Abstract
The yeast TEL1 and MEC1 genes (homologous to the mammalian ATM and ATR genes, respectively) serve partially redundant roles in the detection of DNA damage and in the regulation of telomere length. Haploid yeast tel1 mec1 strains were subcultured nonselectively for ∼200 cell divisions. The subcultured strains had very high rates of chromosome aberrations: duplications, deletions, and translocations. The breakpoints of the rearranged chromosomes were within retrotransposons (Ty or δ-repeats), and these chromosome aberrations nonrandomly involved chromosome III. In addition, we showed that strains with the hypomorphic mec1-21 allele often became disomic for chromosome VIII. This property of the mec1-21 strains is suppressed by a plasmid containing the DNA2 gene (located on chromosome VIII) that encodes an essential nuclease/helicase involved in DNA replication and DNA repair.
IN the yeast Saccharomyces cerevisiae, as in other eukaryotes, the rates of mutation and changes in chromosome structure in wild-type strains are very low. The rate of deletion of the CAN1 gene located near the end of chromosome V in a wild-type haploid strain is ∼4 × 10−10/division (Myung et al. 2001). This low deletion rate is dramatically elevated in certain genetic backgrounds. In strains with mutations in both TEL1 (related to the mammalian ATM gene) and MEC1 (related to the mammalian ATR gene), this deletion rate is ∼10−5/division, an increase of more than four orders of magnitude (Myung et al. 2001; Craven et al. 2002). In addition, tel1 mec1 strains have very elevated rates of chromosome loss and mitotic recombination (Craven et al. 2002). As discussed below, the synergistic effect of the two mutations on genome stability is likely to reflect their shared role in two processes: DNA damage checkpoints and telomere length regulation.
The TEL1 gene was first defined as a mutant that had unusually short poly(G1–3T) telomeric tracts (Lustig and Petes 1986). TEL1 encodes a very large kinase that is related to the human ATM gene (the gene mutated in patients with ataxia telangiectasia) and to the yeast MEC1 gene described below (Greenwell et al. 1995; Morrow et al. 1995). Two recent articles demonstrate that Tel1p is involved in recruiting telomerase to the telomeres. Goudsouzian et al. (2006) showed that recruitment of Est2p (the protein subunit of telomerase) and Est1p (a telomerase cofactor) is substantially reduced in a tel1 mutant. In addition, Tseng et al. (2006) demonstrated that Tel1p and Mec1p phosphorylate Cdc13p, a telomere-binding protein that interacts with Est1p. Thus, it is likely that Tel1p- and Mec1p-dependent phosphorylation of Cdc13p aids in the recruitment of telomerase and cofactors to the chromosome ends.
Although tel1 strains have the same sensitivity to most DNA-damaging agents as wild-type strains, Usui et al. (2001) showed that Tel1p was involved in the repair of unprocessed double-strand DNA breaks (DSBs). In addition, the DNA damage-induced phosphorylation of various proteins involved in the DNA damage checkpoint is substantially reduced in a tel1 strain (D'Amours and Jackson 2001; Usui et al. 2001). In summary, Tel1p has an important role in the regulation of telomere length and a modest role in the DNA damage checkpoint resulting from unprocessed DSBs.
In contrast to Tel1p, Mec1p has a minor role in telomere length regulation (Ritchie et al. 1999) and a major role in the DNA damage checkpoint (Harrison and Haber 2006). Strains with a mec1 mutation are sensitive to both DNA-damaging agents and drugs that block DNA synthesis because the strains lack multiple checkpoint pathways. Mec1p, like Tel1p, is a large kinase and the phosphorylation of many proteins in the DNA damage checkpoint pathways is Mec1p dependent (Harrison and Haber 2006). Overproduction of Tel1p can alleviate some of the sensitivity of mec1 strains to DNA-damaging agents (Morrow et al. 1995). Unlike TEL1, MEC1 is an essential gene, but double-mutant mec1 sml1 strains are viable (Zhao et al. 1998); the sml1 mutation results in elevated dNTP pools. Desany et al. (1998) showed that the lethality associated with the mec1 mutation reflects an inability of cells to complete DNA replication. Consistent with this conclusion, Cha and Kleckner (2002) showed that mec1 strains accumulated broken chromosomes with the DSBs occurring in regions in which DNA replication forks were moving slowly. Presumably as a consequence of elevated levels of DSB formation and inefficient DNA damage checkpoints, mec1 strains have increased rates of chromosome loss and mitotic recombination (Klein 2001; Craven et al. 2002).
As a generalization, the genome-destabilizing properties of the single tel1 and mec1 single mutations are very greatly increased in strains with the double mutation. As discussed above, the double-mutant strain has a greatly elevated rate of loss of the CAN1 gene that is located ∼32 kb from the end of chromosome V (Myung et al. 2001; Craven et al. 2002). In most of these can1 strains, the deletion of CAN1 includes all centromere-distal sequences and the remaining portion of chromosome V is fused to other chromosomal sequences by nonhomologous end joining (NHEJ) (Myung et al. 2001; Craven et al. 2002). In addition to fusions between different chromosomal sequences by NHEJ, which presumably requires two DSBs, the GCR assay used in the Kolodner lab also detects terminal deletions with telomere additions. Such events presumably require only a single DSB. Terminal deletions with telomere additions are not detected in the tel1 mec1 sml1 background (Myung et al. 2001).
As described above, tel1 strains have very short, but stable telomeres, and mec1 strains have telomeres that are only slightly shorter than those of wild-type strains. In strains of the tel1 mec1 genotype, the telomeres are unstable. Although most cells with this genotype die, a fraction survive as a consequence of recombination-dependent amplification of telomeric or subtelomeric repeats (Ritchie et al. 1999). In addition, high levels of telomere–telomere fusions occur in tel1 mec1 strains, but not in either single-mutant strain (Mieczkowski et al. 2003).
The tel1 mec1 strains are also more sensitive to DNA-damaging agents than either single mutant (Morrow et al. 1995), as a consequence of complete loss of the DNA damage checkpoint. Since some proteins involved in the DNA damage checkpoint response (for example, Rad9p and Ies4p; Emili 1998; Morrison et al. 2007) are substrates for both the Tel1p and the Mec1p kinases, this increased sensitivity to DNA damage is expected. In addition, Tel1p contributes to the processing of broken DNA ends that affects the efficiency of the Mec1p-dependent checkpoint response (Mantiero et al. 2007).
Because of the association between very high levels of genome instability and the formation of solid tumors (Lengauer et al. 1998), it is important to understand the nature of the chromosome rearrangements in strains with genome-destabilizing mutations. Below, we describe the use of DNA microarrays and other methods to look for chromosome rearrangements in subcultured tel1 mec1 yeast strains in which the only selection was for cell viability. We found a very high rate of chromosome alterations. Many of these rearrangements occurred on chromosome III and involved homologous recombination between nonallelic Ty elements. Ectopic recombination between nonallelic Ty elements has been observed previously in a number of studies (Dunham et al. 2002; Umezu et al. 2002; Lemoine et al. 2005; Mieczkowski et al. 2006); Umezu et al. (2002) also observed ectopic recombination between the MAT locus and HMR, a rearrangement also detected in our study. In addition, we found that haploid strains of the mec1-21 genotype develop chromosome VIII disomy. This property is related to the gene dosage of DNA2, a gene encoding a DNA replication-associated nuclease/helicase located on chromosome VIII.
MATERIALS AND METHODS
Strain and plasmid constructions:
All strains used in this study were isogenic with W303a (Thomas and Rothstein 1989), except for alterations introduced by transformation or by crosses with isogenic strains. The progenitor W303a strain has the markers a leu2-3,112 his3-11,15 ura3-1 ade2-1 trp1-1 can1-100 rad5-535). Details of the construction of derivatives of this strain are described in supplemental materials.
In some of the haploid strains, we introduced the hypomorphic mec1-21 allele by transformation with the plasmid pMD92. This plasmid was derived from the URA3-containing integrating plasmid YIp5. We used two primers (MEC1-21F 5′ GACACTATAGAGATCTTTGCTAAGATTATGTGTGATG and MEC1-21R 5′ TTCAGACAGGAGATCTACCTAATTCAGGCTTGCCTAC) to amplify an ∼800-bp fragment containing the mec1-21 mutant substitution (G to A at +2644 in the MEC1 gene; Mallory and Petes 2000); DNA isolated from the mec1-21-containing yeast strain JMY303-1d (a spore derived from JMY303; Mallory and Petes 2000) was used as a substrate for the PCR amplification. This fragment was treated with BglII and inserted within the BamHI site of YIp5 to generate pMD92. The restriction enzyme BsrGI was used to linearize the plasmid for transformation. The mec1-21 mutation was introduced into various strains by the standard two-step transplacement procedure (details in supplemental materials).
Genetic methods:
Standard media and genetic procedures were employed. For most of the experiments, each strain was subcultured on solid rich growth medium (YPD) 10 times. Strains were grown at 30° for 2–3 days for each subculturing. For each subculturing, we used a toothpick to scrape cells from regions of relatively heavy cell growth, rather than picking individual colonies. Individual colonies were picked only after the last subculturing.
Microarray and gel analysis:
Each strain was then analyzed by microarrays containing all yeast ORFs [microarrays purchased from Corning (Corning, NY)] or all ORFs and all intergenic regions (Lemoine et al. 2005). Following subculturing described above, we picked a single colony to begin the DNA isolation. The colony was inoculated into 5 ml of liquid growth medium and grown to stationary phase at 30°, representing ∼10 additional cell generations. Protocols for isolating and labeling DNA, and the hybridization conditions for the microarray analysis, are also described in the supplemental data section of Lemoine et al. (2005). The hybridization images were acquired using a GenePix 4000B scanner and were analyzed with GenePix Pro 5.0 software. Subsequent analysis was done with either Gene Spring 5.1 (Silicon Genetics) or CGH Miner (http://www-stat.stanford.edu/∼wp57/CGH-Miner/). We examined chromosomal DNA using the contour-clamped homogeneous electric field (CHEF) Mapper from Bio-Rad (Hercules, CA). Details of the procedures are described in Narayanan et al. (2006) (supplemental material).
RESULTS
On the basis of our previous studies (Craven et al. 2002; Mieczkowski et al. 2003) and those of others (Myung et al. 2001), we expected that haploid strains of the tel1 mec1 genotype would have a very high rate of genomic instability. Consequently, in the experiments described below, we subcultured tel1 mec1 strains and analyzed the subcultured derivatives by DNA microarrays (which detect changes in gene dosage), CHEF gels (which detect alterations in chromosome size), Southern analysis, and PCR analysis.
Pilot experiments demonstrating high rates of instability in tel1 mec1-21 strains:
Our initial study was based on examining tel1 mec1 spores derived from two independent diploid strains (strains 24 and 56) that were heterozygous for a null mutation of tel1 and the hypomorphic mec1-21 allele. Three tel1 mec1-21 spores derived from diploid 24 (MV24-15, MV24-17, and MV24-18) and two tel1 mec1-21 spores (MV56-3 and MV56-5) were examined. Colonies derived from each spore were streaked to solid rich growth medium and allowed to form new colonies. Two colonies derived from each spore were then subcultured separately by streaking to solid growth medium. After 10 subculturings (∼200 cell divisions), we isolated DNA from individual colonies and examined the DNA on microarrays containing all of the yeast genes. This type of analysis (comparative genome hybridization, CGH) detects deletions and duplications with the resolution of about one gene (Dunham et al. 2002; Lemoine et al. 2005). We also did CGH analysis on DNA isolated from each of the original five spore colonies before subcloning.
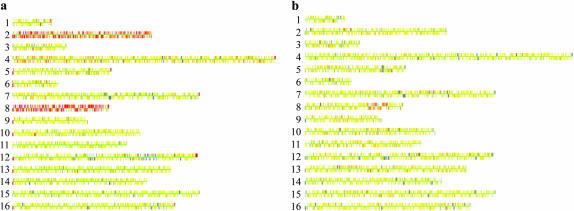
The results of this analysis are summarized in supplemental Table 1. All five spores were disomic for chromosome VIII, even before subculturing. In addition, MV24-5 was disomic for chromosome II, MV24-17 was disomic for chromosome III, and MV24-18 had a duplication of a 35-kb interstitial region of chromosome VII. Upon subculturing, some additional alterations occurred and some of the alterations observed in the original spore cultures were lost (supplemental Table 1). Examples of strains disomic for II and VIII (MV24-15) and having an interstitial duplication of VIII (MV56-5-2) are shown in Figure 1, a and b, respectively.
Figure 1.—
Microarray analysis (CGH) of two tel1 mec1-21 haploid strains derived from diploids MV24 and MV56. Diploids heterozygous for the tel1 and mec1-21 mutations were sporulated and dissected. Cultures derived from spores with the double-mutant genotype were examined before or after vegetative subculturing. DNA was isolated from each subcultured strain, labeled with a Cy5-fluorescent nucleotide, and mixed with DNA from a control strain that was labeled with a Cy3-fluorescent nucleotide. This mixture was hybridized to a microarray containing all of the yeast ORFs, and the ratio of hybridization of the two different samples to each ORF was determined (Dunham et al. 2002). In the Gene Spring 5.1 images used, each chromosome is shown by a series of adjacent very small rectangles representing the ORFs. In the image, yellow indicates the same dosage in the experimental and the control strain; red and blue indicate a duplication and a deletion, respectively, in the experimental strain relative to the control. The large gray region on chromosome XII is an uninformative signal on the microarray. (a) MV24-15. This strain (which was examined prior to subculturing) is disomic for chromosomes II and VIII. (b) MV56-5-2. This strain was vegetatively subcultured 10 times before the DNA was analyzed. The strain has a duplication of a 120-kb region of chromosome VIII that includes the DNA2 gene.
Although we did not examine these chromosome alterations in detail in the pilot experiment, several conclusions were evident. First, the genomic differences between different subcultured isolates of the same haploid strains indicate a high rate of chromosome nondisjunction and rearrangements associated with the tel1 mec1-21 genotype. Second, some of the rearrangements had breakpoints near or within the repetitive Ty or δ-elements. Third, chromosome III appeared prone to rearrangements and chromosome VIII appeared prone to changes in ploidy. The rearrangements involving chromosome III occurred during the subcloning, whereas the disomy of chromosome VIII was present in the original spore cultures. Since the diploids 24 and 56 from which the MV24 and MV56 strains were derived had not been saved, we could not rule out the possibility that the observed disomy in the haploid strains reflected trisomy or tetrasomy of chromosome VIII in the diploids rather than acquired disomy in the haploids. This issue was investigated in other experiments described below.
Haploid strains of the mec1-21 genotype often acquire an extra copy of chromosome VIII:
By two independent matings of the haploids MV40-3b (tel1∷KANMX arg4∷HYG CAN1) and RCY308-7b (MATα mec1-21 RAD5; Craven et al. 2002), we constructed two isogenic diploids, MV58 and MV59. By microarray analysis, we found that MV40-3b was euploid and RCY308-7b was disomic for chromosome VIII. Thus, both MV58 and MV59 were trisomic for chromosome VIII with one copy of VIII containing the arg4∷HYG mutation and two copies containing the wild-type ARG4 allele. Of 24 dissected tetrads derived from MV58 and MV59 that had four viable spores, we found that 16 tetrads had four Arg+ and zero Arg− spores, 3 had three Arg+ and one Arg− spores, and 5 had two Arg+ and two Arg− spores, as expected for the meiotic segregation of three chromosomes. In the tetrads with two Arg+ and two Arg− spores, the Arg− spores should have one copy of chromosome VIII and the Arg+ spores should be disomic.
We selected three Arg− spores for each of three genotypes: tel1, mec1-21, and tel1 mec1-21. Spore colonies were streaked onto rich solid growth medium and new colonies were formed. From each of the original spore colonies, we picked seven isolates that were subcultured separately 10 times. Thus, for each of the three genotypes, we had 21 strains. After the 10th subculturing, we did a CGH microarray analysis on each of these 62 strains. Twenty of the 21 mec1-21 strains and 18 of 21 tel1 mec-21 strains were disomic for chromosome VIII. None of the 21 tel1 strains was disomic for chromosome VIII.
Using an alternative approach, we introduced heterozygous tel1 and mec1-21 mutations into a diploid by transformation rather than by crossing mutant strains; details of the construction are in supplemental materials. The resulting diploid (MV70), when examined by CGH microarrays, was euploid and had no chromosome rearrangements resulting in deletions or duplications. We sporulated MV70 and selected two spores each of the four possible genotypes: wild type (MV70-1d, MV70-2c), mec1-21 (MV70-6d, MV70-7b), tel1 (MV70-1c, MV70-6c), and tel1 mec1-21 (MV70-5a, MV70-11d). Without subculturing, we analyzed cultures of all of the strains by CGH microarrays. No aneuploidy, duplications, or deletions were detectable in any of the wild-type or tel1 strains. All of the mec1-21 and tel1 mec1-21 strains were disomic for chromosome VIII, and one of the tel1 mec1-21 strains (MV70-11d) was also disomic for chromosome III. These results demonstrate that strains with the mec1-21 allele become disomic for chromosome VIII very rapidly.
One interpretation of these results is that the mec1-21 mutation specifically elevates the rate of nondisjunction for chromosome VIII. Alternatively, it is possible that mec1-21 strains with an extra copy of chromosome VIII have a selective growth advantage relative to mec1-21 strains with only one copy of VIII. One of the tel1 mec1-21 strains analyzed in the pilot experiment had an interstitial duplication of VIII (Figure 1b). This region of 120 kb contains ∼60 ORFs. Although most of these ORFs had no obvious functional connection to DNA repair or DNA replication (two processes affected by Mec1p), one gene in this region (DNA2) encodes an essential DNA nuclease/helicase involved in DNA replication and DNA repair (Budd and Campbell 1997, 2000).
To determine whether extra copies of DNA2 (a gene encoding a nuclease/helicase located on chromosome VIII) would suppress the tendency of tel1 mec1-21 strains to become disomic for chromosome VIII, we transformed MV70 with the plasmid pRS316-DNA2, a CEN-containing plasmid with the wild-type DNA2 and URA3 genes (Lee et al. 2000). The resulting diploid transformant (MV71) was sporulated and tetrads were dissected. Twelve plasmid-containing spores (two wild type, two tel1, six mec1-21, and two tel1 mec1-21) were grown into cultures and examined by CGH analysis. All wild-type and tel1 strains had only one copy of chromosome VIII. One of the two plasmid-containing tel1 mec1-21 strains had one copy of VIII and one had a hybridization signal for chromosome VIII, indicating that about half of the cells in the culture had one copy of VIII and about half had two copies. Of the six plasmid-containing mec1-21 strains, five had only one copy of chromosome VIII and one had two copies. We also isolated derivatives of two of the euploid mec1-21 strains that had spontaneously lost the pRS316-DNA2 plasmid. These derivatives acquired disomy for chromosome VIII.
As a control, we transformed MV70 with pRS316 (the URA3 vector without the DNA2 insert). This diploid was sporulated and we examined by CGH the gene dosage of five different mec1-21 haploid strains containing the vector. All five had two copies of chromosome VIII. In summary, an extra copy of DNA2 reduces the tendency of mec1-21 and tel1 mec1-21 strains to acquire an extra copy of chromosome VIII.
Intrachromosomal chromosome rearrangements associated with the tel1 mec1-21 genotype:
In addition to the extra copy of chromosome VIII, the 21 tel1 mec1-21 strains derived from MV58 and MV59 spores often had chromosome rearrangements or aneuploidy involving chromosome III. Most of the rearrangements involved homologous recombination between the 6-kb Ty retrotransposons or the 330-bp long-terminal repeats of these elements (δ's). These rearrangements are summarized in Table 1 and discussed in detail in the supplemental materials. Our conclusions concerning the nature of the rearrangements are based on the pattern of deletions and duplications as determined by microarrays and Southern analysis of separated chromosomal DNA molecules. Some of the rearrangements were characterized in more detail by standard Southern analysis and PCR analysis.
TABLE 1.
Genome alterations in subcultured tel1 mec-21 haploid strains derived from diploids MV58 and MV59
| Strain name | Genome alterations |
|---|---|
| MV58-20a#1 | Disomy for VIII. Two copies of III, one ∼100 kb smaller than wild type and one ∼100 kb larger (products of unequal crossing over between MAT and HMR). No Y′ amplification. |
| MV58-20a#2 | Disomy for VIII. Two copies of III, one wild-type chromosome and one with a deletion between MAT and HMR. Y′ amplification. |
| MV58-20a#3 | Disomy for VIII. Interstitial 35-kb duplication of VII [breakpoints near unannotated Ty elements (A. Gabriel, personal communication) located at YGRWδ19 and YGRWδ21]. Two copies of III, one wild-type chromosome and one isochromosome with duplication of sequences between LAHS and left telomere, and deletion of sequences distal to FS1. No Y′ amplification. |
| MV58-20a#4 | Disomy for VIII. Two copies of III, one normal copy and one isochromosome with duplication of sequences between the LAHS and the left telomere, and deletion of sequences distal to YCRCδ6. Y′ amplification. |
| MV58-20a#5 | Disomy for VIII (normal size). Two copies of III, one normal copy and one isochromosome with duplication between the LAHS and the left telomere, and deletion of sequences distal to FS2. No Y′ amplification. |
| MV58-20a#6 | Intrachromosomal duplication on chromosome IV between two pairs of Ty elements (YDRWTy2-2/YDRCTy1-2 and YDRWTy2-3/YDRCTy1-3). Three chromosomes with sequences from IIIL: (1) normal III, (2) isochromosome with duplication between the LAHS and the left telomere and deletion of sequences distal to YCRCδ6, and (3) translocation between YHRCTy1-1 on VIII and Ty at LAHS on III. No Y′ amplification. |
| MV58-20a#7 | Disomy for VIII. Translocation between III and X with breakpoint on III near the LAHS and breakpoint on X near YJLCδ4, which maps near an unannotated Ty (A. Gabriel, personal communication). Two other uncharacterized chromosome rearrangements involving chromosome III. Y′ amplification. |
| MV59-6a#1 | Disomy for VIII. Two copies of III, one isochromosome resulting from recombination between YCLCδ6 and the LAHS and one circular chromosome resulting from recombination between HML and HMR. Y′ amplification. |
| MV59-6a#2 | Disomy for VIII. Two copies of III, one normal and one with the Hawthorne deletion (loss of region between MAT and HMR). Y′ amplification. |
| MV59-6a#3 | Disomy for VIII. Two copies of III, one normal and one with the Hawthorne deletion (loss of region between MAT and HMR). Y′ amplification. |
| MV59-6a#4 | Disomy for VIII. Y′ amplification. |
| MV59-6a#5 | Disomy for 640-kb III–VIII translocation with breakpoints near LAHS on III and YHRCTy1-1: 440-kb derivative of III with duplication of region between the LAHS and FS1. Y′ amplification. |
| MV59-6a#6 | No disomy for VIII, but chromosome is ∼60 kb larger than the wild type; the origin of the extra DNA is not known. Disomy for III. Y′ amplification. |
| MV59-6a#7 | No disomy for VIII and VIII of normal size. III is ∼100 kb larger than the normal chromosome as a consequence of an unequal crossover, duplicating the region between MAT and HMR. Y′ amplification. |
| MV59-16c#1 | Disomy for VIII: 250-kb III–XII translocation with breakpoints at the LAHS and near YLRCsigma1, which maps at an unannotated Ty (A. Gabriel, personal communication). Interstitial 35-kb duplication of VII (same breakpoints as MV58-20a#3). Y′ amplification. |
| MV59-16c#2 | Disomy for VIII. One VIII of normal size (565 kb) and one ∼640 kb (VIII–XV translocation between YHRCTy1-1 and YOLWTy1-1); 250-kb III–XII translocation with same breakpoints as MV59-16c#1. No Y′ amplification. |
| MV59-16c#3 | Disomy for VIII. One VIII of normal size (565 kb) and one ∼640 kb (VIII–XV translocation between YHRCTy1-1 and YOLWTy1-1). Two copies of III, one normal III and one isochromosome resulting from recombination between YCLCδ6 and the 5′ δ of Ty1 at the LAHS. Interstitial 35-kb duplication of VII (same breakpoints as MV58-20a#3). Y′ amplification. |
| MV59-16c#4 | Disomy for I and VIII. Two copies of III, one normal III and one isochromosome resulting from recombination between YCLCδ6 and the 5′ δ of Ty1 at the LAHS. Y′ amplification. |
| MV59-16c#5 | Uncharacterized alteration duplicating the leftmost 280 kb of VIII. Extensive Y′ amplification. |
| MV59-16c#6 | Disomy for chromosome VIII. Both VIII and III hybridized to regions extending ∼50 kb beyond their normal size, probably the result of extensive Y′ amplification. |
| MV59-16c#7 | Disomy for chromosome VIII. Interstitial 35-kb duplication of VII (same breakpoints as MV58-20a#3). No Y′ amplification. |
These haploid strains were derived from sporulation of diploids heterozygous for the tel1 and mec1-21 mutations, as described in the text. These strains are isogenic with W303a (Thomas and Rothstein 1989). The strains were subcultured 10 times before analysis. Prior to subcloning, none of the strains had chromosome rearrangements, although all were disomic for chromosome VIII.
Among the 21 tel1 mec1-21 strains, there were 20 chromosomes that had detectable deletions or duplications resulting from intrachromosomal homologous recombination between repetitive elements in the chromosome. Two general classes were observed. One class (13 of the 20 intrachromosomal events) reflected unequal crossing over between directly oriented repeats, generating a deletion or a duplication of the sequences located between the repeats. The remaining 7 rearrangements resulted from a different type of recombination in which a break in a repeat located on one arm of the chromosome was repaired, utilizing a repeat located in inverted orientation on the opposite arm by break-induced replication (BIR) (Kraus et al. 2001). We describe two examples of these types of rearrangements in detail below.
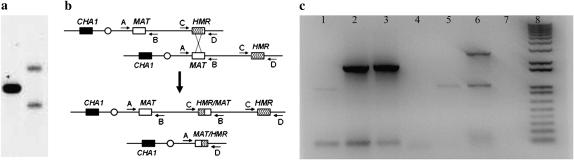
Microarray analysis indicated that the tel1 mec1-21 strain MV58-20a#1 was disomic for chromosomes VIII and III (data not shown). By CHEF gel analysis, although chromosome VIII had the same size as in the progenitor strain, MV58-20a#1 did not have a wild-type-sized chromosome III (∼340 kb), but had one chromosome that was ∼90 kb larger and one that was ∼90 kb smaller (Figure 2a). One explanation of this pattern is that these altered chromosomes reflected an unequal crossover (Figure 2b) between the mating-type locus (MAT) and HMR (a region of homology located ∼90 kb from the mating-type locus). As expected from this model, both chromosomes hybridized to the HIS4 probe derived from the left arm of chromosome III (Figure 2a), but only the larger chromosome hybridized with a probe (THR4) located between MAT and HMR (data not shown). This model also predicts two novel junctions, one containing the left end of the MAT locus and the right end of HMR and a second containing the left end of HMR fused to the right end of MAT. Both junctions were detected by PCR (Figure 2c). Of the 20 chromosome rearrangements observed in the tel1 mec1-21 strains, 7 involved recombination between HML or MAT and HMR (Table 1).
Figure 2.—
Analysis of a tel1 mec1-21 strain (MV58-20a#1) that has two rearranged chromosome IIIs generated by unequal crossing over. By microarray analysis, this strain was disomic for chromosomes III and VIII and had no other altered gene dosage. (a) CHEF gel analysis of chromosome III in a wild-type strain (W303, left lane) and in MV58-20a#1 (right lane). The separated chromosomal DNAs were hybridized to a probe derived from the left end of chromosome III (CHA1). Chromosome III in the wild-type strain was ∼340 kb, whereas the two chromosome III derivatives in MV58-20a#1 were ∼250 and 430 kb. (b) Depiction of the chromosome rearrangements resulting from unequal crossing over between the MAT and HMR repeats that share ∼1 kb of homology. The small arrows show the positions of primers used to diagnose the chromosome rearrangements. The chromosome is not drawn to scale. (c) PCR analysis of the breakpoints resulting from unequal crossing over between MAT and HMR. In lanes 1–4, we analyzed the products of a PCR reaction performed with primers A (BUD5-5Prime-Rev) and D (ARS318-Rev) to detect the MAT–HMR fusion. The PCR reactions contained W303a DNA (lane 1), MV58-20a#1 DNA (lane 2), MV58-20a#3 DNA (lane 3), and water (lane 4). We also did PCR reactions with primers B (ARS317-Upstream-For) and C (TAF2-3Prime-For) to detect the HMR–MAT fusion. The reactions contained DNA from W303a (lane 5), WV58-20a#1 (lane 6), and water (lane 7). Lane 8 contains size standards. The expected sizes of the fusions are 1.9 kb for MAT–HMR and 2.8 kb for HMR–MAT.
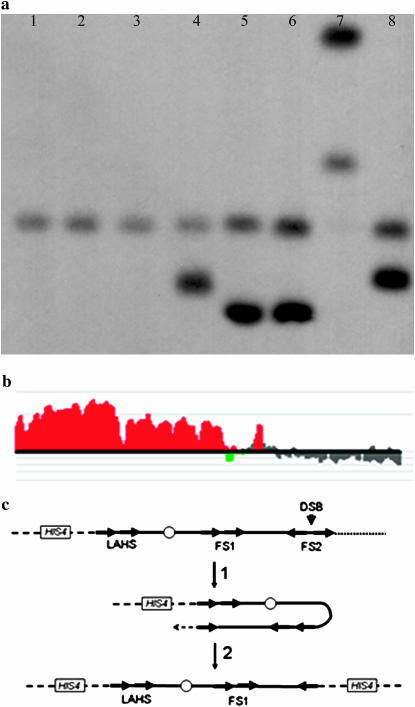
Thirteen of the 20 intrachromosomal rearrangements were a consequence of recombination between retrotransposons, involving two Ty elements, one Ty element and a solo δ (the long-terminal repeat associated with Ty), or two solo δ's. One strain with this type of rearrangement is MV58-20a#5. This strain had two chromosomes that hybridized to a probe derived from the left end of chromosome III, one of normal size (340 kb) and one ∼260 kb (Figure 3a, lane 8). In addition, by microarray analysis, one region of chromosome III was present in one copy (FS2 to right telomere), one region in two copies (left-arm hotspot, LAHS, to FS2), and one region in three copies (LAHS to left telomere, Figure 3b). Ty elements were located at the boundaries of these regions of different gene dosages. FS1 and FS2 are direct and inverted pairs, respectively, of Ty elements that are hotspots for chromosome rearrangements in strains with low levels of α-DNA polymerase (Lemoine et al. 2005). The LAHS is a region defined as a hotspot for the insertion of transposable elements; in W303, there is a Ty1 element centromere distal to a Ty2 element.
Figure 3.—
Analysis of tel1 mec1-21 strains (MV58-20a#5) with intrachromosomal rearrangements of chromosome III. (a) CHEF gel analysis of chromosome III in wild-type strains and in strains with the tel1 mec1-21 mutations. Separated chromosomal DNAs were hybridized to HIS4, a probe located on the left arm of chromosome III. The normal-sized chromosome III in W303 (lane 1) and two isogenic wild-type strains (lanes 2 and 3) derived from RCY278 (described in supplemental materials) is ∼340 kb. The tel1 mec1-21 strains examined include MV58-20a#3 (lane 4), MV59-16c#3 (lane 5), MV59-16c#4 (lane 6), MV59-6a#5 (lane 7), and MV58-20a#5 (lane 8); all except MV59-6a#5 have a normal chromosome III in addition to the rearranged III. The smaller-than-normal chromosome IIIs in lanes 4, 5, 6, and 8 could reflect DSBs in repetitive elements (Ty or δ) on the right arm of III that were repaired by a BIR event using a Ty or a δ on the left arm as a template (Figure 3c). In MV59-6a#5, the 640-kb chromosome represents a translocation between chromosomes III and VIII, whereas the 440-kb chromosome has an internal duplication on III. These rearrangements are described in detail in Table 1 and in supplemental materials. (b) Level of gene duplication in different regions of chromosome III by DNA microarray analysis in MV58-20a#5. The data from the microanalysis were analyzed using CGH Miner. The region of chromosome III between the Ty elements of the LAHS and the left telomere was in three copies per genome, the region between the LAHS and FS2 was in two copies, and the region between FS2 and the right telomere was in one copy. (c) Depiction of the mechanism for the generation of an isochromosome duplicating the region between the LAHS and the left telomere (indicated by a dashed line). A double-strand break in the centromere-proximal Ty element of FS2 could be repaired by a BIR event involving a Ty element in the LAHS. Ty elements are shown as arrows.
To explain the microarray and CHEF data, we suggest that MV58-20a#5 was initially disomic for chromosome III. One of these two copies had a DSB near or within the centromere-proximal Ty1 element of FS2. This DSB was repaired by a BIR event utilizing the Ty1 element of the LAHS as a template (Figure 3c). The result of this mechanism would be an isochromosome of 260 kb. It should be noted that the 260 kb hybridizes to the HIS4 gene used as hybridization probe with twice the intensity of the wild-type 340-kb chromosome (Figure 3a, lane 8), as expected since the 260-kb chromosome has two copies of the HIS4 gene. The predicted structure was also confirmed by standard Southern analysis as described in supplemental materials.
It is striking that 15 of the 20 intrachromosomal rearrangements involve chromosome III, one of the smallest yeast chromosomes. There are two likely explanations for this result. First, there may be some physical feature of chromosome III (for example, the high density of repetitive elements) that predisposes it to rearrangements by homologous recombination. Alternatively, since many of the rearrangements result in a duplication or a triplication of sequences between the LAHS and the left telomere, it is possible that extra doses of genes in this region confer a selective growth advantage to tel1 mec1-21 haploids. These alternatives are discussed further below.
Yeast strains lacking telomerase amplify telomeric [poly(G1–3T)] and subtelomeric (Y′) repeats by homologous recombination (Teng and Zakian 1999). Amplification of subtelomeric repeats is also observed in tel1 mec1-21 strains (32). From our microarray analysis, amplification of Y′ elements was evident in 16 of the 21 tel1 mec1-21 strains (Table 1). This amplification is detected as a signal of increased gene dosage at the ends of the Y′-bearing chromosomes (Figure 1a).
Interchromosomal chromosome rearrangements associated with the tel1 mec1-21 genotype:
In addition to the 20 intrachromosomal rearrangements, we observed seven interchromosomal events (translocations) (Table 1). Ty elements were observed at all breakpoints, indicating that the rearrangements were generated by ectopic homologous recombination. Four of the five types of translocations involved the Ty1 element located at the LAHS on chromosome III. Thus, both inter- and intrachromosomal rearrangements nonrandomly involve chromosome III. We discuss only one of the translocations in detail.
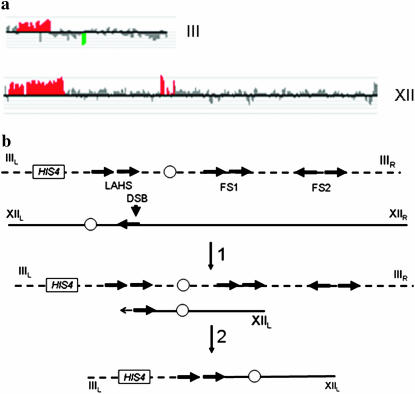
In MV59-16c#2, microarray analysis indicated a duplication of sequences from chromosome III between the LAHS and the left telomere and a 168-kb duplication of sequences on chromosome XII (Figure 4a). The breakpoint on XII mapped very close to an unannotated Ty2 element identified by A. Gabriel (personal communication). The predicted size of a translocation between the Ty elements on III and the Ty element on XII is ∼250 kb. By CHEF gel analysis, we found that a probe derived from the left arm of chromosome III hybridized to two chromosomes, one normal-sized III (340 kb) and one chromosome of ∼250 kb (data not shown); a probe from the right arm of III hybridized only to the longer chromosome. A more detailed Southern and PCR analysis (described in supplemental materials) demonstrated that the III–XII translocation was a consequence of a break in or near a δ-element associated with Ty2 on chromosome XII that was repaired by recombination with the δ-element of Ty1 on chromosome III (Figure 4b); although the translocation could also be generated by a DSB at or near the LAHS on III followed by a BIR event involving the Ty2 element on XII, BIR events that extend through the centromere occur inefficiently (Morrow et al.1997). Other translocations in our study also occur as a consequence of homologous recombination between repetitive elements (Table 1 and supplemental materials).
Figure 4.—
Analysis of a tel1 mec1-21 strain (MV59-16c#2) that has a III–XII translocation. (a) Gene dosage on chromosomes III and XII (based on CGH Miner analysis of microarrays). The boundaries for the gene dosage changes on III and XII are at the positions of the LAHS and an unannotated Ty element on XII, respectively. (b) Formation of a III–XII translocation by homologous recombination between Ty elements located on chromosomes III and XII.
Chromosome rearrangements are much less frequent in single-mutant mec1-21 and tel1 strains than in the double mutant:
The 21 mec1-21 strains examined in our study were derived from three mec1-21 spores, MV58-5b (#1-7), MV58-13b (#1-7), and MV59-18c (#1-7). As described above, after subculturing the strains 10 times, all strains except one (MV58-5b#7) were disomic for chromosome VIII. The MV58-5b#7 strain was disomic for chromosome XIV. In addition, strain MV58-13b#6 was disomic for both III and VIII, and strain MV59-18c#3 was disomic for both II and VIII. Only one chromosome rearrangement was detected. In MV58-5b#2, a 35-kb region of chromosome VII located between two like-oriented δ-/Ty elements (YGRWδ19/Ty and YGRWδ21/Ty) was duplicated; this same rearrangement was observed in four of the tel1 mec1-21 strains. No amplification of Y′ was observed for any of the mec1-21 strains.
The 21 subcultured tel1 strains were derived from three spore cultures, MV58-3d#1-7, MV59-2d#1-7, and MV59-10a#1-7. No aneuploidy or chromosome rearrangements were observed in any of the 21 strains. Interestingly, on the basis of the microarrays, 8 of the 21 tel1 strains had some degree of Y′ amplification, although the level of amplification was considerably less than that observed in most tel1 mec1-21 strains. In summary, in contrast to the tel1 mec1-21 genotype in which chromosome rearrangements were very common, only one of 42 strains of the mec1-21 and tel1 genotypes had a chromosome rearrangement.
DISCUSSION
Genome stability is usually examined using selective assays that are specific for certain loci or certain chromosomal regions. This constraint exists for two reasons. First, in general, the level of genome instability is not high enough to be readily analyzed by nonselective methods. Second, until recently, methods did not exist to allow a whole-genome analysis of genetic instability. In our study, we have used DNA microarrays and CHEF gel analysis to study the genetic instability associated with mutations in the two related genes TEL1 and MEC1 in subcultured derivatives. Our conclusions are different from those predicted by previous studies in which more selective methods have been employed. Even in our experiments, however, it is possible that the types of chromosome rearrangements that were detected were influenced by selection for those that had the least deleterious consequences for cell growth.
In two previous assays of genome stability in the single mutants and the tel1 mec1 double mutants, the rates of large deletions that include the CAN1 locus were examined (Myung et al. 2001; Craven et al. 2002). Both studies found that haploid strains with both tel1 and mec1 mutations had a level of deletions that was much higher than that observed in either single-mutant strain. There are no essential genes on chromosome V that are centromere distal to CAN1 and PCM1 (located ∼12 kb from CAN1) is the first essential centromere-proximal gene. In the can1 deletions derived from the tel1 mec1 sml1 (Myung et al. 2001) or the tel1 mec1-21 (Craven et al. 2002) strains, deletion derivatives of chromosome V with a breakpoint between CAN1 and PCM1 were fused to a chromosome segment derived from a nonhomologous chromosome by nonhomologous end joining.
In contrast, in the present study, most chromosome rearrangements involve homologous recombination between dispersed repeated genes. This difference has a simple explanation. The genome instability assay used by the previous studies selects for a breakpoint in a 12-kb region that has no repetitive DNA elements. Thus, any deletion that occurs in this region will be repaired by telomere capping (which is very inefficient in strains with a tel1 deletion; Goudsouzian et al. 2006; Tseng et al. 2006) or by nonhomologous end joining with DNA fragments derived from other chromosomes (as observed). It is likely, therefore, that the spectrum of chromosome alterations assayed by Myung et al. (2001) or Craven et al. (2002) would be dramatically altered and the rate of the events would be dramatically elevated by insertion of a Ty element between CAN1 and PCM1.
Several other points should be made concerning the chromosome rearrangements that are mediated by recombination between retrotransposons. First, it is not clear whether the retrotransposons are preferred sites for DSBs in the tel1 mec1 strains. It is possible that DSBs are random, but only the DSBs occurring within repetitive elements result in rearrangements. It is also possible that DSBs in nonrepetitive genomic regions could be processed by exonucleases to generate a chromosomal fragment with a Ty element at the end. Vanhulle et al. (2007) recently showed that processing of DSBs in nonrepetitive DNA on chromosome III can result in Ty–Ty-mediated rearrangements. Second, we do not know the source of the recombination-initiating DSBs in the tel1 mec1 strains. There are at least three plausible sources: (1) breaks resulting from exonucleolytic degradation of chromosomes that lack telomere “caps” (Hackett and Greider 2003), (2) breaks resulting from processing of dicentric chromosomes reflecting telomere–telomere fusions (Mieczkowski et al. 2003), and (3) breaks resulting from stalled DNA replication forks (Cha and Kleckner 2002). Third, our results support previous observations that recombination between dispersed retrotransposons is important in the evolution of the yeast genome, occurring both in nature and in the lab (reviewed by Mieczkowski et al. 2006). Fourth, although DSBs generated in a variety of ways result in retrotransposon-mediated chromosome rearrangements, there are quantitative differences in the frequencies of the rearrangements. For example, in strains with low levels of α-DNA polymerase, a preferred site for translocations was the inverted pair of Ty elements on the right arm of chromosome III (FS2; Lemoine et al. 2005). Although we also found a rearrangement involving FS2 in our study, a solo δ also on the right arm of III (YCLCδ) was more frequently observed at rearrangement breakpoints.
Both the tel1 mec1-21 and the mec1-21 strains became disomic for chromosome VIII and this tendency to become disomic for chromosome VIII was reduced in strains with an extra copy of DNA2 on a plasmid. Since Dna2p is a helicase and exo/endonuclease with many roles (Okazaki fragment processing, DNA repair, chromatin remodeling, and telomere maintenance; Budd et al. 2005), a simple interpretation of this effect is not yet possible. On the basis of the essential role of Mec1p in DNA replication (Desany et al. 1998) and the relatively small role of Mec1p in telomere length regulation (Ritchie et al. 1999), however, we suggest that the extra dose of Dna2p helps alleviate problems of DNA replication in the mec1-21 strains. Strains with a mec1 mutation also have an elevated rate of loss of chromosome V (Klein 2001; Craven et al. 2002), although this rate is low (5 × 10−5/division) compared to the rate of disomy for chromosome VIII observed in the present study. Haploid strains in a different genetic background and with a different mec1 mutation (a mec1 deletion, in which the lethality of the deletion is suppressed by overproduction of RNR1) become disomic for chromosome IV, rather than chromosome VIII (Gasch et al. 2001). We do not know whether the difference between our observations and this previous study reflects the type of mec1 mutation or some other feature of the genetic background.
Hughes et al. (2000) reported that ∼8% of 300 yeast strains from the deletion collection were aneuploid for one or more chromosomes. Although most of these aneuploid strains were not analyzed in any detail, Hughes et al. pointed out that, in several strains, the duplicated chromosome has a gene that was structurally related to the deleted gene. For two of the strains, they demonstrate that the derivative with the extra chromosome grew better than the euploidy progenitor.
In addition to disomy of chromosome VIII, tel1 mec1-21 strains also have a high frequency of chromosome rearrangements involving chromosome III. Since many of these rearrangements result in an elevated dosage of genes located between the LAHS and the telomere (a region of 80 kb containing ∼40 genes), it is possible that an extra dose of one or more genes in this region compensates for growth defects of the tel1 mec1-21 strain. A promising candidate gene in this region is MRC1, since Mrc1p is a substrate of the Mec1p kinase and is located at the replication fork (Katou et al. 2003; Osborn and Elledge 2003). In addition, the Mrc1p has a role in telomere capping (Tsolou and Lydall 2007). Thus far, however, we have not been able to suppress the accumulation of chromosome III rearrangements in tel1 mec1-21 strains with a plasmid-borne copy of MRC1 (our unpublished data).
Cells derived from solid tumors often exhibit a very high level of aneuploidy and chromosome rearrangements, and it is likely that an early step in the development of some tumors is a mutation that results in elevated rates of genetic instability (Lengauer et al. 1998). Our analysis of the instability in tel1 mec1-21 strains argues that the same mutant background can produce both elevated rates of aneuploidy and elevated rates of translocations. In addition, our results and those of Hughes et al. (2000) argue that the gene amplifications that occur in tumor cells may promote tumorigenesis by two different mechanisms: some amplifications (“accelerators”) may directly stimulate tumor growth, whereas others (“compensators”) may alleviate the negative effects of the original genome-destabilizing mutant.
Acknowledgments
We thank G. Gawel, M. Dominska, and P. Greenwell for help with the analysis of the translocations. This research was supported by National Institutes of Health grant GM52319 (T.P.) and National Science Foundation grant MCB-0417088 (K.L.).
References
- Budd, M. E., and J. L. Campbell, 1997. A yeast replicative helicase, Dna2 helicase, interacts with yeast FEN-1 nuclease in carrying out its essential function. Mol. Cell. Biol. 17 2136–2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budd, M. E., and J. L. Campbell, 2000. The pattern of sensitivity of yeast dna2 mutants to DNA damaging agents suggests a role in DSB and postreplication repair pathways. Mutat. Res. 459 173–186. [DOI] [PubMed] [Google Scholar]
- Budd, M. E., A. H. Y. Tong, P. Polaczek, X. Peng, C. Boone et al., 2005. A network of multi-tasking proteins at the DNA replication fork preserves genome stability. PLoS Genet. 1 634–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha, R. S., and N. Kleckner, 2002. ATR homolog Mec1 promotes fork progression, thus averting breaks in replication slow zones. Science 297 602–606. [DOI] [PubMed] [Google Scholar]
- Craven, R. J., P. W. Greenwell, M. Dominska and T. D. Petes, 2002. Regulation of genome stability by TEL1 and MEC1, yeast homologs of the mammalian ATM and ATR genes. Genetics 161 493–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amours, D., and S. P. Jackson, 2001. The yeast Xrs2 complex functions in S phase checkpoint regulation. Genes Dev. 15 2238–2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desany, B. A., A. A. Alcasabas, J. B. Bachant and S. J. Elledge, 1998. Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes Dev. 12 2956–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham, M. J., H. Badrane, T. Ferea, J. Adams, P. O. Brown et al., 2002. Characteristic genome rearrangements in experimental evolution of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 99 16144–16149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emili, A., 1998. MEC1-dependent phosphorylation of Rad9p in response to DNA damage. Mol. Cell 2 183–189. [DOI] [PubMed] [Google Scholar]
- Gasch, A. P., M. Huang, S. Metzner, D. Botstein, S. J. Elledge et al., 2001. Genomic expression responses to DNA-damaging agents and the regulatory role of the yeast ATR homolog Mec1p. Mol. Biol. Cell 12 2987–3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudsouzian, L. K., C. T. Tuzon and V. A. Zakian, 2006. S. cerevisiae Tel1p and Mre11p are required for normal levels of Est1p and Est2p telomere association. Mol. Cell 24 603–610. [DOI] [PubMed] [Google Scholar]
- Greenwell, P. W., S. L. Kronmal, S. E. Porter, J. Gassenhuber, B. Obermaier et al., 1995. TEL1, a gene involved in controlling telomere length in S. cerevisiae, is homologous to the human ataxia telangiectasia gene. Cell 82 823–829. [DOI] [PubMed] [Google Scholar]
- Hackett, J. A., and C. W. Greider, 2003. End resection initiates genomic instability in the absence of telomerase. Mol. Cell. Biol. 23 8450–8461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison, J. C., and J. E. Haber, 2006. Surviving the breakup: the DNA damage checkpoint. Annu. Rev. Genet. 40 209–235. [DOI] [PubMed] [Google Scholar]
- Hughes, T. R., C. J. Roberts, H. Dai, A. R. Jones, M. R. Meyer et al., 2000. Widespread aneuploidy revealed by DNA microarray expression profiling. Nat. Genet. 25 333–337. [DOI] [PubMed] [Google Scholar]
- Katou, Y., Y. Kanoh, M. Bando, H. Noguchi, H. Tanaka et al., 2003. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 424 1078–1083. [DOI] [PubMed] [Google Scholar]
- Klein, H. L., 2001. Spontaneous chromosome loss in Saccharomyces cerevisiae is suppressed by DNA damage checkpoint functions. Genetics 159 1501–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus, E., W. Y. Leung and J. E. Haber, 2001. Break-induced replication: a review and an example in budding yeast. Proc. Natl. Acad. Sci. USA 98 8255–8262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, K. H., D. W. Kim, S. H. Bae, J. A. Kim, G. H. Ryu et al., 2000. The endonuclease activity of the yeast Dna2 enzyme is essential in vivo. Nucleic Acids Res. 28 2873–2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemoine, F. J., N. P. Degtyareva, K. Lobachev and T. D. Petes, 2005. Chromosomal translocations in yeast induced by low levels of DNA polymerase: a model for chromosome fragile sites. Cell 120 587–598. [DOI] [PubMed] [Google Scholar]
- Lengauer, C., K. Kinzler and B. Vogelstein, 1998. Genetic instabilities in human cancers. Nature 396 643–649. [DOI] [PubMed] [Google Scholar]
- Lustig, A. J., and T. D. Petes, 1986. Identification of yeast mutants with altered telomere structure. Proc. Natl. Acad. Sci. USA 83 1398–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory, J. C., and T. D. Petes, 2000. Protein kinase activity of Tel1p and Mec1p, two Saccharomyces cerevisae proteins related to the human ATM protein kinase. Proc. Natl. Acad. Sci. USA 97 13749–13754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantiero, D., M. Clerici, G. Lucchini and M. P. Longhese, 2007. Dual role for Saccharomyces cerevisiae Tel1 in the checkpoint response to double-strand breaks. EMBO Rep. 8 380–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mieczkowski, P. A., J. O. Mieczkowska, M. Dominska and T. D. Petes, 2003. Genetic regulation of telomere-telomere fusions in the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 100 10854–10859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mieczkowski, P. A., F. J. Lemoine and T. D. Petes, 2006. Recombination between retrotransposons as a source of chromosome rearrangements in the yeast Saccharomyces cerevisiae. DNA Repair 5 1010–1020. [DOI] [PubMed] [Google Scholar]
- Morrison, A. J., J.-A. Kim, M. D. Person, J. Highland, J. Xiao et al., 2007. Mec1/Tel1 phosphorylation of the INO80 chromatin remodeling complex influences DNA damage checkpoint responses. Cell 130 499–511. [DOI] [PubMed] [Google Scholar]
- Morrow, D. M., D. A. Tagle, Y. Shiloh, F. S. Collins and P. Hieter, 1995. TEL1, an S. cerevisiae homolog of the human gene mutated in ataxia telangiectasia, is functionally related to the yeast checkpoint gene MEC1. Cell 82 831–840. [DOI] [PubMed] [Google Scholar]
- Morrow, D. W., C. Connelly and P. Hieter, 1997. Break copy duplication: a model for chromosome fragment formation in Saccharomyces cerevisiae. Genetics 147 371–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung, K., A. Datta and R. D. Kolodner, 2001. Suppression of spontaneous chromosomal rearrangements by S phase checkpoint functions in Saccharomyces cerevisiae. Cell 104 397–408. [DOI] [PubMed] [Google Scholar]
- Narayanan, V., P. A. Mieczkowski, H.-M. Kim, T. D. Petes and K. S. Lobachev, 2006. The pattern of gene amplification is determined by the chromosomal location of hairpin-capped breaks. Cell 125 1283–1296. [DOI] [PubMed] [Google Scholar]
- Osborn, A. J., and S. J. Elledge, 2003. Mrc1 is a replication fork component whose phosphorylation in response to DNA replication stress activates Rad53. Genes Dev. 17 1755–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie, K. B., J. C. Mallory and T. D. Petes, 1999. Interactions of TLC1 (which encodes the RNA subunit of telomerase), TEL1, and MEC1 in regulating telomere length in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 19 6065–6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng, S. C., and V. A. Zakian, 1999. Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol. Cell. Biol. 19 8083–8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, B. J., and R. Rothstein, 1989. Elevated recombination rates in transcriptionally active DNA. Cell 56 619–630. [DOI] [PubMed] [Google Scholar]
- Tseng, S.-F., J.-J. Lin and S.-C. Teng, 2006. The telomerase-recruitment domain of the telomere binding protein Cdc13 is regulated by Mec1p/Tel1p-dependent phosphorylation. Nucleic Acids Res. 34 6327–6336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsolou, A., and D. Lydall, 2007. Mrc1 protects uncapped budding yeast telomeres from exonuclease Exo1. DNA Repair 6 1607–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezu, K., M. Hiraoka, M. Mori and H. Maki, 2002. Structural analysis of aberrant chromosomes that occur spontaneously in diploid Saccharomyces cerevisiae: retrotransposon Ty1 plays a crucial role in chromosomal rearrangements. Genetics 160 97–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usui, T., H. Ogawa and J. H. Petrini, 2001. A DNA damage response pathway controlled by Tel1 and the Mre11 complex. Mol. Cell 7 1255–1266. [DOI] [PubMed] [Google Scholar]
- VanHulle, K., F. J. Lemoine, V. Narayanan, B. Downing, K. Hull et al., 2007. Inverted DNA repeats channel repair of distant double-strand breaks into chromatid fusions and chromosomal rearrangements. Mol. Cell. Biol. 27 2601–2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, X., E. G. Muller and R. Rothstein, 1998. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol. Cell 2 329–340. [DOI] [PubMed] [Google Scholar]