Abstract
Several tests have been proposed to detect departures of nucleotide variability patterns from neutral expectations. However, very different kinds of evolutionary processes, such as selective events or demographic changes, can produce similar deviations from these tests, thus making interpretation difficult when a significant departure of neutrality is detected. Here we study the effects of demography and recombination upon neutrality tests by analyzing their power under sudden population expansions, sudden contractions, and bottlenecks. We evaluate tests based on the frequency spectrum of mutations and the distribution of haplotypes and explore the consequences of using incorrect estimates of the rates of recombination when testing for neutrality. We show that tests that rely on haplotype frequencies—especially Fs and ZnS, which are based, respectively, on the number of different haplotypes and on the r2 values between all pairs of polymorphic sites—are the most powerful for detecting expansions on nonrecombining genomic regions. Nevertheless, they are strongly affected by misestimations of recombination, so they should not be used when recombination levels are unknown. Instead, class I tests, particularly Tajima's D or R2, are recommended.
AN increasing number of statistical tests (Tajima 1989a; Fu and Li 1993; Fu 1997; Fay and Wu 2000; Ramos-Onsins and Rozas 2002) have been developed to detect departures of DNA sequence variability from the expectations of the neutral theory of evolution (Kimura 1968). Most of the research in this area is based upon the Wright–Fisher model (Fisher 1930; Wright 1931; Hein et al. 2005), which assumes populations of constant size that are panmictic and nonrecombining. Moreover, the Wright–Fisher model provided the founding of coalescent theory (Kingman 1982a,b, 2000; Hudson 1990; Donnelly and Tavare 1995; Fu and Li 1999), which was fundamental for developing neutrality tests and furthering their study. Even if these models are quite prone to mathematical treatment, analytic derivations are often unreachable and the significance of departures from neutrality and the statistical power of the tests are estimated by computer simulations based on the coalescent process (Wall 1999; Ramos-Onsins and Rozas 2002; Depaulis et al. 2003).
The detection of departures from the null hypothesis of neutrality points to the violation of one or more of its assumptions. These deviations can be due to selective and/or demographic events. For example, selective sweeps or population growth can produce longer external branches in the genealogy that result in an excess of recent mutations over neutral predictions. In contrast, population subdivision or balancing selection will result in longer internal branches and, consequently, in an excess of old over recent mutations. In summary, different kinds of processes can produce similar genealogies and therefore confound the interpretation of tests.
Much effort has been devoted to ascertaining the power of different statistical tests to reject the null hypothesis of neutrality when it is actually false, as well as to defining properly which tests perform best in each scenario. Ramos-Onsins and Rozas (2002) studied the power of 17 statistical tests under sudden or logistic population-expansion models. The tests were classified in three categories on the basis of the information they used. Class I tests are based on the frequency spectrum of mutations, class II on the haplotype distribution, and class III on the distribution of pairwise differences. Depaulis et al. (2003) studied the power of seven statistics under bottlenecks (both severe and moderate) and hitchhiking with positively selected mutations. More recently, the power of several tests has been studied under exponential population growth and bottlenecks (Sano and Tachida 2005) and population structure and hitchhiking (Jensen et al. 2005). However, the effect of intragenic recombination has been considered in only a limited number of studies, and, in particular, the joint effect of recombination and population expansions on the statistical power of neutrality tests has not, to the best of our knowledge, been explored.
The neutral model with no recombination, which is commonly used as the null hypothesis, has larger variance in genealogy length than the same model including recombination (Hein et al. 2005). Such larger variance makes the assumption of no recombination a conservative assumption for many statistical tests. In particular, tests based on the frequency spectrum of mutations are likely to be conservative on recombining regions (Tajima 1989a; Fu and Li 1993; Fu 1996). On the other hand, tests based on haplotype or linkage disequilibrium (LD) are expected to be strongly affected by recombination, since it will break down existing haplotypes and generate new ones, thus decreasing LD. Moreover, as recombination can also smooth the mismatch distribution, it is likely that statistical tests based on this distribution will have little power (Ramos-Onsins and Rozas 2002). Finally, recombination can mimic the effect of some demographic models, such as population growth. It is therefore of general interest to distinguish the individual effects of recombination and population growth on DNA sequence variation and on neutrality tests (Schierup and Hein 2000).
The study of population expansions is also of great interest since their effects on genealogies (and, thus, on many neutrality statistics) are similar to those of other selective or demographic events. Among the former, selective sweeps caused by positively selected variants, as well as background selection against deleterious mutations, lead to an excess of low-frequency variants (Charlesworth et al. 1993; Przeworski 2002). On the other hand, recent bottlenecks can also mimic the effects of an expansion (Tajima 1989a,b), so these phenomena can be quite difficult to disentangle. In spite of such difficulty, considerable progress is being made to distinguish between expansions and selective sweeps (Jensen et al. 2005; Williamson et al. 2005) or between bottlenecks and positive selection (Haddrill et al. 2005).
Here we use coalescent simulations to test the power of 16 statistical tests to detect population expansions, contractions, and bottlenecks under different recombination levels. The selected tests belong to the first two categories described by Ramos-Onsins and Rozas (2002). We pay special attention to the problem of misestimation of recombination rates and study how the use of incorrect recombination rates when simulating neutral samples can affect the power and the false-positive rates of tests. We have found that statistics based on haplotype diversity are the most powerful tests for detecting population expansions on nonrecombining regions. In contrast, since they are very sensitive to recombination, their use should be avoided when there is recombination.
MATERIALS AND METHODS
Statistics:
We have considered two classes of statistics: statistics based on the frequency spectrum of mutation (class I) and statistics based on linkage disequilibrium and haplotype distribution (class II). No statistics based on the distribution of pairwise differences (e.g., the mismatch distribution) have been used, as they were shown to perform very poorly in the study by Ramos-Onsins and Rozas (2002). A summary of all statistics can be found in Table 1.
TABLE 1.
Definition of the neutrality statistics used
| Test | Definition | Reference |
|---|---|---|
| Class I | ||
| Tajima's D | Comparison of estimates of the no. of segregating sites and the mean pairwise difference between sequences | Tajima (1989a) |
| Fu and Li's D (DF) and D* | Comparison of the number of derived singleton mutations and the total number of derived nucleotide variants (the asterisk indicates “without an outgroup”) | Fu and Li (1993) |
| Fu and Li's F and F* | Comparison of the number of derived singleton mutations and the mean pairwise difference between sequences (the asterisk indicates “without an outgroup”) | Fu and Li (1993) |
| Fay and Wu's H | Comparison of the number of derived segregating sites at low and high frequencies and the number of variants at intermediate frequencies | Fay and Wu (2000) |
| R2 | Comparison of the difference between the number of singleton mutations and the average number of nucleotide differences | Ramos-Onsins and Rozas (2002) |
| Class II | ||
| Fu's Fs | Based on Ewens' sampling distribution, taking into account the number of different haplotypes in the sample | Fu (1997) |
| Dh | Based on the number of different haplotypes in the sample | Nei (1987) |
| EHH average | Weighted average for all core haplotypes of the position at which the haplotype homozygosity decays to ≤0.5 | Based on Sabeti et al. (2002) |
| EHH maximum | The position corresponding to the core haplotype that decays to ≤0.5 at a greater distance | Based on Sabeti et al. (2002) |
| Wall's B | Counts the number of pairs of adjacent segregating sites that are congruent (if the subset of the data consisting of the two sites contains only two different haplotypes) | Wall (1999) |
| Wall's Q | Adds the number of partitions (two disjoint subsets whose union is the set of individuals in the sample) induced by congruent pairs to Wall's B | Wall (2000) |
| Kelly's ZnS | Average of the squared correlation of the allelic identity between two loci over all pairwise comparisons | Kelly (1997) |
| Rozas' ZA | Average of the squared correlation of the allelic identity between two loci over adjacent pairwise comparisons | Rozas et al. (2001) |
| Rozas' ZZ | Comparison between ZnS and ZA | Rozas et al. (2001) |
Class I statistics:
Class I statistics use information on the frequency of mutations and are based on the differences between estimators of the population mutation rate θ = 4Nμ, where N is the effective population size and μ is the mutation rate. From this class, we present results for Tajima's D (Tajima 1989a), Fu and Li's D, F, D*, and F* (Fu and Li 1993), and Fay and Wu's H (Fay and Wu 2000). We have also included the R2 statistic (Ramos-Onsins and Rozas 2002), which is based on the difference between the number of singletons per sequence and the average number of nucleotide differences.
Class II statistics:
Class II includes statistics based on the haplotype distribution. They are expected to be the most affected by recombination. Within this class, we have studied the following statistics: Fu's Fs (Fu 1997), the unbiased haplotype diversity estimate Dh (Nei 1987, Equation 8.5), Wall's B and Q (Wall 1999), Kelly's ZnS (Kelly 1997), Rozas' ZA and ZZ (Rozas et al. 2001), and two statistics based on the extended haplotype homozygosity (EHH) (Sabeti et al. 2002).
EHH statistics are a complex family of heuristic methods for which no consensus summary statistic has yet been developed. We have computed two EHH-based statistics by taking the first three SNPs of each sequence as a core haplotype (that is, as the locus of interest) and then considering the distance from each core at which EHH decays to ≤0.5. Two values are given: (1) the EHH average, corresponding to the weighted average for all core haplotypes of the distance at which EHH decays to ≤0.5 and (2) the EHH maximum, the distance corresponding to the core haplotype that decays to ≤0.5 at a greater distance. If a simulated segment finishes without EHH reaching a value ≤0.5, taking the chromosome length as L, we arbitrarily consider that the position will be at 2L.
Coalescent simulations:
We tested the statistical power of the statistics under different demographic models by running neutral coalescent simulations using the algorithm described by Hudson (1990) and implemented in the ms package (Hudson 2002). This program generates coalescent trees for a given sample size, recombination rate, and a demographic scenario, implementing an infinite-sites mutation model that leads to biallelic sites.
There is an intense debate on how to best perform simulations and, specifically, on the suitability of running coalescent simulations by fixing either the number of segregating sites (S) or the population mutation rate θ = 4Nμ (Hudson 1993; Wall and Hudson 2001; Depaulis et al. 2001, 2005). Conditioning on θ has the disadvantage that its value has to be estimated. Furthermore, even if its true value could be known, it produces broader confidence intervals, thus reducing the power of tests (Depaulis et al. 2005). On the other hand, although forcing a given number of segregating sites in all trees without considering their particularities (such as branch length) is also unrealistic, conditioning on the number of segregating sites has the advantage that S is a parameter that can be observed in the sample. To solve this problem, several strategies have been proposed to obtain realistic samples conditioning on both θ and S (Hudson 1993; Depaulis et al. 2001, 2003, 2005; Wall and Hudson 2001; Przeworski 2002). Different authors agree that simulated parameters are more accurate if the simulations are conditioned on S taking into account the uncertainty of θ (Tavare et al. 1997; Pritchard et al. 1999).
However, obtaining neutral models fixing the number of segregating sites—which can be directly obtained from the sample—or estimating θ from S are still widely used by researchers (Macdonald and Long 2005; Soejima et al. 2005; Stajich and Hahn 2005; Tarazona-Santos and Tishkoff 2005). Taking into account this popularity, we have conditioned our simulations on S and the θ estimator θW after proper validation of this approach (see below). For simulations conditioned on the number of segregating sites, S values were set to 10, 100, and 400. This corresponds to the rounded minimum, average, and maximum segregating sites found in the genes resequenced by SeattleSNPs (http://pga.gs.washington.edu/; Crawford et al. 2005), the largest ongoing human resequencing project, which currently contains sequences of a length of 3.5–71 kb for >300 genes obtained from 23 European–American and 24 African–American individuals. For simulations conditioned on θW (Watterson 1975), θW values correspond to the S values used in the previous simulations. All the values have been fixed assuming a panmictic and stationary neutral population, which could cause incorrect power estimations for statistics, depending on the number of segregating sites.
To ascertain the validity of our approach, results for simulations fixing S in expansions have been compared with results obtained considering the uncertainty of θ and using the rejection algorithm (Tavare et al. 1997). Comparisons have been performed for all S values studied and for the minimum (0) and maximum (10−7) recombination values. Comparison shows that the differences in estimates of nominal rejection level between the two methods are very small. In fact, in 96% of the cases they are <5%, and in no case do they reach values >15%. Moreover, these differences become even smaller with increasing recombination rates (results not shown). In summary, we use a methodology that is accurate for neutral simulations (Depaulis et al. 2001; Wall and Hudson 2001; Ramos-Onsins et al. 2007) and for all our expansion models (Ramos-Onsins et al. 2007; materials and methods). However, our approximate method can produce deviations when computing statistical power under other alternative models, such as the contraction and bottleneck models (Ramos-Onsins et al. 2007). The magnitude of these deviations depends on the particular statistic and the parameters of the model. Deviation from the exact strategy has been evaluated partially, and our results indicate that, in the studied cases, the deviation in the statistical power is not large.
Recombination:
Recombination rates were set to r = 10−10, r = 10−8, and r = 10−7 per nucleotide pair; as simulations are scaled in units as 4N generations, assuming Ne = 10,000 for humans (Takahata et al. 1995), this would correspond to population recombination rates of R = 4Nr equal to 4 × 10−6, 4 × 10−4, and 4 × 10−3 per nucleotide, respectively. These values correspond to the rounded minimum, average, and maximum values estimated by Kong et al. (2002) for the human genome. Simulations without recombination were also performed. To calculate the crossover probability, we have assumed sequence lengths of 3000, 21,000, and 72,000 bp, as these lengths correspond to the minimum, average, and maximum rounded lengths of the resequenced human genes in SeattleSNPs, and Ne = 10,000.
Demography:
We have simulated four basic demographic models: stationarity, sudden population growth, sudden population contraction, and population bottleneck, with emphasis on the two first cases. All scenarios consider different recombination rates. For each scenario, we ran 10,000 simulations. The significance level was set to 0.05, and the critical value for each statistic was obtained from the empirical distribution of the corresponding neutral model. As mutations were simulated under an infinite-sites model (which implies no recurrent mutation), the outgroup (for those statistics that require it) was set to a string of 0's, where 0 is the ancestral state as coded by ms.
The sudden population growth model (Rogers and Harpending 1992) assumes that a population of size N0 in equilibrium experienced a sudden growth and reached maximum size (Nmax) Te generations before present. In the simulations, time is scaled in units of 4Ne generations. Changes in population size are performed according to the standard procedures in the ms program. Expansion times have been set to range from 0.0075 to 0.2. In humans, taking again Ne = 10,000 and a generation time of 20 years, the simulated expansion times would range from 6000 to 160,000 years ago. The latter can be taken as the earliest estimate for a human expansion (Jobling et al. 2004). Since some statistics showed significant power at that earlier expansion time, and to produce results applying to other species, some additional expansion times were added at Te = 0.4, Te = 0.6, Te = 0.8, and Te = 1 (equivalent, respectively, to 16,000, 24,000, 32,000, and 40,000 generations and, in human terms, 320,000, 480,000, 640,000, and 800,000 years ago). The degree of expansion (De = Nmax/N0) has been set to De = 10 and De = 100.
The sudden contraction model considers a population of constant size N0, which has instantaneously contracted to a size Nmin; the contraction degree is defined as De = Nmin/N0. This model has been simulated as the opposite of our expansion model and thus the same Ne and Te have been used. Contraction degree has been set to Dc = 0.1 and Dc = 0.01. We have performed simulations for S = 10 and S = 100.
The bottleneck model has been simulated as in Voight et al. (2005). It assumes a population of size NA, which has been suddenly reduced to a second size, b · NA for Tdur generations Tstart generations ago, where b is the bottleneck severity. Immediately afterward, the population has instantaneously recovered its original size, NA. As in the expansion model, NA has been set to 10,000. According to the results by Voight et al. (2005), we have simulated five bottleneck severities (b): 0.4, 0.1, 0.05, 0.01, and 0.005, and for each we have simulated durations of 0.01, 0.02, 0.03, and 0.04 (400, 800, 1200, and 1600 generations). Tstart has been set to 0.02, 0.04, 0.08, and 0.12 (800, 1600, 3200, and 4800 generations, respectively). Additional points have been simulated to better illustrate this model in figures.
RESULTS
Exploring parameter space:
We have tested the power of 16 statistics under the different conditions of sample size (n), number of segregating sites (S), neutral mutation parameter (θ), demography, and recombination rates. Although a wide range of parameters have been studied, only the most interesting cases are shown. Full results are provided as supplemental data A–D. Supplemental data A and B show all results with correct recombination rates in the neutral model for the expansion model (A) and the contraction and bottleneck (B). Supplemental data C and D show results for mispecified recombination rates in the neutral model for expansions (C) and contractions and bottlenecks (D). As expected, larger sample sizes improve the power of the statistics, so we have plotted all power curves with n = 100; curves for other sample sizes can be found in the supplemental data files. Also, we have fixed the degree of expansion, De, to 10 in all expansion figures, as De = 100 produced uniformly maximal power for all statistics. For figures involving contraction, the degree of contraction, Dc, has been fixed to 0.1, and bottlenecks are shown with a severity of b = 0.01. Other parameter values fixed in some figures have also been chosen to avoid saturation.
The power of statistics has been shown for the tail for which each statistic shows power under its corresponding model. Since a population-growth event will cause an excess of derived low-frequency nucleotide variants and of the number of haplotypes and a reduction of linkage disequilibrium values, this tail corresponds in expansions to the left one for all statistics except for Dh (in the simulations fixing S) and Fay and Wu's H. The same happens for most bottlenecks, but not for population contractions or recently finished bottlenecks where tests show power at the right tail of the distribution (except Dh and H). Only some of the statistics have been represented to make graphs clearer. Of all Fu and Li's statistics, only F* is shown, as it is the one with greatest power (although for simulations based on S, it has the same power as F). However, considering that in the contraction model D and D* are more powerful, D has been chosen for these simulations. Of Wall's B and Q, we represent only the statistic that shows more power under each circumstance, as with the EHH average and EHH maximum. We also leave ZZ out, as it is simply the difference between ZnS and ZA and therefore can be easily inferred from them. In expansions, only Te from 0.0075 to 0.2 and Te = 1 has been represented, as in general at Te = 0.4 tests have reached a power of ∼0.5.
Statistical power for different demographic events:
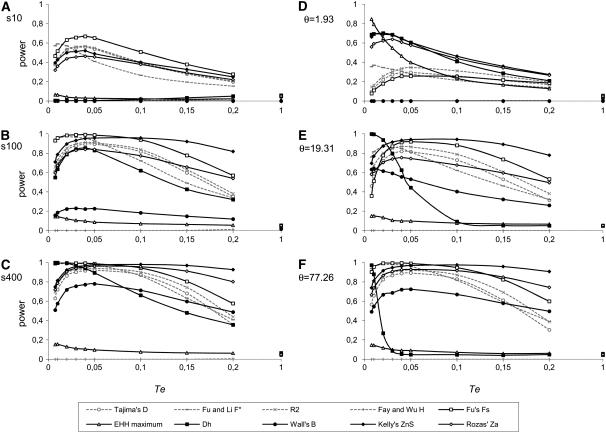
Figure 1 shows the statistical power of the tests under study in the absence of recombination and for different times since the expansion. Expansion ages range from Te = 0.0075 (300 generations) to Te = 1 (40,000 generations). Our results reproduce those from Ramos-Onsins and Rozas (2002). For all statistics, power increases with the number of segregating sites (Figure 1, A–C), especially in class II statistics, which are based on haplotype structure. In fact, as shown in Ramos-Onsins and Rozas (2002), Fu's Fs is the best-performing statistic for large sample sizes; however, ZnS and ZA statistics become more powerful when considering 100 or more segregating sites, especially to detect expansions older than Te = 0.05 (2000 human generations). This can be explained by a saturation effect, such as the saturation of the number of haplotypes due to a high number of segregating sites with respect to n (Nei 1987). Figure 1, D–F, shows the effect of increasing θ upon statistical power. Simulations based on θ show the same patterns as when conditioning on S; the main differences reside in the behavior of Dh, which does not show power at the right tail of the distribution (as we would expect; see Table 2), but rather at the left tail. Moreover, the pattern of Dh's power is also particular, as it has its maximum at very recent times and a decay that becomes more pronounced as θ increases. For very low values of θ, most genealogical trees have zero or only one mutation, which greatly affects the power of any haplotype-based statistic. In both kinds of simulations (fixing either S or θ), the lowest values (S = 10 and θ = 1.93) show power curves that are different from those obtained for a higher polymorphism level, the reason for this behavior being that low polymorphism leads to imprecise statistics.
Figure 1.—
Power of the test depending on the time elapsed since the expansion, without recombination. N = 100, De = 10. (A) S = 10. (B) S = 100. (C) S = 400. (D) θ = 1.93. (E) θ = 19.31. (F) θ = 77.26.
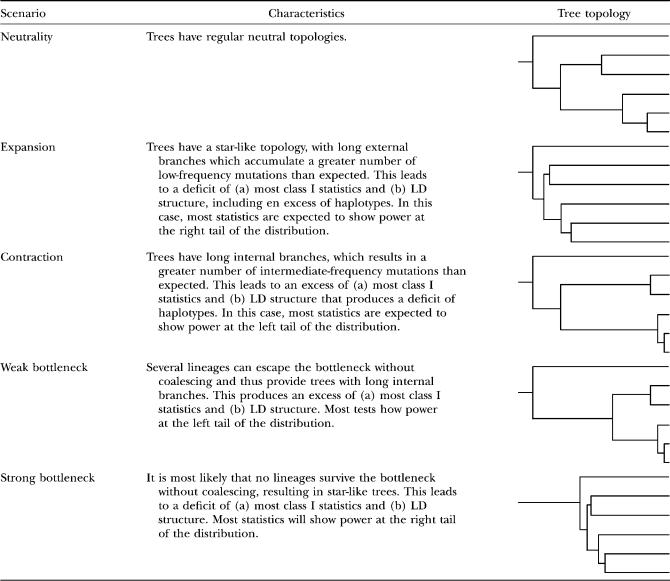
TABLE 2.
Description of tree topologies in different demographic scenarios
As for the effect of the elapsed time since the expansion, in both normal and high-polymorphism scenarios most well-performing statistics (statistics with powers that are normally >0.4, which excludes EHH estimators and Fay and Wu's H) reach their maximum power between Te = 0.04 and Te = 0.05. Exceptions to this rule are Fu and Li's tests (D, F, D*, and F*; only F* is shown in Figure 1), which have maximum power at shorter times (Te = 0.01–0.02), and ZnS, which does not reach its maximum power until Te = 0.1. Power decays with time since the expansion, but does it differently for class I and class II statistics: while class I statistics decay rapidly after reaching their maximum power, class II statistics have gentler slopes (again considering only well-performing statistics in medium and high-polymorphism scenarios). However, Fu's Fs and R2 always decay in a very similar way. At Te = 1 all statistics tend to 0.05 for simulations based on S, thus reaching the nominal type I error rate.
As expected (see Table 2), in the sudden contraction model the power of the neutrality tests behaves opposite than in expansions. The most powerful tests are Fu and Li's D* and Fs. Maximum power to detect population contractions is influenced mainly by the number of segregating sites and can be found between Tc = 0.1 (S = 100) and Tc = 0.4 (S = 10). More details can be found in the supplemental Results and supplemental Figure 1.
Bottlenecks have different outcomes depending on whether or not several lineages have survived the bottleneck stage without coalescing. Thus, the effects of severity and the age of the perturbation are nonmonotonic and tests have power at both tails of the distribution (see Table 2). For old (Tstart = 0.04–0.08), strong (b = 0.05 or stronger), and long-lasting bottlenecks (after which most lineages will have coalesced), the most powerful statistics are R2, Fs, and ZnS. However, they lack the power to detect bottlenecks that have just finished; as in this case, the fact that all sequences have coalesced during the bottleneck produces a very shallow genealogy and thus the statistics will behave similarly. On the contrary, in the case of weak, recent (Tstart = 0.02–0.04) and recently finished bottlenecks, the most powerful tests are class I tests and Fu's Fs. More details can be found in the supplemental Results and in supplemental Figure 2.
The very different genealogy shapes that can be produced by bottlenecks (Table 2) dramatically affect the variance of the power of certain tests, especially in intermediate bottlenecks, which, depending on the run, can lead to either shallow or deep genealogies. To study this effect, we have calculated the variance of the statistics for each set of bottleneck parameters. We focused on bottleneck severity and studied how it modifies variance when other parameters are equal (supplemental data E). Variance patterns are quite similar for all n and S simulated values, and in general take maximum values around severities of b = 0.05, ranging from b = 0.01 to b = 0.1. This moment of maximum variance depends mainly on the time of onset and duration of the bottleneck, approaching b = 0.01 in recent and short bottlenecks and b = 0.1 in old and long-lasting ones. Moreover, some tests are more sensitive to changes in the bottleneck parameters Tstart and Tdur. Fu and Li's tests and Fs tend to reach maximum variance values for less severe bottlenecks later than other tests (that is, for older and more long-lasting bottlenecks), while R2 and Fay and Wu's H behave in an opposite way. Variance patterns seem to correlate inversely to the power of the statistics seen for the different bottleneck severities, as tests show maximum powers for severities ≥0.05 while maximum variances correspond to severities ≤0.05. This could be due to the fact that less severe bottlenecks leave a much weaker signature.
Recombination:
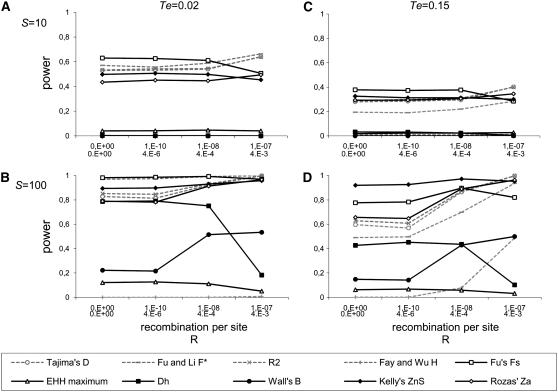
Figure 2 shows the effects of recombination rate on statistical power in population expansions. While for S = 10 recombination has hardly any effect, for higher values of S the power increases dramatically between low recombination levels (0 and 10−10) and high recombination levels (10−8 and 10−7). For low recombination values (Figure 2), in most cases power is not affected, although under some circumstances (e.g., Fu and Li's D and D* for S = 100; data not shown) power can be lower for r = 10−10. In contrast, most statistics improve their power under high recombination, with the exceptions of Dh, EHH, and, for ancient expansions, of Fs and ZnS, which have a tendency to decrease their power in the interval between r = 10−8 to r = 10−7. It is noteworthy that, for ancient expansions and high recombination values, Fay and Wu's H shows increased power, reaching values >0.9 for S = 400 (results not shown). Simulations based on θ show similar patterns to those based on S (results not shown). As for population contractions and bottlenecks, the changes in the power of the tests in the presence of recombination are very similar to those found in expansions (more details in the supplemental Results and supplemental Figures 3 and 4, respectively).
Figure 2.—
Power of the test in the expansion model depending on recombination rates. N = 100, De = 10. (A) S = 10, Te = 0.02. (B) S = 100, Te = 0.02. (C) S = 10, Te = 0.15. (D) S = 100, Te = 0.15.
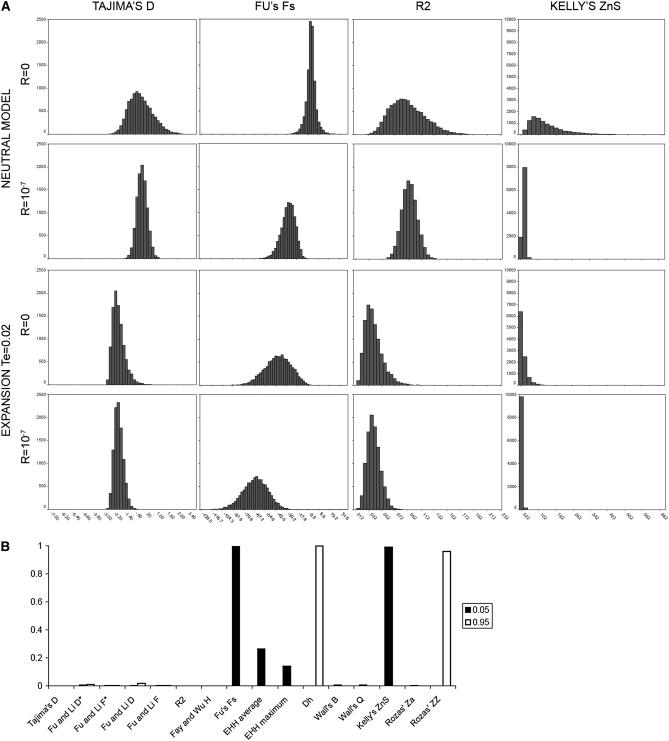
Recombination shuffles nucleotide variation, increasing the number of haplotypes through the creation of new recombinant ones. Thus, it is expected to strongly affect the mean of haplotype-based tests and also to reduce the variance of both kinds of statistics (Figure 3A). Changes in the power curve reflected in Figure 2 may therefore be due to two nonmutually exclusive factors: (i) an actual increase in the amount of information in the sequence that detects population expansions when recombination is acting and (ii) recombination shifting the distribution of neutrality statistics. To assess the relative weights of these two explanations, we have compared the constant-population model without recombination with the constant-population model with high recombination. As shown in Figure 3B (see also supplemental data F), Fs and ZnS are very sensitive to recombination (left tail of the distribution), while Dh and ZZ are sensitive at the right tail. Both EHH estimators are also able to detect recombination (left tail) but with less power (<0.3). Thus, these statistics are liberal for detecting a population expansion acting on a recombining sequence and may often produce false positives.
Figure 3.—
(A) distribution of the values of Tajima's D, Fu and Li's F, R2, and Kelly's ZnS under the neutral model and the expansion model at Te = 0.02 without recombination and with r = 10−7. (B) Power of the tests to detect recombination in the null model for the right and left tails of the distribution. (A and B) S = 100, n = 100.
Unknown or misspecified recombination:
Under some circumstances, recombination rates cannot be taken into account when testing for demographic or selective events. Indeed, the default option in Arlequin 3.0 (Excoffier et al. 2005) or DnaSP (Rozas et al. 2003), the two standard software packages for genetic analysis, is to compute the significance of neutrality tests without recombination, although DnaSP will also produce null distributions with any population recombination rate supplied by the user. When estimates of recombination rates are available, it is important to consider that they may be over- or underestimates. For example, Kong et al. (2002) measured recombination rates in human pedigrees at intervals of median length ∼350 kb. Considering that it has been estimated that there is a hotspot every 50 kb (Myers et al. 2005), such intervals will most likely contain recombination hotspots, and therefore the recombination rate estimated for the whole interval will be much greater than the real one for most parts of the region. On the other hand, hotspots may also result in a recombination reaching saturation and thus to an underestimate of the average recombination rate in the region.
To investigate the potential errors caused by over- or underestimation of recombination, we compared the statistical power of tests when true recombination values were assumed vs. the power of the same tests assuming erroneous rates. Figure 4A shows the difference between the apparent power of a test (that is, comparing the constant-size null hypothesis without recombination with the population-expansion alternative hypothesis using the true recombination value) and its real power (that is, comparing the null hypothesis and the alternative hypotheses using the true recombination value). When actual recombination rates are small (10−10) and thus underestimation is not too serious (e.g., using as a null hypothesis the constant nonrecombining model when the actual rate is r = 10−10), the true and the apparent statistical power are not appreciably different. However, for larger underestimates (when the real rates are r = 10−8 or 10−7), class I statistical tests become conservative (with an increase in type II error) whereas some class II statistics (Dh, Fu's Fs, EHH, and ZnS) become liberal (with an increase in type I error). In population contractions, class I statistics behave as in expansions, while most class II statistics are liberal. As expected, strong bottlenecks tend to behave as expansions while weak ones tend to behave as contractions (see Table 2; more details are in the supplemental Results and supplemental Figures 5 and 6 for contractions and bottlenecks, respectively).
Figure 4.—
Error made by statistics when testing for expansions when recombination is under- or overestimated in the null model. S = 100, n = 100, De = 10, Te = 0.15. The cartoon shows how this error is calculated: first, we calculated the real power of the statistic, comparing the null hypothesis with the alternative hypothesis with the same recombination values. Afterward, we calculated the probability of rejecting the null hypothesis when recombination has been erroneously estimated, that is, when the recombination rate used to generate the null hypothesis is different from that of the alternative hypothesis. The error made is the latter (apparent power) minus the real power. (A) The apparent power of the null hypothesis was produced without recombination. (B) The apparent power of the null hypothesis has a recombination rate of 10−7.
Figure 4B shows the difference between the apparent power of the tests when assuming an overestimated recombination of 10−7 on the constant-size null hypothesis and their true power (that is, using true recombination values to compare the null hypothesis and alternative hypotheses). The effect of recombination overestimation shows the opposite pattern, and therefore all tests are liberal with the exception of Dh, Fu's Fs, EHH, and ZnS, which become conservative. The difference in behavior between these class II and class I statistics (Figure 3) can be due to the fact that the former are highly dependent on the actual recombination rates (Wall 2000). As in the case of underestimation of recombination, when recombination is overestimated in a scenario of population contraction, class I tests behave similarly than in expansions. This is not the case for class II tests, which became liberal. Again, strong bottlenecks behave similarly to expansions while weak ones behave more like contractions (Table 2; more details are in the supplemental Results and supplemental Figures 5 and 6 for contractions and bottlenecks, respectively).
DISCUSSION
Power of neutrality tests:
We have examined statistical power in detecting a sudden population expansion, a sudden contraction, or a bottleneck analyzing DNA polymorphism data by means of a wide range of statistics. The most powerful tests are those belonging to class II, that is, those based on haplotype frequencies. Within those, Fu's Fs and ZnS perform best although, for small sample sizes, R2 is also recommended. Class II tests, however, are strongly affected by recombination, particularly Fs and ZnS, which not only lose power under high recombination rates, but also become significant with recombination events when no expansion has taken place and the population has remained constant. For this reason, when recombination is suspected to be acting on a sample, and especially if there is risk of over- or underestimating it, it is not advisable to use class II tests and, thus, R2, Tajima's D or Fu and Li's tests should be used instead. A similar situation, with class II statistics being more powerful than class I, can be found for contractions, although tests have power at the opposite tail of the distribution and they start to perform well at larger times and for longer S. However, in the case of bottlenecks, class I tests are best as a general rule, with the exception of Fs and ZnS in some particular cases.
Combinations of the different tests and the tail at which they show power can therefore provide a rough idea about the demographic event acting over a population and about the time and strength of this event, as well as about other factors such as recombination.
The effect of recombination on the power of the tests:
As discussed above, recombination is expected to have some effects on the power of statistical tests based on the allele frequency spectrum and to reduce the power of tests that rely on haplotype-based statistics (see, for example, Quesada et al. 2006 and references therein). We performed a detailed series of simulations suggesting that high recombination rates generally improve the power of most statistical tests, although some class II tests (Dh, EHH, Fs, and ZnS) lose power for the maximum recombination rates considered in this study, especially for old expansions. Furthermore, even when recombination is considered in the null constant-population models, those tests have great power to “detect recombination” in a sample. This efficiency in recognizing recombination can be easily explained as recombination modifies the mean and reduces the width of the distribution of class II statistics. Therefore, careful attention should be paid to the interpretation of those tests when recombination is suspected to have shaped the genealogy of the sampled sequences. We have also examined the effect of using mistaken recombination rates, because their estimation can be a problem for most organisms. In the case of humans, actual recombination maps made through the genotyping of 5136 microsatellite markers for 146 families, with a total of 1257 meiotic events, are available at an ∼350-kb resolution (Kong et al. 2002). Beyond that level, current efforts are aimed at pinpointing hotspots and recombination deserts as inferred from linkage disequilibrium patterns (McVean et al. 2004; Fearnhead and Smith 2005; Myers et al. 2005). Wall (1999) observed that larger sequence sizes with high recombination decreased the power of tests and suggested that this was due to the difference between the recombination rates of the null and the alternative hypotheses. Our results show that severe over- or underestimations of recombination have large impacts on power. When recombination is underestimated, the power of tests decreases in expansion and bottlenecks, supporting the results in Wall (1999). In contrast, when recombination is overestimated, type I errors increase, leading to a spurious gain in the power of the tests. Since class II ZnS, Fu's Fs, Dh, and EHH statistics are very sensitive to recombination, they have an opposite behavior. Considering all recombination results together, a conservative recombination rate should be used in the null hypothesis when using class II statistics—if recombination could be acting over the sample, for example, it would be recommended to use a lower bound or no recombination if there is a deficit of haplotypes or an upper bound if there is an excess.
Recommendations for test usage arising from our results are summarized in Tables 3–5.We hope that they help to produce guidelines for a rational choice among the wide variety of neutrality tests available.
TABLE 3.
Sudden expansion model decision table
| No recombination
|
Recombination (108)
|
|||
|---|---|---|---|---|
| Te | Strong (De = 0.01) | Weak (De = 0.1) | Strong (De = 0.01) | Weak (De = 0.1) |
| Small S (≈10), small n (≈20) | ||||
| ≤0.01 | Fs, R2, D* | Fs, R2, ZnS, D* | R2, F, D*, F* | R2 |
| 0.01–0.15 | R2, Fs | Fs, R2, ZnS | R2 | R2 |
| ≥0.15 | Fs, R2, ZnS | Fs, R2, ZnS | R2 | R2 |
| Small S (≈10), large n (≈100) | ||||
| ≤0.01 | Fs, R2, D, F, F* | F and F* | R2, D, F, D*, F* | DF |
| 0.01–0.05 | Fs, R2, D | Fs | R2, D | DF |
| ≥0.05 | Fs | Fs | R2, D | DF |
| Large S (≈100 or more), small n (≈20) | ||||
| ≤0.01 | Fs, R2, D, DF, F, D*, F*, ZnS, ZA, Dha | Fs, Dha | R2, D, DF, F, D*, F* | R2 |
| 0.01–0.03 | Fs, R2, D, DF, F, D*, F*, ZnS, ZA | Fs | R2, D, DF, F, D*, F* | R2 |
| 0.03–0.05 | R2, D, ZnS | R2, ZnS | R2, D, DF, F, D*, F* | R2 |
| >0.05 | ZnS | ZnS | R2 | R2 |
| Large S (≈100 or more), large n (≈100) | ||||
| ≤0.02 | Fs, R2, D, DF, F, D*, F*, ZnS, ZA | Fs, DF, F, D*, and F* | R2, D, DF, F, D*, F* | DF, F, D*, F* |
| 0.02–0.05 | Fs, R2, D, DF, F, D*, F*, ZnS | Fs | R2, D, DF, F, D*, F* | DF, F, D*, F* |
| 0.02–0.15 | Fs, R2, D, ZnS | Fs, ZnS | R2, D | R2, D |
| ≥0.15 | ZnS | ZnS | R2, D | R2, D |
Power is at the left tail of the distribution unless otherwise indicated. DF, Fu and Li's D.
Power is at the right tail of the distribution.
TABLE 4.
Sudden contraction model decision table
| No recombination
|
Recombination (108)
|
|||
|---|---|---|---|---|
| Tc | Strong (Dc = 0.01) | Weak (Dc = 0.1) | Strong (Dc = 0.01) | Weak (Dc = 0.1) |
| Small S (≈10), small n (≈20) | ||||
| ≤0.10 | D* | D* | D* | D* |
| 0.10–0.40 | Fs, D* | Fs, D* | D* | D* |
| 0.40–0.80 | Fs, ZnS, ZA | Fs, ZnS, ZA | D* | D* |
| >0.80 | Fs, ZnS, ZA, B, Q | Fs, ZnS, ZA, B, Q, Dha | D* | D* |
| Small S (≈10), large n (≈100) | ||||
| ≤0.20 | DF | DF | DF | DF |
| 0.20–0.60 | ZnS | ZnS, B | DF | DF |
| ≥0.60 | ZnS, ZA, B, Q, Fs, DF | ZnS, ZA, B, Q, Fs, DF | DF | DF |
| Large S (≈100 or more), small n (≈20) | ||||
| ≤0.10 | Fs, Dha | Fs, Dha | DF | DF |
| 0.10–0.40 | Fs, Dha | Fs, Dha | DF, F, F* | DF, F, F* |
| ≥0.40 | Dha | Dha | DF, D* | D* |
| Large S (≈100 or more), large n (≈100) | ||||
| ≤0.02 | DF, D* | DF, D* | DF, D* | DF, D* |
| 0.02–0.10 | Fs | Fs | F, F* | F, F* |
| 0.10–0.40 | Fs, Dha | Fs | F, F*, R2, D | F, F* |
| 0.40–0.80 | Fs, ZnS, ZA, B, Q, Dha | ZnS, ZA | DF, D* | DF |
| >0.80 | Fs, DF, D*, ZnS, ZA, B, Q | ZnS, ZA, B, Q | DF, D* | F, F* |
Power is at the right tail of the distribution unless otherwise indicated. DF, Fu and Li's D.
Power is at the left tail of the distribution.
TABLE 5.
Bottleneck decision table
| No recombination
|
Recombination (r8)
|
|||
|---|---|---|---|---|
| Tstart | Strong (b = 0.01) | Weak (b = 0.1) | Strong (b = 0.01) | Weak (b = 0.1) |
| Small S (≈10), small n (≈20) | ||||
| 0.02 | D, F, D*, F*, R2, Fs | D*a, Fsa | Da, Fa, D*a, F*a, R2a | Da, Fa, D*a, F*a, R2a |
| 0.04 | D, R2, Fs | Fsa, ZnSa, ZAa | Da, R2a | Da, Fa, D*a, F*a, R2a |
| 0.08–0.12 | R2, Fs | D, D*, F*, R2, Fs, ZnS | R2 | D, R2 |
| Small S (≈10), large n (≈100) | ||||
| 0.02 | D, F, F*, R2, Fs, Dh | Da, DFa, Fsa | D*, Dh | Da, DFa, R2a |
| 0.04 | D, R2, Fs | Fsa, Ba, ZnSa, ZAa | D, D*, R2 | Da, DFab, R2a |
| 0.08 | D, R2, Fs | D, F, R2, Fs, Dh | D, R2 | DF |
| 0.12 | Fs | D, R2, Fs | D, R2 | DF |
| Large S (≈100 or more), small n (≈20) | ||||
| 0.02 | D, DF, F, D*, F*, R2, Fs | D*a, Fsa | Da, DFa, Fa, D*a, F*a, R2a | DFa, Fa, D*a, F*a |
| 0.04 | D, DF, F, D*, F*, R2, Fs, ZnS, ZA | Fsab, Ba, Qa, ZnSa, ZAa | Da, DFa, Fa, D*a, F*a, R2a | Da, DFa, Fa, F*a, R2a |
| 0.08 | D, R2, ZnS, ZA | R2, Fs | D, DF, F, D*, F*, R2 | D, R2 |
| 0.12 | R2, ZnS | R2 | D, R2 | D, R2 |
| Large S (≈100 or more), large n (≈100) | ||||
| 0.02 | D, F, D*, F*, R2 | D*a, Fsa | DFa, D*a | Da, DFab, Fab, D*ab, F*ab, R2a |
| 0.04 | D, DF, F, D*, F*, R2, Fs, ZnS, ZA | Fsa, Ba, Qa, ZnSa, ZAa | Da, DFa, Fa, D*a, F*a, R2a | Da, DFab, Fab, D*ab, F*ab, R2a |
| 0.08 | D, DF, F, D*, F*, R2, ZnS, ZA | Fs | D, DF, F, D*, F*, R2 | DF, F, D*, F*, R2 |
| 0.12 | D, R2, ZnS | Fs | D, F, F*, R2 | D, F, F*, R2 |
Power is at the left tail of the distribution unless otherwise indicated. DF, Fu and Li's D.
Power is at the right tail of the distribution.
Only for finished bottlenecks.
Acknowledgments
We thank J. Alegre and R. Sangrós for their helpful advice and support in programming, Tomàs Marquès-Bonet for his comments, and G. Berniell for her revision of the manuscript. This research was funded by grant BFU2006-15413-C02-01 to A.N. and BFU2004-02002 to F.C. from the Spanish Ministry of Science and Technology, by the Genoma España/Genome Canada joint R+D+I projects, and by the National Institute of Bioinformatics (http://www.inab.org), a platform of Genoma España. S.E.R-O. acknowledges the support of Generalitat de Catalunya (Distinció per la Promoció de la Recerca Universitària) to Montserrat Aguadé.
References
- Charlesworth, B., M. T. Morgan and D. Charlesworth, 1993. The effect of deleterious mutations on neutral molecular variation. Genetics 134 1289–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford, D. C., D. T. Akey and D. A. Nickerson, 2005. The patterns of natural variation in human genes. Annu. Rev. Genomics Hum. Genet. 6 287–312. [DOI] [PubMed] [Google Scholar]
- Depaulis, F., S. Mousset and M. Veuille, 2001. Haplotype tests using coalescent simulations conditional on the number of segregating sites. Mol. Biol. Evol. 18 1136–1138. [DOI] [PubMed] [Google Scholar]
- Depaulis, F., S. Mousset and M. Veuille, 2003. Power of neutrality tests to detect bottlenecks and hitchhiking. J. Mol. Evol. 57(Suppl. 1): 190–200. [DOI] [PubMed] [Google Scholar]
- Depaulis, F., S. Mousset and M. Veuille, 2005. Detecting selective sweeps with haplotype tests, pp. 34–54 in Selective Sweep, edited by D. Nurminsky. Landes Bioscience, Georgetown, TX.
- Donnelly, P., and S. Tavare, 1995. Coalescents and genealogical structure under neutrality. Annu. Rev. Genet. 29 401–421. [DOI] [PubMed] [Google Scholar]
- Excoffier, L., G. Laval and S. Schneider, 2005. Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol. Bioinform. Online 1 47–50. [PMC free article] [PubMed] [Google Scholar]
- Fay, J. C., and C. I. Wu, 2000. Hitchhiking under positive Darwinian selection. Genetics 155 1405–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearnhead, P., and N. G. Smith, 2005. A novel method with improved power to detect recombination hotspots from polymorphism data reveals multiple hotspots in human genes. Am. J. Hum. Genet. 77 781–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher, R. A., 1930. The Genetical Theory of Natural Selection. Clarendon Press, Oxford.
- Fu, Y. X., 1996. New statistical tests of neutrality for DNA samples from a population. Genetics 143 557–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, Y. X., 1997. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147 915–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, Y. X., and W. H. Li, 1993. Statistical tests of neutrality of mutations. Genetics 133 693–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, Y. X., and W. H. Li, 1999. Coalescing into the 21st century: an overview and prospects of coalescent theory. Theor. Popul. Biol. 56 1–10. [DOI] [PubMed] [Google Scholar]
- Haddrill, P. R., K. R. Thornton, B. Charlesworth and P. Andolfatto, 2005. Multilocus patterns of nucleotide variability and the demographic and selection history of Drosophila melanogaster populations. Genome Res. 15 790–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein, J., M. H. Schierup and C. Wiuf, 2005. Gene Genealogies, Variation and Evolution: A Primer in Coalescent Theory. Oxford University Press, London/New York/Oxford.
- Hudson, R. R., 1990. Gene genealogies and the coalescent process, pp. 1–44 in Oxford Surveys in Evolutionary Biology, edited by J. Antonovics and D. Futuyama. Oxford University Press, Oxford.
- Hudson, R. R., 1993. The how and why of generating gene genealogies, pp. 23–36 in Mechanisms of Molecular Evolution, edited by N. Takahata and A. G. Clark. Sinauer Associates, Sunderland, MA.
- Hudson, R. R., 2002. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics 18 337–338. [DOI] [PubMed] [Google Scholar]
- Jensen, J. D., Y. Kim, V. B. DuMont, C. F. Aquadro and C. D. Bustamante, 2005. Distinguishing between selective sweeps and demography using DNA polymorphism data. Genetics 170 1401–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobling, M. A., M. E. Hurles and C. Tyler-Smith, 2004. Human Evolutionary Genetics: Origins, Peoples and Disease. Garland Science, New York.
- Kelly, J. K., 1997. A test of neutrality based on interlocus associations. Genetics 146 1197–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, M., 1968. Genetic variability maintained in a finite population due to mutational production of neutral and nearly neutral isoalleles. Genet. Res. 11 247–269. [DOI] [PubMed] [Google Scholar]
- Kingman, J. F. C., 1982. a On the genealogy of large populations. J. Appl. Probab. 19A 27–43. [Google Scholar]
- Kingman, J. F. C., 1982. b The coalescent. Stoch. Proc. Appl. 13 235–248. [Google Scholar]
- Kingman, J. F., 2000. Origins of the coalescent: 1974–1982. Genetics 156 1461–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong, A., D. F. Gudbjartsson, J. Sainz, G. M. Jonsdottir, S. A. Gudjonsson et al., 2002. A high-resolution recombination map of the human genome. Nat. Genet. 31 241–247. [DOI] [PubMed] [Google Scholar]
- Macdonald, S. J., and A. D. Long, 2005. Identifying signatures of selection at the enhancer of split neurogenic gene complex in Drosophila. Mol. Biol. Evol. 22 607–619. [DOI] [PubMed] [Google Scholar]
- McVean, G. A. T., S. R. Myers, S. Hunt, P. Deloukas, D. R. Bentley et al., 2004. The fine-scale structure of recombination rate variation in the human genome. Science 304 581–584. [DOI] [PubMed] [Google Scholar]
- Myers, S., L. Bottolo, C. Freeman, G. McVean and P. Donnelly, 2005. A fine-scale map of recombination rates and hotspots across the human genome. Science 310 321–324. [DOI] [PubMed] [Google Scholar]
- Nei, M., 1987. Molecular Evolutionary Genetics. Columbia University Press, New York.
- Pritchard, J. K., M. T. Seielstad, A. Perez-Lezaun and M. W. Feldman, 1999. Population growth of human Y chromosomes: a study of Y chromosome microsatellites. Mol. Biol. Evol. 16 1791–1798. [DOI] [PubMed] [Google Scholar]
- Przeworski, M., 2002. The signature of positive selection at randomly chosen loci. Genetics 160 1179–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada, H., S. E. Ramos-Onsins, J. Rozas and M. Aguadé, 2006. Positive selection versus demography: evolutionary inferences based on an unusual haplotype structure in Drosophila simulans. Mol. Biol. Evol. 23 1643–1647. [DOI] [PubMed] [Google Scholar]
- Ramos-Onsins, S. E., and J. Rozas, 2002. Statistical properties of new neutrality tests against population growth. Mol. Biol. Evol. 19 2092–2100. [DOI] [PubMed] [Google Scholar]
- Ramos-Onsins, S. E., S. Mousset, T. Mitchell-Olds and W. Stephan, 2007. Population genetic inference using a fixed number of segregating sites: a reassessment. Genet. Res. 89 231–244. [DOI] [PubMed] [Google Scholar]
- Rogers, A. R., and H. Harpending, 1992. Population growth makes waves in the distribution of pairwise genetic differences. Mol. Biol. Evol. 9 552–569. [DOI] [PubMed] [Google Scholar]
- Rozas, J., M. Gullaud, G. Blandin and M. Aguade, 2001. DNA variation at the rp49 gene region of Drosophila simulans: evolutionary inferences from an unusual haplotype structure. Genetics 158 1147–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas, J., J. C. Sanchez-DelBarrio, X. Messeguer and R. Rozas, 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19 2496–2497. [DOI] [PubMed] [Google Scholar]
- Sabeti, P. C., D. E. Reich, J. M. Higgins, H. Z. Levine, D. J. Richter et al., 2002. Detecting recent positive selection in the human genome from haplotype structure. Nature 419 832–837. [DOI] [PubMed] [Google Scholar]
- Sano, A., and H. Tachida, 2005. Gene genealogy and properties of test statistics of neutrality under population growth. Genetics 169 1687–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schierup, M. H., and J. Hein, 2000. Consequences of recombination on traditional phylogenetic analysis. Genetics 156 879–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soejima, M., H. Tachida, M. Tsuneoka, O. Takenaka, H. Kimura et al., 2005. Nucleotide sequence analyses of human complement 6 (C6) gene suggest balancing selection. Annu. Hum. Genet. 69 239–252. [DOI] [PubMed] [Google Scholar]
- Stajich, J. E., and M. W. Hahn, 2005. Disentangling the effects of demography and selection in human history. Mol. Biol. Evol. 22 63–73. [DOI] [PubMed] [Google Scholar]
- Tajima, F., 1989. a Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima, F., 1989. b The effect of change in population size on DNA polymorphism. Genetics 123 597–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata, N., Y. Satta and J. Klein, 1995. Divergence time and population size in the lineage leading to modern humans. Theor. Popul. Biol. 48 198–221. [DOI] [PubMed] [Google Scholar]
- Tarazona-Santos, E., and S. A. Tishkoff, 2005. Divergent patterns of linkage disequilibrium and haplotype structure across global populations at the interleukin-13 (IL13) locus. Genes Immun. 6 53–65. [DOI] [PubMed] [Google Scholar]
- Tavare, S., D. J. Balding, R. C. Griffiths and P. Donnelly, 1997. Inferring coalescence times from DNA sequence data. Genetics 145 505–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voight, B. F., A. M. Adams, L. A. Frisse, Y. Qian, R. R. Hudson et al., 2005. Interrogating multiple aspects of variation in a full resequencing data set to infer human population size changes. Proc. Natl. Acad. Sci. USA 102 18508–18513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall, J. D., 1999. Recombination and the power of statistical tests of neutrality. Genet. Res. 74 65–79. [Google Scholar]
- Wall, J. D., 2000. A comparison of estimators of the population recombination rate. Mol. Biol. Evol. 17 156–163. [DOI] [PubMed] [Google Scholar]
- Wall, J. D., and R. R. Hudson, 2001. Coalescent simulations and statistical tests of neutrality. Mol. Biol. Evol. 18 1134–1135. [DOI] [PubMed] [Google Scholar]
- Watterson, G. A., 1975. On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 7 256–276. [DOI] [PubMed] [Google Scholar]
- Williamson, S. H., R. Hernandez, A. Fledel-Alon, L. Zhu, R. Nielsen et al., 2005. Simultaneous inference of selection and population growth from patterns of variation in the human genome. Proc. Natl. Acad. Sci. USA 102 7882–7887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, S., 1931. Evolution in Mendelian populations. Genetics 16 97–159. [DOI] [PMC free article] [PubMed] [Google Scholar]