchimura, Y., M. Nakao and H. Saitoh, 2004. a Generation of SUMO-1 modified proteins in E. coli: towards understanding the biochemistry/structural biology of the SUMO-1 pathway. FEBS Lett. 564 85–90. [DOI] [PubMed] [Google Scholar]
- Uchimura, Y., M. Nakamura, K. Sugasawa, M. Nakao and H. Saitoh, 2004. b Overproduction of eukaryotic SUMO-1 and SUMO-2-conjugated proteins in Escherichia coli. Anal. Biochem. 331 204–206. [DOI] [PubMed] [Google Scholar]
- Weeks, D. P., 1992. Chlamydomonas: an increasingly powerful model plant cell system. Plant Cell 4 871–878. [Google Scholar]
- Weissman, A. M., 2001. Themes and variations on ubiquitylation. Nat. Rev. Mol. Cell Biol. 2 169–178. [DOI] [PubMed] [Google Scholar]
- Wu, T. D., and C. K. Watanabe, 2005. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 21 1859–1875. [DOI] [PubMed] [Google Scholar]
- Xiang, Y., J. Zhang and D. P. Weeks, 2001. The Cia5 gene controls formation of the carbon concentrating mechanism in C. reinhardtii. Proc. Natl. Acad. Sci. USA 98 5341–5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh, E. T. H., L. Gong and T. Kamitani, 2000. Ubiquitin-like proteins: new wines in new bottles. Gene 248 1–14. [DOI] [PubMed] [Google Scholar]