Abstract
The unicellular green alga Chlamydomonas reinhardtii is used extensively as a model to study eukaryotic photosynthesis, flagellar functions, and more recently the production of hydrogen as biofuel. Two of these processes, photosynthesis and hydrogen production, are highly dependent on iron–sulfur (Fe–S) enzymes. To understand how Fe–S proteins are assembled in Chlamydomonas, we have analyzed its recently sequenced genome for orthologs of genes involved in Fe–S cluster assembly. We found a total of 32 open reading frames, most single copies, that are thought to constitute a mitochondrial assembly pathway, mitochondrial export machinery, a cytosolic assembly pathway, and components for Fe–S cluster assembly in the chloroplast. The chloroplast proteins are also expected to play a role in the assembly of the H-cluster in [FeFe]-hydrogenases, together with the recently identified HydEF and HydG proteins. Comparison with the higher plant model Arabidopsis indicated a strong degree of conservation of Fe–S cofactor assembly pathways in the green lineage, the pathways being derived from different origins during the evolution of the photosynthetic eukaryote. As a haploid, unicellular organism with available forward and reverse genetic tools, Chlamydomonas provides an excellent model system to study Fe–S cluster assembly and its regulation in photosynthetic eukaryotes.
IRON–SULFUR (Fe–S) clusters are versatile cofactors of enzymes involved in essential biological processes including photosynthesis, respiration, and metabolism (Imsande 1999; Beinert 2000). Chlamydomonas is estimated to have at least 30 enzymes that depend on Fe–S as a cofactor. This number is based on the available literature and Chlamydomonas orthologs of Fe–S proteins identified in plants and metazoa (Balk and Lobréaux 2005; Lill and Mühlenhoff 2008; and annotations in the Chlamydomonas genome). To briefly summarize, the thylakoid membranes of the chloroplast harbor components of photosynthetic electron transfer, including the [2Fe–2S] Rieske protein in the cytochrome b6f complex and three [4Fe–4S] clusters in Photosystem I (PSI). The electrons are then passed onto [2Fe–2S] ferredoxin and ferredoxin-thioredoxin reductase, which has a [4Fe–4S] cluster. In contrast to cyanobacteria and higher plants, the NDH complex, thought to contain three clusters, is missing in Chlamydomonas (Peltier and Cournac 2002). A possible role of the NDH complex in siphoning off electrons from PSI to avoid redox damage could be equivalent to the role of the soluble [FeFe]-hydrogenases HydA1 and HydA2 in Chlamydomonas, which contain a specialized Fe–S cluster, [4Fe–4S] + [FeFe], called the H-cluster (Peters et al. 1998). The chloroplast stroma also contains numerous catalytic Fe–S enzymes involved in nitrogen and sulfur assimilation (nitrite reductase, sulfite reductase), amino acid biosynthesis (glutamate synthase, isopropylmalate isomerase), and cofactor biosynthesis (chlorophyll a/b oxygenase, the thiamin biosynthetic protein THIC). Mitochondria are the other cell compartment in which Fe–S proteins are abundant, such as aconitase and the respiratory complexes, especially respiratory complex I with eight Fe–S clusters. In the cytosol and nucleus, Fe–S proteins are less abundant, but nevertheless essential for transcription, translation, and DNA repair (Lill and Mühlenhoff 2008). Several novel Fe–S proteins have recently been discovered in yeast and mammals, including the ABC protein Rli1, DNA primase, and the helicase Rad3 (FancJ), and are also found in the Chlamydomonas genome. More Fe–S proteins are doubtlessly waiting to be discovered, as Fe–S binding cannot always be predicted from the primary amino acid sequence.
The importance of Fe–S clusters for photosynthesis stimulated early studies in the assembly of these metal cofactors in chloroplasts using nuclear-encoded, imported ferredoxin as a model protein (Takahashi et al. 1986; Li et al. 1990). By adding 35S-labeled cysteine to lysed spinach chloroplasts, it was shown that cysteine is the source of sulfur for the [2Fe–2S] cluster of ferredoxin (Takahashi et al. 1991b). Moreover, the process required energy in the form of ATP or GTP, and reducing equivalents in the form of NADPH (Takahashi et al. 1991a). However, it was not until the identification of a cysteine desulphurase activity and its corresponding gene in the nitrogen fixing Azotobacter vinelandii (Zheng et al. 1993) that the genetic components and therefore the biochemical pathways underlying Fe–S cluster assembly started to become unraveled, primarily in Escherichia coli and Baker's yeast Saccharomyces cerevisiae (Johnson et al. 2005; Lill and Mühlenhoff 2008).
The building blocks for most Fe–S clusters are simply iron and sulfide (Beinert 2000). Intracellular free iron and sulfide concentrations are thought to be extremely low or zero, respectively, as both are toxic to the cell. In all Fe–S cluster assembly pathways studied to date, sulfide is provided by a pyridoxal phosphate-dependent cysteine desulfurase enzyme, which catalyzes the transfer of sulfur from l-cysteine in the form of persulfide to a scaffold protein (Mihara and Esaki 2002). How iron is delivered to the scaffold is less clear, but it is thought to involve the frataxin protein in nonphotosynthetic bacteria and mitochondria. After assembly on the scaffold protein, the nascent Fe–S cluster is transferred to the target apo-protein, which is facilitated by Hsp70-type chaperones and/or glutaredoxins.
In bacteria, three systems have been identified for Fe–S cluster assembly, the nitrogen fixation (NIF), the iron sulfur cluster (ISC), and the sulfur mobilization (SUF) systems (Johnson et al. 2005). While the NIF system is dedicated to the assembly of the intricate Fe–S cofactors of nitrogenase, the ISC system is regarded as the “housekeeping” pathway of Fe–S cluster assembly. In eukaryotes, the ISC components are found predominantly in the mitochondria, most likely because this system functions better under low-oxygen conditions that exist inside this organelle. The SUF system appears to function under conditions of oxidative stress or iron limitations (Nachin et al. 2003; Outten et al. 2004). It is therefore not surprising that the SUF system is found in Arabidopsis chloroplasts (Ye et al. 2006b), the cell compartment in which oxygenic photosynthesis takes place.
The mitochondria also play a part in the assembly of cytosolic and nuclear Fe–S proteins, as demonstrated in yeast (Kispal et al. 1999), mammalian cell lines (reviewed in Lill and Mühlenhoff 2008), and Arabidopsis (J. Balk, unpublished data). The half ABC transporter Atm1 in yeast (ABCB7 in mammals) is thought to export an as yet unknown product of the ISC machinery that is required for the cytosolic Fe–S protein assembly machinery (CIA) consisting of at least four proteins, including the P-loop NTPases Nbp35 and Cfd1, the hydrogenase-like protein Nar1, and the WD40 protein Cia1 (Lill and Mühlenhoff 2008).
The Fe–S cluster assembly proteins have not only attracted the interest of biochemists, but also that of evolutionary biologists. Fe–S clusters are thought to be ancient catalysts, as reduced iron and sulfur were abundant when life emerged. Fe–S clusters are endowed with unique chemical properties required for processes that are crucial to life on earth, such as photosynthesis and nitrogen fixation. Characterization of Fe–S assembly genes in “primitive” eukaryotes has helped to identify “degenerate” mitochondria and has contributed to reshuffling these organisms in the phylogenetic tree (Van Der Giezen and Tovar 2005; Embley and Martin 2006).
The genes involved in Fe–S protein biogenesis in photosynthetic eukaryotes have thus far primarily been characterized in Arabidopsis thaliana. Localization studies, reverse genetics and recombinant protein techniques have enabled the construction of a working model in which Fe–S cluster assembly is highly compartmentalized in the mitochondria, plastids, and cytosol (for reviews, see Balk and Lobréaux 2005; Kessler and Papenbrock 2005; Ye et al. 2006b). Despite the extensive genetic resources and tools developed for Arabidopsis, the study of basic biochemical pathways is hampered by additional levels of complexity in multicellular organisms with highly specialized tissues. A single-celled photosynthetic organism like Chlamydomonas would clearly overcome these limitations and would have the additional benefit of fast-forward genetics. Indeed, Chlamydomonas has already been used successfully to find novel genes involved in Fe–S protein assembly, as demonstrated by the identification of the [FeFe]-hydrogenase maturation proteins HydEF and HydG (Posewitz et al. 2004). As these genes are only the tip of a specialized pathway, we have searched the recently sequenced Chlamydomonas genome (Merchant et al. 2007) for orthologs of Fe–S protein assembly genes known in bacteria, yeast, and Arabidopsis. We have identified a near-complete set of genes with similarity to the bacterial ISC and SUF genes, the cyanobacterial/plastid NFUs and HCF101, as well as the mitochondrial export and cytosolic cluster assembly genes found in yeast. Comparison with Arabidopsis showed that the assembly pathways are highly conserved in the green lineage, but that interesting differences exist providing insights into the evolution of photosynthetic eukaryotes. The results of this analysis form an important basis for future studies on the assembly of Fe–S proteins including PSI and [FeFe]-hydrogenases.
RESULTS AND DISCUSSION
To analyze which pathways of Fe–S protein assembly exist in Chlamydomonas, we first searched the genome for the basic components for Fe–S cluster assembly, cysteine desulfurases (Table 1), and Fe–S scaffold proteins (Figure 1). We also searched for orthologs of other known or suspected Fe–S assembly proteins identified in E. coli, S. cerevisiae, A. thaliana, and Synechocystis. We then tried to assign each protein to a cell compartment on the basis of (i) experimentally confirmed localization of the eukaryotic ortholog and (ii) the presence of an N-terminal targeting sequence and prediction software. The results are summarized in Table 2 and detailed below. For further information on the individual proteins, please see reviews (Balk and Lobréaux 2005; Barras et al. 2005; Johnson et al. 2005; Lill and Mühlenhoff 2008).
TABLE 1.
Cysteine desulfurases in Chlamydomonas
| Chlamydomonas model | EST evidence | Sequence in active site | Similar to | Enzyme | Organism | Group |
|---|---|---|---|---|---|---|
| NFS1 | Yes | SSGSACTS | SSGSACTS | NifS | Azotobacter vinelandii | I |
| SSGSACTS | IscS | Escherichia coli | I | |||
| SSGSACTS | NFS1 | Saccharomyces cerevisiae | I | |||
| SSGSACTS | NFS1 | Arabidopsis thaliana | I | |||
| SSGSACSS | Slr0387 | Synechocystis | I | |||
| CSD2 | No | SAGAACHS | SAGAACHS | 95927 | Volvox carteri | I |
| SAGAACNS | Sll0704 | Synechocystis | I | |||
| SUFS | No | RSGHHCA | RTGHHCA | SufS/CsdB | Escherichia coli | II |
| RSGHHCA | SUFS/CpNifS | Arabidopsis thaliana | II | |||
| RSGHHCT | Slr0077 | Synechocystis | II | |||
| CSDH | No | No conserved cysteine | ZP_00111076 | Nostoc punctiforme | II |
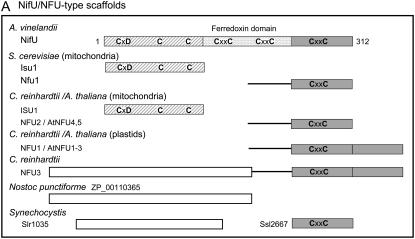
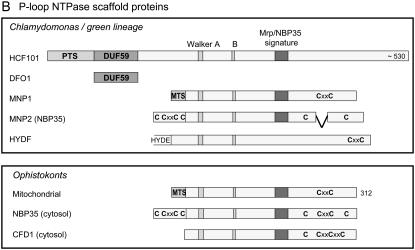
Figure 1.—
Domain homology of Fe–S scaffold proteins found in Chlamyodomonas to well-studied orthologs in bacteria, plants, and yeast. Bar diagrams represent predicted scaffold proteins, specific domains, and amino acid motifs. The lengths of the bars are to scale within each box, with the length in amino acids indicated in the top bar (protein). (A) NifU orthologs; (B) P-loop NTPases. PTS, plastid targeting sequence; MTS, mitochondrial targeting sequence.
TABLE 2.
Orthologs of Fe–S cluster assembly proteins in Chlamydomonas, Arabidopsis, bacteria, and yeast
| C. reinhardtii | C. r. protein ID | A. thaliana | A. t. localization | A. t. AGI gene model | E. coli | Synechocystis | S. cerevisiae | Molecular functionor protein domaina | |
|---|---|---|---|---|---|---|---|---|---|
| ISC | NFS1 | 118253 | NFS1 | Mit | At5g65720 | IscS | Slr0387 | Nfs1 | Cysteine desulfurase |
| ISD11 | 184200 | ISD11 | Mitb | At5g61220 | Isd11 | Unknown | |||
| ISU1 | 187899 | ISU1, -2, -3 | Mit | At4g22220, At3g01020 At4g04080 | IscU | Isu1, -2 | Fe–S scaffold | ||
| ISCA1 | 196772 | Not annotated | Mitb | At2g16710 | IscA | Isa1, -2 | Fe–S scaffold | ||
| IBA57c | 187731d | Not annotated | Mitb | At4g12130 | Unknown | ||||
| NFU2 | 205851e 205852e | NFU4, NFU5 | Mit | At3g20970, At1g51390 | NP_312283 | Nfu1 | Fe–S scaffold | ||
| MFDX | 154720 | ADX1, -2 = MFDX1, -2 | Mit | At4g05450 | Fdx | Yah1 | Ferredoxin | ||
| ARH1 | 134840 | ADXR = MFDR | Mit | At4g32360 | Arh1 | Reductase | |||
| MNP1 | 133548 | HCF101-L1 | Mitf | At4g19540 | Mrp | P-loop NTPasea | |||
| Not specialized?g | Not specialized?g | HscA | Ssq1 | Chaperone (HSP70) | |||||
| HSC20 | 144583 | Not annotated | Mitb | At5g06410 | HscB | Jac1 | Cochaperone | ||
| MGE1 | 132633 | Not annotated | Mitb | At5g55200 | Mge1 | Nucleotide release factor | |||
| FTX1 | 196778 | FH | Mit | At4g03240 | CyaY | Yfh1 | Iron chaperone | ||
| GRX5 | 195767 | Not annotated | Mitb | At3g15660 | Grx | Grx5 | Glutaredoxin | ||
| Export | CDS1h | 133289 | ATM1 | Mit | At4g28630 | Atm1 | Half ABC transporter | ||
| ATM2 | 105113 | ATM2 | Mit | At4g28620 | Atm1 | Half ABC transporter | |||
| ATM3 | 99736 | ATM3 = STA1 | Mit | At5g58270 | Atm1 | Half ABC transporter | |||
| ERV1i | 114232 | ERV1 | Mit | At1g49880 | Erv1 | Sulfhydryl oxidase | |||
| CIA | Absent | Absent | Mrp | Cfd1 | P-loop NTPasea | ||||
| MNP2 | 206480 | HCF101-L2 | Cytosolf | At5g50960 | Nbp35 | P-loop NTPasea | |||
| IOP1 (HYD3) | 206492d | NAR1 | Cytosolb | At4g16440 | Nar1 | Hydrogenase-likea | |||
| CIA1 | 158085 | Not annotated | Cytosolb | At2g26060 | Cia1 | WD40 protein | |||
| SUF | SUFA | 100292 | SUFA = ISCA | Plastid | At1g10500 | SufA | Slr1417 | Fe–S scaffold | |
| SUFB | 132600d | SUFB = NAP1 | Plastid | At4g04770 | SufB | Slr0074 | ABC/ATPase | ||
| SUFC | 289 | SUFC = NAP7 | Plastid | At3g10670 | SufC | Slr0075 | ABC/ATPase | ||
| SUFD | 154386 | SUFD = NAP6 | Plastid | At1g32500 | SufD | Slr0076 | ABC/ATPase | ||
| SUFE | 156669 | SUFE1, -2, -3 (N-term.)j | Plastid | At4g26500, At1g67810, At5g50210 | SufE | Slr1419 | Unknown | ||
| SUFS | 206481 | NFS2 = CpNifS | Plastid | At1g08490 | SufS/CsdB | Slr0077 | Cysteine desulfurase | ||
| NFU | NFU1 | 206476 | NFU1, -2, -3 | Plastid | At4g01940, At5g49940, At4g25910 | NP_312283 | Ssl2667 | Fe–S scaffold | |
| NFU3 | 188996 | NFU1, -2, -3 | Plastid | Ssl2667 | Fe–S scaffold | ||||
| HCF101 | HCF101 | 102098d | HCF101 | Plastid | At3g24430 | Slr0067 | DUF59 + P-loop NTPasea | ||
| Hydrogenase maturation | HYDEF | 128256 | Absent | Radical SAM + GTPasea | |||||
| HYDG | 196226 | Absent | Radical SAMa | ||||||
| Other | CSD2 | 167884 | Absent | Sll0704 | Cysteine desulfurase | ||||
| DFO1 | 206478 | Not annotated | At1g68310 | DUF59 (unknown)a | |||||
| ISCA2 | 162474d | Not annotated | Mit/plastidb | At5g03905 | ErpA? | Fe–S scaffold |
Organisms: C. r., Chlamydomonas reinhardtii; A. t., the higher plant Arabidopsis thaliana; E. coli, the proteobacterium Escherichia coli; cyanobacterium Synechocystis sp. PCC 6803; and S. cerevisiae, Baker's yeast, Saccharomyces cerevisiae.
Function inferred from conserved protein domain(s). DUF59, domain of unknown function 59 (Pfam PF01883); radical SAM, radical S-adenosylmethionine enzyme.
Predicted.
IBA57, iron–sulfur cluster assembly factor for biotin synthase- and aconitase-like mitochondrial proteins, Mr 57 kDa. (Gelling et al. 2008).
The sequence is incomplete or not deemed correct because of annotation issues.
The two models are generated by alternative splicing, which appears to be conserved in Volvox. The significance of this remains to be investigated.
K. Bych and J. Balk, unpublished results.
The Hsp70 proteins HscA from E. coli and Ssq1 from yeast are specialized for Fe–S cluster assembly, but do not share the same genetic origin. The yeast Yarrowia lipolytica does not have a specialized Hsp70, and this situation may be the case for most eukaryotes (Schilke et al. 2006).
CDS1, Cadmium sensitive 1, is the only Chlamydomonas gene characterized experimentally (Hanikenne et al. 2005).
Two genes with high similarity to ERV1 are present in Chlamydomonas. This may reflect the situation in S. cerevisiae, where ERV1 is a sulfhydryl oxidase of the mitochondrial inner membrane space, whereas ERV2 resides in the endoplasmic reticulum.
In Arabidopsis, SUFE3 is a fusion protein of NADA and a SUFE domain (Murthy et al. 2007).
The core components—cysteine desulfurases and scaffolds:
Cysteine desulfurases:
Using standard BLAST tools, we identified in the Chlamydomonas genome four open reading frames with similarity to cysteine desulfurases involved in Fe–S cluster assembly in other organisms (Table 1). On the basis of sequence features, in particular the amino acids around the active site in the C terminus, cysteine desulfurases can be divided into group I or group II (see Table 1) (Mihara and Esaki 2002). The two groups are also thought to differ functionally, to the extent in which oxygen is tolerated. For example, the group-II enzymes from E. coli and A. thaliana function well under conditions of oxidative stress or in proximity of oxygenic photosynthesis, respectively, whereas the group-I enzymes generally function in strict anaerobes or in low-oxygen environments like the mitochondrial matrix (Outten et al. 2004; Johnson et al. 2005; Ye et al. 2006b). Two of the Chlamydomonas proteins, annotated as NFS1 and CSD2, belong to group I, whereas SUFS and CSDH belong to group II. The Chlamydomonas NFS1 sequence has high similarity over its full length to AvNifS, EcIscS, and yeast NFS1 (70% identity, 85% similarity), indicating that this enzyme is part of the ISC pathway in the mitochondria. The mitochondrial enzyme is also thought to provide sulfur for biotin, thiamine, lipoate, and thiolated tRNAs (Lill and Mühlenhoff 2008). In the case of biotin and lipoate, the sulfur is not directly provided by NFS1/IscS, but by a noncatalytic Fe–S cluster in BioB and LipA, respectively (Picciocchi et al. 2003; Berkovitch et al. 2004; Cicchillo and Booker 2005). The CrSUFS enzyme, on the other hand, is similar to Arabidopsis SUFS/CpNifS (60% identity, 75% similarity) which is the major sulfur donor to Fe–S proteins in the plastids (Van Hoewyk et al. 2007). The Chlamydomonas CSDH protein (ID 206474) has high sequence similarity to SUFS (42%), but is lacking the conserved cysteine in the C terminus, and it is therefore not certain whether CSDH is a functional cysteine desulfurase. Interestingly, Chlamydomonas encodes a second group-I enzyme, CSD2, that is not found in the higher plants Arabidopsis and rice. It bears homology to Synechocystis Sll0704/Csd2, which can drive Fe–S cluster assembly in vitro (Kato et al. 2000). If CrCSD2 is indeed of the same evolutionary origin as Sll0704, it is probably located in the plastids and as an oxygen-sensitive group-I enzyme, may function under anaerobic conditions, which occur more frequently in algae than in land plants.
U-type scaffold proteins:
Scrutiny of the Chlamydomonas genome identified four gene products with homology to domains of the A. vinelandii NifU Fe–S scaffold protein (Figure 1A). NifU has a modular structure, with the modules often occurring as separate, single-domain proteins in other organisms (Mühlenhoff and Lill 2000; Johnson et al. 2005). The Chlamydomonas ISU1 protein is similar to the N terminus of AvNifU and has three conserved cysteines that have been shown to be critical for binding a labile Fe–S cluster in bacterial IscU and yeast Isu1 proteins. The ISU scaffold proteins in yeast and Arabidopsis are localized in mitochondria, suggesting that Chlamydomonas ISU1 is also located in this organelle.
The NFU proteins share sequence similarity to the C terminus of AvNifU. The Arabidopsis NFU proteins are found in mitochondria and plastids and can be distinguished by domain structure: the plastid proteins have a C-terminal duplication of the entire NFU domain that lacks the CxxC motif (Figure 1A) (Léon et al. 2003). In Chlamydomonas, the single-domain NFU2 is therefore likely to reside in the mitochondria, whereas NFU1 and NFU3 are predicted to localize to the chloroplast. The function of the mitochondrial NFUs is not clear, as the deletion of the yeast NFU1 gene has no phenotype, unless in combination with mutations in SSQ1, encoding the HSP70-type chaperone specifically adapted to Fe–S cluster transfer. In contrast, the plastid NFU2 protein in Arabidopsis has been demonstrated to play a key role in Fe–S protein assembly in chloroplasts. nfu2 knock-out mutants had decreased protein levels and/or activities of PSI, ferredoxin, and sulfite reductase, although glutamate synthase and the Rieske protein were unaffected (Touraine et al. 2004; Yabe et al. 2004). The CrNFU3 protein carries an extra N-terminal domain that is found as a separate protein in cyanobacteria. This N-terminal domain has no recognizable protein motifs, but does have weak similarity to monothiol glutaredoxins, also implicated in Fe–S cluster assembly (Herrero and De La Torre-Ruiz 2007). A similar two-domain version of NFU3 is found in the red alga Cyanidioschyzon merolae (gene CMP295C).
A-type scaffold proteins:
In addition to the U-type scaffolds ISU1 and NFU1–3, the Chlamydomonas genome contains three so-called A-type scaffolds, annotated as SUFA, ISCA1, and ISCA2 (Table 2). Members of this protein family have three conserved cysteines and are usually associated with Fe–S cluster assembly operons in bacteria, but their function has remained elusive, as deletion mutants do not have a clear phenotype. Sequence alignments of the Chlamydomonas A-type proteins with (cyano)bacterial and Arabidopsis orthologs showed that CrSUFA clustered with the Arabidopsis SUFA and Synechocystis Slr1417 proteins, whereas the CrISCA1 protein clustered with E. coli IscA and the mitochondrial Isa1 in yeast. The latter protein is required for the function of biotin synthase (Mühlenhoff et al. 2007). The CrISCA2 protein appears to represent a third group together with the uncharacterized Arabidopsis protein AT5G03905. These proteins may be required for Fe–S assembly on IspG (Protein ID 55268, HDS), an enzyme of isoprenoid biosynthesis in the chloroplast, as suggested by the recent identification and characterization of the ErpA scaffold protein in E. coli (Loiseau et al. 2007).
P-loop NTPase scaffold proteins:
A recent study (Netz et al. 2007) has provided evidence for another type of scaffold protein in yeast. The P-loop NTPases Nbp35 and Cfd1 were shown to transfer Fe–S clusters in vitro, supporting in vivo data that the proteins are required for the assembly of cytosolic and nuclear Fe–S clusters (Roy et al. 2003; Netz et al. 2007). Chlamydomonas has four P-loop NTPases that could be involved in Fe–S cluster assembly: three proteins of the Mrp/NBP35 class and HydF (fused to HydE, Figure 1B). The presence (or absence) of targeting sequences suggests that MNP2 (NBP35) is localized in the cytosol, MNP1 in the mitochondria, and HCF101 in the plastid. Both MNP2 and HCF101 lack the conserved CxxC motif that is usually found in the C terminus of Mrp/NBP35 proteins. Mutants in high chlorophyll fluorescence (HCF)101 have been characterized in Arabidopsis and have a defect in the assembly of PSI, as well as instability of ferredoxin-thioredoxin reductase, but not ferredoxin (Lezhneva et al. 2004; Stöckel and Oelmüller 2004). HCF101 has an extra domain at its N terminus, domain of unknown function (DUF)59 (pfam PF01883) not found in the other Mrp/NBP35-like proteins. This domain is also found as a separate, single-domain Chlamydomonas protein, DUF59-only (DFO)1 protein.
The HydF protein, as part of the HydEF fusion protein, was identified in screens for Chlamydomonas mutants defective in hydrogenase maturation (Posewitz et al. 2004). The HydF ortholog in Thermotoga maritima has been characterized at the biochemical level (Brazzolotto et al. 2006). The recombinant TmHydF protein displayed GTPase activity and bound an Fe–S cluster, but its molecular function in the maturation of the H-cluster and its CO/CN ligands remains to be determined.
Fe–S protein assembly machineries and compartmentalization:
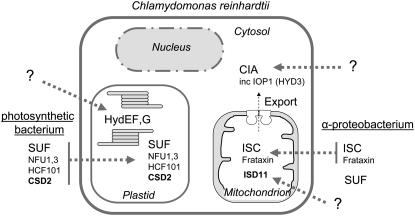
The above inventarization of cysteine desulfurases and scaffold proteins yielded a wealth of well-conserved Fe–S protein assembly components. Grouping these proteins on the basis of sequence and predicted localization resulted in a complex model for Fe–S cluster assembly in Chlamydomonas, compartmentalized in the mitochondria, cytosol, and chloroplast (Table 2 and Figure 2).
Figure 2.—
Fe–S protein assembly machineries in Chlamydomonas based on genome analysis and their evolutionary origin. See text and Table 2 for details.
The mitochondrial ISC system and export:
Chlamydomonas is anticipated to contain a complete ISC system in the mitochondria, including the group I cysteine desulfurase NFS1, its helper protein ISD11, the scaffold protein ISU1, the ferredoxin MFDX, the A-type scaffold ISCA1, and the NFU2 protein. The ISC system in bacteria and Baker's yeast contains a dedicated Hsp70-type chaperone, but it is not certain whether this is the case in all eukaryotes (Schilke et al. 2006). The mitochondria are also predicted to harbor the frataxin protein, FTX1, thought to deliver iron to the ISU1 scaffold protein; a glutaredoxin related to Grx5 in yeast which is involved in cluster transfer from ISU1 to target Fe–S proteins; and the P-loop NTPase MNP1, which is required for the assembly of respiratory complex I (K. Bych, S. Kerscher, D. J. A. Netz, A. J. Pierik, K. Zwicker, M. A. Huynen, R. Lill, U. Brandt and J. Balk, unpublished results).
The mitochondrial ISC pathway in Chlamydomonas is likely to be required for both mitochondrial as well as cytosolic Fe–S proteins, because of the presence of several ATM transporters. One of these mitochondrial half ABC transporters, CDS1, was identified in a screen for cadmium-sensitive mutants, and its mitochondrial localization was confirmed (Hanikenne et al. 2005). A link between cadmium detoxification and the mitochondrial ABC transporters has also been observed for Arabidopsis (Kim et al. 2006). Cadmium may interfer with transport of the ISC-dependent compound required for cytosolic Fe–S proteins, either by binding to crucial thiol groups or by depleting glutathione, which is involved in the transport process (Sipos et al. 2002).
The cytosolic CIA system:
Of four cytosolic proteins known to be involved in Fe–S protein maturation in Baker's yeast, three could be identified in Chlamydomonas. These are the P-loop NTPase MNP2 (Nbp35 in yeast), the WD40 protein CIA1, and the hydrogenase-like protein IOP1 (previously annotated as HYD3, similar to Nar1 in yeast; Balk et al. 2004; Huang et al. 2007). The three yeast proteins form protein interactions in vivo, with Nbp35 acting upstream of Nar1, which acts upstream of Cia1 (Balk et al. 2005; Netz et al. 2007), but their precise molecular functions are as yet unknown. An ortholog of the yeast P-loop NTPase Cfd1 was not found in Chlamydomonas, nor in any of the other green lineage organisms sequenced to date (Figure 1B). As a result, MNP2 is likely to function as a homodimer, rather than a Nbp35-Cfd1 heterotetramer (Netz et al. 2007).
Chloroplast–SUF plus additional scaffolds:
A complete set of six SUF proteins can be found in the Chlamydomonas genome. By similarity with Arabidopsis the SUF proteins are likely to reside in the chloroplast, where they facilitate Fe–S cluster assembly under aerobic conditions. The cysteine desulfurase activity of SUFS/CpNifS is enhanced by SUFE, which facilitates persulfide transfer (Outten et al. 2003; Ye et al. 2006a). SufBCD forms a nonintrinsic ABC protein, and also stimulates the activity of SUFS (Outten et al. 2003). The Arabidopsis SUFE1 has been reported to be dual-targeted to mitochondria and chloroplasts (Xu and Møller 2006), but it remains to be seen whether the same holds true for other SUFE proteins in the green lineage. In addition to the SUF pathway, the Chlamydomonas chloroplast is predicted to harbor several scaffold proteins, NFU1, NFU3, and HCF101. On the basis of characterization of the Arabidopsis nfu2 and hcf101 mutants, these proteins may serve specific Fe–S proteins (see above).
Chloroplast–HydEFG:
Chlamydomonas contains at least two proteins specifically dedicated to the maturation of [FeFe]-hydrogenases (Posewitz et al. 2004). Although the localization of HydEF and HydG gene products has not been confirmed to date, they are likely to reside in the chloroplast, colocalizing with the HydA1 and HydA2 hydrogenases. HydEFG constitute a complex molecular machinery, consisting of two radical SAM enzymes (HydE and HydG), and an Fe–S-containing P-loop GTPase (HydF). The proteins are conserved in organisms with functional Fe-only hydrogenases and are required for assembly of the intricate H-cluster, consisting of a di-iron center liganded by CN and CO groups. The di-iron center is bridged to a [4Fe–4S] cluster via a cysteine sulfur (Böck et al. 2006). The latter cluster, as well as the clusters on HydE, -F, and -G, is likely to be provided by the generic Fe–S cluster assembly machinery in the chloroplast.
Evolution:
Fe–S clusters are ancient cofactors that are key to the activity of a number of highly conserved enzymes. It is therefore not surprising that the assembly pathways of Fe–S clusters are also conserved from bacteria to higher eukaryotes. The mitochondrial ISC pathway is most likely derived from its α-proteobacterial ancestor, having lost the SUF pathway (present in E. coli). The cyanobacterial ancestor would have possessed an assembly pathway more adapted to oxygenic photosynthesis (i.e., no ISU proteins), which has survived in the plastids of the photosynthetic eukaryotes. The origin of the CIA proteins is unknown. IOP1 bears striking sequence similarity to [FeFe]-hydrogenases, but is not thought to function as such (Meyer 2007). First of all, it lacks a critical double cysteine motif. Second, our Chlamydomonas genome analysis suggests that the HydEFG proteins are not in the same cell compartment as IOP1, excluding a role for HydEFG in the assembly of an H-cluster on IOP1. The eukaryotic IOP1 (Nar1) proteins have a monophyletic origin, whereas the HydEFG and the HydAs have been acquired later in separate events by only a handful of eukaryotes (Meyer 2007).
Comparing Chlamydomonas and Arabidopsis reveals a remarkable conservation of Fe–S protein maturation in the green lineage. The organisms contain almost the same set of components of the ISC, export, CIA, and chloroplast machinery. Exceptions are the cysteine desulfurase CSD2 and the N-terminal domain of the NFU3 protein in Chlamydomonas, which are not found in higher plants. Another difference between the two green organisms is a difference in copy number (Table 2). In several cases Chlamydomonas has one gene copy, whereas Arabidopsis has two to three, for instance for ISU, MFDX, mitochondrial, and plastid NFUs, and SUFE. These gene copies may be specialized for specific tissues, which is corroborated by data on the Arabidopsis ISUs and SUFEs (Léon et al. 2005; Murthy et al. 2007). Of course, additional copies may still be found in the remaining part of the Chlamydomonas genome that has not yet been sequenced/assembled. In Chlamydomonas, multiple copies were found for the A-type scaffolds, NFUs, and ABC transporters of the mitochondria (ATMs). For the A-type scaffolds and NFUs, there are good reasons to assume that they are targeted to different organelles and/or perform different functions (discussed above). Whether the same is true for the three mitochondrial ABC transporters remains to be investigated.
Comparison of Chlamydomonas and Arabidopsis with nongreen eukaryotes shows a few features that appear to be specific for the green lineage. These include the absence of a cytosolic Cfd1 ortholog and alterations to the C-terminal domain of Nbp35, as well as duplication of the NFU domain (without the CxxC motif) in plastids. It will be interesting to investigate what the significance of these adaptations is.
Future research questions:
From our comparative genome analysis, it is clear that Chlamydomonas has a very elaborate and interesting set of >30 genes involved in Fe–S assembly. Other than the HydEFG genes, only the ABC transporter CDS1 has been investigated experimentally (Hanikenne et al. 2005), but not in the context of Fe–S protein maturation. The current set of putative Fe–S cluster assembly genes (Table 2) provides a working model for future investigations including (i) to confirm the proposed function of each gene; (ii) to identify the chloroplast proteins that assist HydEFG in the maturation of [FeFe]-hydrogenase, such that they can be targeted for increased hydrogen production; and (iii) to investigate the coordination and regulation of the compartmentalized pathways, as well as iron distribution.
Acknowledgments
We acknowledge the use of the Chlamydomonas and Volvox genome resources. We thank Martin Croft for discussion on annotation issues; Attila Molnár, Mark van der Giezen, and the reviewers for critical reading of the manuscript; and Olivier Vallon for correcting the gene models discussed in this article. J.G. is supported by an F. E. Fritsch studentship and J.B. by a university research fellowship from the Royal Society.
References
- Balk, J., and S. Lobréaux, 2005. Biogenesis of iron-sulfur proteins in plants. Trends Plant Sci. 10 324–331. [DOI] [PubMed] [Google Scholar]
- Balk, J., A. J. Pierik, D. J. Aguilar Netz, U. Mühlenhoff and R. Lill, 2004. The hydrogenase-like Nar1p is essential for maturation of cytosolic and nuclear iron-sulphur proteins. EMBO J. 23 2105–2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balk, J., D. J. Aguilar Netz, K. Tepper, A. J. Pierik and R. Lill, 2005. The essential WD40 protein Cia1 is involved in a late step of cytosolic and nuclear iron-sulfur protein assembly. Mol. Cell. Biol. 25 10833–10841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barras, F., L. Loiseau and B. Py, 2005. How Escherichia coli and Saccharomyces cerevisiae build Fe/S proteins. Adv. Microb. Physiol. 50 41–101. [DOI] [PubMed] [Google Scholar]
- Beinert, H., 2000. Iron-sulfur proteins: ancient structures, still full of surprises. J. Biol. Inorg. Chem. 5 2–15. [DOI] [PubMed] [Google Scholar]
- Berkovitch, F., Y. Nicolet, J. T. Wan, J. T. Jarrett and C. L. Drennan, 2004. Crystal structure of biotin synthase, an S-adenosylmethionine-dependent radical enzyme. Science 303 76–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böck, A., P. W. King, M. Blokesch and M. C. Posewitz, 2006. Maturation of hydrogenases. Adv. Microb. Physiol. 51 1–71. [DOI] [PubMed] [Google Scholar]
- Brazzolotto, Z., J. K. Rubach, J. Gaillard, S. Gambarelli, M. Atta et al., 2006. The [Fe-Fe]-hydrogenase maturation protein HydF from Thermatoga maritima is a GTPase with an iron-sulfur cluster. J. Biol. Chem. 281 769–774. [DOI] [PubMed] [Google Scholar]
- Cicchillo, R. M., and S. J. Booker, 2005. Mechanistic investigations of lipoic acid biosynthesis in Escherichia coli: both sulfur atoms in lipoic acid are contributed by the same lipoyl synthase polypeptide. J. Am. Chem. Soc. 127 2860–2861. [DOI] [PubMed] [Google Scholar]
- Embley, T. M., and W. Martin, 2006. Eukaryotic evolution, changes and challenges. Nature 440 623–630. [DOI] [PubMed] [Google Scholar]
- Gelling, C., I. W. Dawes, N. Richhardt, R. Lill and U. Mühlenhoff, 2008. Mitochondrial Iba57p is required for Fe/S cluster formation on aconitase and activation of radical SAM enzymes. Mol. Cell. Biol. 28 1851–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanikenne, M., P. Motte, W. C. S. Wu, T. Wang, R. Loppes et al., 2005. A mitochondrial half-size ABC transporter is involved in cadmium tolerance in Chlamydomonas reinhardtii. Plant Cell Environ. 28 863–873. [Google Scholar]
- Herrero, E., and M. A. de la Torre-Ruiz, 2007. Monothiol glutaredoxins: a common domain for multiple functions. Cell. Mol. Life Sci. 64 1518–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, J., D. Song, A. Flores, Q. Zhao, S. M. Mooney et al., 2007. IOP1, a novel hydrogenase-like protein that modulates hypoxia-inducible factor1-alpha activity. Biochem. J. 401 341–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imsande, J., 1999. Iron-sulfur clusters: formation, perturbation, and physiological functions. Plant Physiol. Biochem. 37 87–97. [Google Scholar]
- Johnson, D. C., D. R. Dean, A. D. Smith and M. K. Johnson, 2005. Structure, function, and formation of biological iron-sulfur clusters. Annu. Rev. Biochem. 74 247–281. [DOI] [PubMed] [Google Scholar]
- Kato, S., H. Mihara, T. Kurihara, T. Yoshimura and N. Esaki, 2000. Gene cloning, purification, and characterization of two cyanobacterial NifS homologs driving iron-sulfur cluster formation. Biosci. Biotechnol. Biochem. 64 2412–2419. [DOI] [PubMed] [Google Scholar]
- Kessler, D., and J. Papenbrock, 2005. Iron-sulfur cluster biosynthesis in photosynthetic organisms. Photosynth. Res. 86 391–407. [DOI] [PubMed] [Google Scholar]
- Kim, D. Y., L. Bovet, S. Kushnir, E. W. Noh, E. Martinoia et al., 2006. AtATM3 is involved in heavy metal resistance in Arabidopsis. Plant Physiol. 140 922–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kispal, G., P. Csere, C. Prohl and R. Lill, 1999. The mitochondrial proteins Atm1p and Nfs1p are essential for biogenesis of cytosolic Fe/S proteins. EMBO J. 18 3981–3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Léon, S., B. Touraine, C. Ribot, J. F. Briat and S. Lobréaux, 2003. Iron-sulphur cluster assembly in plants: distinct NFU proteins in mitochondria and plastids from Arabidopsis thaliana. Biochem. J. 371 823–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Léon, S., B. Touraine, J.-F. Briat and S. Lobréaux, 2005. Mitochondrial localization of Arabidopsis thaliana Isu Fe-S scaffold proteins. FEBS Lett. 28 1930–1934. [DOI] [PubMed] [Google Scholar]
- Lezhneva, L., K. Amann and J. Meurer, 2004. The universally conserved HCF101 protein is involved in assembly of [4Fe-4S]-cluster-containing complexes in Arabidopsis thaliana chloroplasts. Plant J. 37 174–185. [DOI] [PubMed] [Google Scholar]
- Li, H.-M., S. M. Theg, C. M. Bauerle and K. Keegstra, 1990. Metal-ion-center assembly of ferredoxin and plastocyanin in isolated chloroplasts. Proc. Natl. Acad. Sci. USA 87 6748–6752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lill, R., and U. Mühlenhoff, 2008. Maturation of iron-sulfur proteins in eukaryotes: mechanisms, connected processes, and diseases. Annu. Rev. Biochem. 77 (in press). [DOI] [PubMed]
- Loiseau, L., C. Gerez, M. Bekker, S. Ollagnier-de-Choudens, B. Py et al., 2007. ErpA, an iron-sulfur (Fe-S) protein of the A-type essential for respiratory metabolism in Escherichia coli. Proc. Natl. Acad. Sci. USA 104 13626–13631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant, S. S., S. E. Prochnik, O. Vallon, E. H. Harris, S. J. Karpowicz et al., 2007. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318 245–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, J., 2007. [FeFe] hydrogenases and their evolution: a genomic perspective. Cell. Mol. Life Sci. 64 1063–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihara, H., and N. Esaki, 2002. Bacterial cysteine desulfurases: their function and mechanisms. Appl. Microbiol. Biotechnol. 60 12–23. [DOI] [PubMed] [Google Scholar]
- Mühlenhoff, U., and R. Lill, 2000. Biogenesis of iron-sulfur proteins in eukaryotes: a novel task of mitochondria that is inherited from bacteria. Biochim. Biophys. Acta 1459 370–382. [DOI] [PubMed] [Google Scholar]
- Mühlenhoff, U., M. J. Gerl, B. Flauger, H. M. Pirner, S. Balser et al., 2007. The ISC proteins Isa1 and Isa2 are required for the function but not the de novo synthesis of the Fe/S clusters of biotin synthase in Saccharomyces cerevisiae. Eukaryot. Cell 6 495–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy, U. N. M., S. Ollagnier-de-Choudens, Y. Sanakis, S. E. Abdel-Ghany, C. Rousset et al., 2007. Characterization of Arabidopsis thaliana SufE2 and SufE3: functions in chloroplast iron-sulfur cluster assembly and Nad synthesis. J. Biol. Chem. 282 18254–18264. [DOI] [PubMed] [Google Scholar]
- Nachin, L., L. Loiseau, D. Expert and Y. Barras, 2003. SufC: an unorthodox cytoplasmic ABC/ATPase required for [Fe-S] biogenesis under oxidative stress. EMBO J. 22 427–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netz, D. J., A. J. Pierik, M. Stümpfig, U. Mühlenhoff and R. Lill, 2007. The Cfd1-Nbp35 complex acts as a scaffold for iron-sulfur protein assembly in the yeast cytosol. Nat. Chem. Biol. 3 278–286. [DOI] [PubMed] [Google Scholar]
- Outten, F. W., M. J. Wood, F. M. Muñoz and G. Storz, 2003. The SufE protein and the SufBCD complex enhance SufS cysteine desulfurase activity as part of a sulfur transfer pathway for Fe-S cluster assembly in Escherichia coli. J. Biol. Chem. 278 45713–45719. [DOI] [PubMed] [Google Scholar]
- Outten, F. W., O. Djaman and G. Storz, 2004. A suf operon requirement for Fe-S cluster assembly during iron starvation in Escherichia coli. Mol. Microbiol. 52 861–872. [DOI] [PubMed] [Google Scholar]
- Peltier, G., and L. Cournac, 2002. Chlororespiration. Annu. Rev. Plant Biol. 53 523–550. [DOI] [PubMed] [Google Scholar]
- Peters, J. W., W. N. Lanzilotta, B. J. Lemon and L. C. Seefeldt, 1998. X-ray crystal structure of the Fe-only hydrogenase (CpI) from Clostridium pasteurianum at 1.8 angstrom resolution. Science 282 1853–1858. [DOI] [PubMed] [Google Scholar]
- Picciocchi, A., R. Douce and C. Alban, 2003. The plant biotin synthase reaction. Identification and characterization of essential mitochondrial accessory protein components. J. Biol. Chem. 278 24966–24975. [DOI] [PubMed] [Google Scholar]
- Posewitz, M. C., P. W. King, S. L. Smolinski, L. Zhang, M. Seibert et al., 2004. Discovery of two novel radical S-adenosylmethionine proteins required for the assembly of an active [Fe] hydrogenase. J. Biol. Chem. 279 25711–25720. [DOI] [PubMed] [Google Scholar]
- Roy, A., N. Solodovnikova, T. Nicholson, W. Antholine and W. E. Walden, 2003. A novel eukaryotic factor for cytosolic Fe-S cluster assembly. EMBO J. 22 4826–4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilke, B., B. Williams, H. Knieszner, S. Pukszta, P. D'Silva et al., 2006. Evolution of mitochondrial chaperones utilized in Fe-S cluster biogenesis. Curr. Biol. 16 1660–1665. [DOI] [PubMed] [Google Scholar]
- Sipos, K., H. Lange, Z. Fekete, P. Ullmann, R. Lill et al., 2002. Maturation of cytosolic iron-sulfur proteins requires glutatione. J. Biol. Chem. 277 26944–26949. [DOI] [PubMed] [Google Scholar]
- Stöckel, J., and R. Oelmüller, 2004. A novel protein for photosystem I biogenesis. J. Biol. Chem. 279 10243–10251. [DOI] [PubMed] [Google Scholar]
- Takahashi, Y., A. Mitsui, T. Hase and H. Matsubara, 1986. Formation of the iron-sulfur cluster of ferredoxin in isolated chloroplasts. Proc. Natl. Acad. Sci. USA 83 2434–2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, Y., A. Mitsui, Y. Fujita and H. Matsubara, 1991. a Roles of ATP and NADPH in formation of the Fe-S cluster of spinach ferredoxin. Plant Physiol. 95 104–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, Y., A. Mitsui and H. Matsubara, 1991. b Formation of the Fe-S cluster of ferredoxin in lysed spinach chloroplasts. Plant Physiol. 95 97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touraine, B., J. P. Boutin, A. Marion-Poll, J. F. Briat, G. Peltier et al., 2004. Nfu2: a scaffold protein required for [4Fe-4S] and ferredoxin iron-sulphur cluster assembly in Arabidopsis chloroplasts. Plant J. 40 101–111. [DOI] [PubMed] [Google Scholar]
- van der Giezen, M., and J. Tovar, 2005. Degenerate mitochondria. EMBO Rep. 6 525–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hoewyk, D., S. E. Abdel-Ghany, C. M. Cohu, S. K. Herbert, P. Kugrens et al., 2007. Chloroplast iron-sulfur cluster protein maturation requires the essential cysteine desulfurase CpNifS. Proc. Natl. Acad. Sci. USA 104 5686–5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, X. M., and S. G. Møller, 2006. AtSufE is an essential activator of plastidic and mitochondrial desulfurases in Arabidopsis. EMBO J. 25 900–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabe, T., K. Morimoto, S. Kikuchi, K. Nishio, I. Terashima et al., 2004. The Arabidopsis chloroplastic NifU-like protein CnfU, which can act as an iron-sulfur cluster scaffold protein, is required for biogenesis of ferredoxin and photosystem I. Plant Cell 16 993–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, H., S. E. Abdel-Ghany, T. D. Anderson, E. A. Pilon-Smits and M. Pilon, 2006. a CpSufE activates the cysteine desulfurase CpNifS for chloroplastic Fe-S cluster formation. J. Biol. Chem. 281 8958–8969. [DOI] [PubMed] [Google Scholar]
- Ye, H., M. Pilon and E. Pilon-Smits, 2006. b CpNifS-dependent iron-sulfur cluster biogenesis in chloroplasts. New Phytol. 171 285–292. [DOI] [PubMed] [Google Scholar]
- Zheng, L., R. H. White, V. L. Cash, R. F. Jack and D. R. Dean, 1993. Cysteine desulfurase activity indicates a role for NIFS in metallocluster biosynthesis. Proc. Natl. Acad. Sci. USA 90 2754–2758. [DOI] [PMC free article] [PubMed] [Google Scholar]