Abstract
Arabidopsis thaliana INOSITOL TRANSPORTER1 (INT1) is a member of a small gene family with only three more genes (INT2 to INT4). INT2 and INT4 were shown to encode plasma membrane–localized transporters for different inositol epimers, and INT3 was characterized as a pseudogene. Here, we present the functional and physiological characterization of the INT1 protein, analyses of the tissue-specific expression of the INT1 gene, and analyses of phenotypic differences observed between wild-type plants and mutant lines carrying the int1.1 and int1.2 alleles. INT1 is a ubiquitously expressed gene, and Arabidopsis lines with T-DNA insertions in INT1 showed increased intracellular myo-inositol concentrations and reduced root growth. In Arabidopsis, tobacco (Nicotiana tabacum), and Saccharomyces cerevisiae, fusions of the green fluorescent protein to the C terminus of INT1 were targeted to the tonoplast membranes. Finally, patch-clamp analyses were performed on vacuoles from wild-type plants and from both int1 mutant lines to study the transport properties of INT1 at the tonoplast. In summary, the presented molecular, physiological, and functional studies demonstrate that INT1 is a tonoplast-localized H+/inositol symporter that mediates the efflux of inositol that is generated during the degradation of inositol-containing compounds in the vacuolar lumen.
INTRODUCTION
In plants, sugars and sugar alcohols play an essential role as transport and storage forms of fixed carbon, they represent the main energy source, they are the initial substrates for numerous biosynthetic reactions, and they may be accumulated in response to several kinds of stress. Their biosynthesis occurs in fully developed and mature source leaves, from where they are allocated via the phloem to the different sink tissues and organs of higher plants.
The relative amounts of sugars and sugar alcohols that are transported in the phloem vary between different plant species. In many plants, sucrose is the exclusive or by far dominant form of assimilated carbon (e.g., in Arabidopsis thaliana or tobacco [Nicotiana tabacum]). Other species, however, transport only low concentrations of sucrose in combination with raffinose family oligosaccharides, such as raffinose, stachyose, or verbascose (Sauer, 2007), and a third group transports sucrose together with sugar alcohols, such as sorbitol (e.g., Plantago major; Ramsperger-Gleixner et al., 2004), mannitol (e.g., Apium graveolens; Noiraud et al., 2001), or myo-inositol (e.g., ice plant [Mesembryanthemum crystallinum]; Nelson et al., 1998). Interestingly, this last group of plants, which transports sucrose in combination with linear or cyclic polyols, seems to benefit from this specialized metabolism and different mode of phloem transport under conditions of drought or salinity stress, when polyols delivered from the phloem can be accumulated as osmolytes (Bohnert et al., 1995).
The long-distance allocation and partitioning of linear and cyclic polyols depends on the presence of specialized biosynthetic machineries in the source leaves of the respective plant species and on the activity of transport proteins that are able to load mannitol, sorbitol, or inositol into the phloem of these plants. Genes or cDNAs encoding transporters for polyol substrates were first cloned from plants that translocate polyols in their phloem (Chauhan et al., 2000; Noiraud et al., 2001; Gao et al., 2003; Ramsperger-Gleixner et al., 2004). However, plants like Arabidopsis that do not transport sugar alcohols in their phloem were shown to possess several genes encoding the very same types of transporters, and, surprisingly enough, at least some of the encoded proteins localize to the Arabidopsis vascular tissue (Klepek et al., 2005; Schneider et al., 2006, 2007).
Arabidopsis has six genes with significant similarity to genes encoding transporters for linear polyols in polyol-translocating plants (PLT genes; Klepek et al., 2005) and four genes for putative transporters for cyclic polyols (INT genes; Schneider et al., 2006). So far, the functional characterizations of one of the PLT proteins, PLT5 (Klepek et al., 2005; Reinders et al., 2005), and of two of the INT proteins, INT2 and INT4 (Schneider et al., 2006, 2007), have been published. All three proteins were shown to localize to the plasma membrane and to catalyze the transport of linear (PLT5) or cyclic (INT2 and INT4) polyols. However, whereas INT2 and INT4 turned out to be highly specific for the transport of different inositol epimers (Schneider et al., 2006, 2007), PLT5 was shown to accept different substrates, including linear polyols of different chain lengths (C3 to C6), pentoses, hexoses, and even myo-inositol (Klepek et al., 2005; Reinders et al., 2005).
The monosaccharide-transporter(-like) gene family of Arabidopsis has >50 members, and most of the genes analyzed to date encode plasma membrane transporters (Büttner, 2007). One of these proteins, At1g16150, has significant homology with a putative hexose transporter found in the inner chloroplast envelope from spinach (Spinacea oleracea; Weber et al., 2000) and may also localize to chloroplast membranes of Arabidopsis. Another protein, encoded by At1g79820, was localized to the Golgi membrane system (Wang et al., 2006), and only recently, two tonoplast-localized glucose transporters, TMT1 (At1g20840; Wormit et al., 2006) and VGT1 (At3g03090; Aluri and Büttner, 2007), were identified and functionally characterized.
So far, no transport proteins for polyols have been characterized from plant endomembranes. Although Chauhan et al. (2000) reported on the vacuolar localization of two putative Na+/myo-inositol symporters from ice plant (ITR1 and ITR2), later analyses by Schneider et al. (2006, 2007) demonstrated that these two proteins most likely represent orthologs of At INT2 and At INT4, which were characterized as plasma membrane–localized H+/inositol symporters. Meanwhile, the sequence of a third, less closely related protein from ice plant has been deposited in publicly accessible databases (ITR3). Sequence comparisons suggest that this protein is the ortholog of the last, yet uncharacterized, putative myo-inositol transporter from Arabidopsis, INT1. In fact, in an approach to characterize the vacuolar proteome of Arabidopsis, INT1 was listed as a potentially tonoplast-localized protein (Carter et al., 2004); however, the same list also contained the well-characterized, plasma membrane–localized hexose transporter STP1 (At1g11260; Sauer et al., 1990; Sherson et al., 2000).
Here, we present a detailed characterization of INT1, a member of the Arabidopsis INT family. We report on the characterization of the functional and physiological properties of this protein by heterologous expression of its cDNA in baker's yeast (Saccharomyces cerevisiae) and by analyses of two independent int1 mutant lines, on the subcellular localization of an INT1-GFP (for green fluorescent protein) fusion, and of the tissue specificity of the INT1 promoter. Finally, we present patch-clamp analyses of isolated plant vacuoles that characterize INT1 as a tonoplast-localized transporter that is capable of mediating H+ symport of myo-inositol. The physiological role of INT1 is discussed.
RESULTS
The Arabidopsis INT1 Protein Sequence
Cloning of a full-length INT1 cDNA has been described previously (Schneider et al., 2006). Analyses of these sequence data showed that the INT1 gene (At2g43330) is interrupted by four introns and encodes an mRNA with an open reading frame (ORF) of 1527 bp. The INT1 protein (509 amino acids) has a predicted molecular mass of 54.2 kD, a pI of 5.05, and 12 predicted transmembrane helices. It is significantly smaller than the two previously characterized Arabidopsis inositol transporters, INT2 and INT4, that have molecular masses of 62.9 kD (pI = 8.70) and 63.4 kD (pI = 8.25), respectively. This size difference is mainly due to an elongated, Cys-rich loop that is found between the predicted transmembrane helices IX and X. The Cys-rich region (69 amino acids) of this loop is present in INT2 and INT4 but missing from INT1.
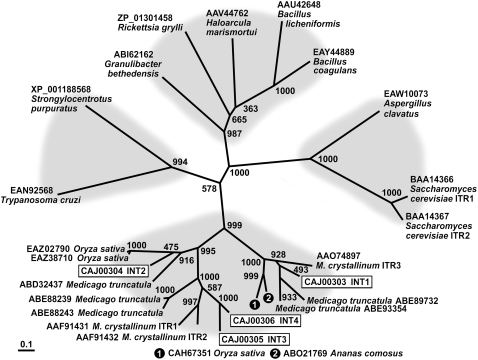
Within the monosaccharide-transporter(-like) superfamily of Arabidopsis, INT1 clearly clusters together with the three other INT proteins (Büttner, 2007). The phylogenetic tree shown in Figure 1, however, which was calculated with all currently available INT homologs from higher plants and with a selection of related sequences from other organisms, shows that INT1 belongs to an independent subgroup of proteins that is clearly separated from a second subgroup of plant inositol transporters. INT2 to INT4 from Arabidopsis and the previously described inositol transporters from ice plant (Chauhan et al., 2000) are part of this second subgroup. Obviously, transporters of the INT1 type are also found in other plant species, including rice (Oryza sativa; CAH67351), ice plant (ITR3; AAO74897), barrel clover (Medicago truncatula; ABE89732 and ABE93354), and pineapple (Ananas comosus; ABO21769). Finally, this tree demonstrates that INT-related proteins exist in archaea, bacteria, fungi, and animals. From these nonplant transporters, only the inositol transporters from baker's yeast (ITR1 and ITR2) were functionally characterized (Nikawa et al., 1991).
Figure 1.
Phylogenetic Tree of the INT Family of Arabidopsis Inositol Transporters and Related Transport Proteins from Other Species Using Maximum Likelihood.
The phylogenetic tree was calculated from confirmed or predicted inositol transporter sequences. The tree reveals four different clusters: metazoa and protozoa on the left, fungi on the right, bacteria and archaea at the top, and plants, with two clearly distinguishable subgroups, at the bottom. Bootstrap samples of 1000 samplings are shown at each branch. GenBank accession numbers and names of the organisms are given plus, where available, the published name of the protein. Arabidopsis INT proteins are boxed.
INT1 Mediates the Accumulation of myo-Inositol in S. cerevisiae
The functional properties of the previously described inositol transporters from Arabidopsis, INT4 (Schneider et al., 2006) and INT2 (Schneider et al., 2007), had been studied by complementation of a baker's yeast mutant (S. cerevisiae strain D458-1B; Nikawa et al., 1991) deficient in myo-inositol uptake (Δitr1) and biosynthesis (Δino1). Wild-type yeast strains possess two genes for inositol transporters, ITR1 and ITR2. Whereas ITR2 is constitutively expressed and the encoded protein catalyzes myo-inositol uptake at only low rates, ITR1 expression is induced by low medium concentrations of inositol, and ITR1-driven myo-inositol uptake occurs at high rates (Nikawa et al., 1993; Miyashita et al., 2003). Both proteins, ITR1 and ITR2, are plasma membrane–localized (Nikawa et al., 1991), and like INT2 and INT4 from Arabidopsis, they possess a long loop between their predicted transmembrane helices IX and X. A yeast transporter of the INT1 type has not been identified.
An INT1 cDNA was expressed in the sense and antisense orientations in the D458-1B yeast mutant. When the resulting yeast strains SSY36 (sense orientation) and SSY37 (antisense orientation) were grown on media with different myo-inositol concentrations, no functional complementation of the Δitr1 mutation was seen on media with low concentrations of myo-inositol (Figure 2A). By contrast, the growth of SSY36 cells appeared to be even slightly slower than the growth of SSY37. This might be the consequence of (1) partially leaky or destabilized membranes, which might result from the insertion of a foreign or potentially misfolded membrane protein; (2) an exporter activity of INT1 in the plasma membrane that would further reduce the already low cytoplasmic concentrations in this mutant; or (3) the targeting of INT1 to an internal membrane system and the removal of myo-inositol from the cytoplasm.
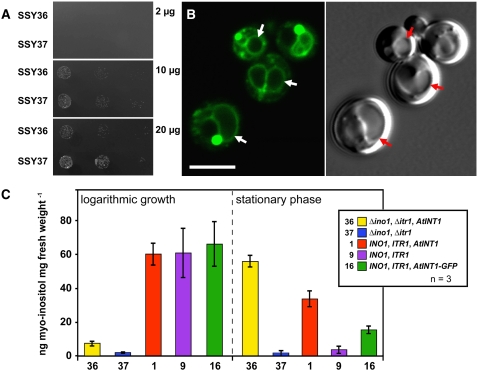
Figure 2.
Characterization of the Δitr1 Δino1 Yeast Double Mutant Expressing the INT1 cDNA in the Sense (SSY36) or Antisense (SSY37) Orientation.
(A) Growth of SSY36 and SSY37 on minimal media supplemented with the indicated concentrations of myo-inositol.
(B) Subcellular localization of INT1-GFP in SSY16 cells. The images show yeast cells with one or more vacuoles that were photographed either under GFP excitation light (confocal section; left) or under white light (Nomarski section; right). Vacuoles of some cells are marked with arrows. Bar = 5 μm.
(C) Ion chromatographic analyses of myo-inositol concentrations determined in whole-cell ethanol extracts of SSY1 (1), SSY9 (9), SSY16 (16), SSY36 (36), and SSY37 (37) cells that were grown in liquid medium with 10 μg/mL myo-inositol and harvested either during the logarithmic growth phase or in the stationary phase. The genotypes of the different yeast strains are shown in the inset.
To discriminate between these different possibilities, we analyzed the intracellular localization of INT1 in the yeast wild-type strain SEY2102 (Emr et al., 1983), which carried a construct for an INT1 protein fused to the N terminus of the GFP. This INT1-GFP–expressing strain was named SSY16. Figure 2B demonstrates that INT1-GFP localizes preferentially to the yeast vacuoles (arrows). Labeling of the plasma membrane was not detected. This suggested that INT1 might be a tonoplast protein. In some cells, additional dot-like structures showed strong GFP labeling. These structures might be aggresomes that are known to occur sometimes upon overexpression of foreign proteins (García-Mata et al., 1999).
We next determined the total concentrations of myo-inositol in various yeast strains at different growth stages (Figure 2C). In particular, we studied the SEY2102 (wild type)–derived strains SSY1 (INO1, ITR1, INT1), SSY9 (INO1, ITR1), and SSY16 (INO1, ITR1, INT1-GFP) and the D458-1B (mutant)–derived strains SSY36 (Δino1, Δitr1, INT1) and SSY37 (Δino1, Δitr1). During logarithmic growth in liquid medium supplemented with myo-inositol (10 μg/mL; all strains harvested at an OD600 of 2.0), all wild-type–derived strains showed comparable and high intracellular concentrations of myo-inositol, whereas both mutant-derived strains showed significantly lower concentrations. Interestingly, however, in all analyses (Figure 2C), the INT1-expressing strain SSY36 had at least fivefold higher concentrations of myo-inositol than the SSY37 control.
This difference became even more obvious when similar analyses were performed on stationary-phase cells. In these analyses, all cells that expressed INT1 or INT1-GFP showed strongly increased myo-inositol concentrations, whereas both controls (wild-type SSY9 and mutant SSY37) did not. This demonstrated that INT1 and INT1-GFP are functional myo-inositol transporters in yeast and that their activity is responsible for the observed accumulation of myo-inositol. The low myo-inositol concentration observed in stationary-grown SSY9 cells may reflect either reduced myo-inositol biosynthesis as a consequence of reduced glucose availability (Granot and Snyder, 1991) or reduced activity of the yeast transporters ITR1 and ITR2.
To test whether the observed myo-inositol accumulation (Figure 2C) results from sequestration within the yeast vacuoles, uptake analyses were performed with [3H]myo-inositol on isolated yeast vacuoles. As a control for the integrity and functionality of the vacuoles, we determined the ATP-dependent uptake of [14C]Lys. This amino acid is imported into yeast vacuoles by a Lys/H+ antiporter that is energized by the vacuolar H+-ATPase (Sato et al., 1984). Although [14C]Lys uptake could be measured, transport rates for [3H]myo-inositol were too low for further analyses of INT1 transport activities.
INT1 Localizes to the Tonoplast in Arabidopsis and Tobacco
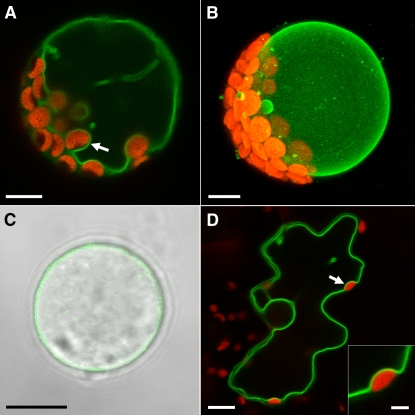
The subcellular localization of INT1 was also studied in planta by transient expression of a construct (pSS21) that encoded an INT1 protein with a C-terminal GFP fusion. pSS21 was used for polyethylene glycol–mediated transformation of Arabidopsis mesophyll protoplasts and for biolistic transformation of tobacco epidermis cells. In Arabidopsis protoplasts (Figures 3A and 3B) and in tobacco epidermis cells (Figure 3D), INT1-GFP–derived fluorescence was seen exclusively in tonoplast membranes. After careful lysis of the plasma membranes of Arabidopsis protoplasts by osmotic shock, intact and fluorescent vacuoles could be observed (Figure 3C).
Figure 3.
Subcellular Localization of INT1-GFP in the Tonoplast Membranes of Arabidopsis Protoplasts or of Tobacco Epidermis Cells.
(A) Confocal section of an intact Arabidopsis protoplast expressing the INT1-GFP fusion construct. The arrow shows the tonoplast membrane passing a chloroplast on the inner side.
(B) Projection of 40 confocal sections taken from an Arabidopsis protoplast expressing the INT1-GFP fusion construct.
(C) Confocal section of a hatched Arabidopsis vacuole after lysis of the plasma membrane (fluorescence and white light images merged).
(D) Confocal section of a tobacco epidermis cell bombarded with the INT1-GFP construct. The arrow marks the tonoplast membrane at the inner side of a chloroplast (enlarged in the inset).
Red fluorescence in (A), (B), and (D) shows chlorophyll autofluorescence. Bars = 8 μm in (A) to (D) and 2 μm in the inset of (D).
These results demonstrated that INT1 is a protein of Arabidopsis tonoplast membranes. In combination with the accumulation data obtained for INT1-GFP–expressing yeast cells (Figure 2C) that demonstrate that INT1-GFP is a functional myo-inositol transporter, a mistargeting of the INT1-GFP fusion due to the C-terminal tag is quite unlikely.
Tissue- and Organ-Specific Expression of INT1
For analyses of the tissue specificity of INT1 expression, we generated and analyzed INT1 promoter/GUS (for β-glucuronidase) and INT1 promoter/GFP plants. A 1210-bp promoter fragment was used to drive the expression of GUS or GFP in plants that had been selected for BASTA resistance after transformation with the plasmids pSS14 (INT1 promoter/GUS) and pSS29 (INT1 promoter/GFP). For both constructs, 20 transformed lines were analyzed.
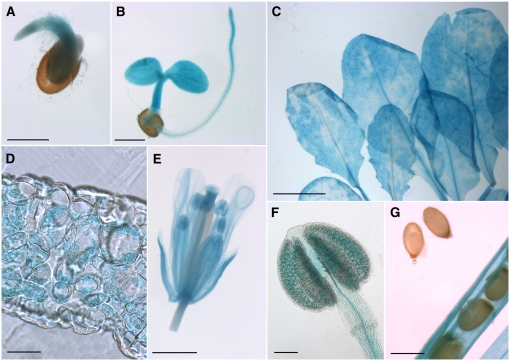
None of the INT1 promoter/GFP plants showed detectable GFP fluorescence, suggesting that the activity of the INT1 promoter is rather low. This was supported by published microarray analyses (http://www.bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi) and by the results obtained from the INT1 promoter/GUS lines (Figure 4). GUS histochemical staining was found in almost all cells and all tissues at all developmental stages. Germinating (Figure 4A) or young seedlings (Figure 4B), rosette leaves (Figure 4C), and flowers (Figure 4E) showed comparable intensities of GUS staining. In cross sections of rosette leaves, this staining was found in the mesophyll but was lacking in both epidermis cell layers (Figure 4D). Flowers showed less intense staining in petals than in most other floral tissues (Figure 4E), and GUS staining seen in anthers was restricted mainly to pollen grains. Surprisingly, no GUS staining was seen in developing seeds at any developmental stage (Figure 4G; see Supplemental Figure 1 online). In principle, these results confirmed data from publicly accessible microarray analyses that also described low and ubiquitous expression of INT1 in all tissues and organs (http://www.genevestigator.ethz.ch/ and http://bbc.botany.utoronto.ca). The only difference observed was the complete absence of GUS staining in seeds, which was not confirmed by the microarray data.
Figure 4.
Analyses of GUS Histochemical Staining in INT1 Promoter/GUS Plants.
(A) and (B) Germinating (A) and young (B) seedling with GUS staining in the cotyledons, the hypocotyl, and the root.
(C) Rosette leaves showing uneven, cloudy GUS staining.
(D) Cross section of a rosette leaf with GUS staining in the mesophyll.
(E) Fully developed flower with staining in all floral organs.
(F) Closed anther with weak GUS staining in all cells and stronger staining in the pollen grains.
(G) Silique with almost mature seeds. No GUS staining is detected in the seeds, but it is detected in all other tissues of the silique.
Bars = 50 μm in (D), 100 μm in (F), 0.5 mm in (A), (B), and (G), 1 mm in (E), and 5 mm in (C).
Generation of Anti-INT1 Antisera
For an independent confirmation of the GUS analyses presented in Figure 4, we aimed to generate INT1-specific antibodies and to perform immunolocalization analyses. To this end, a fusion construct (pSS11-AK1) was made of the last 31 C-terminal amino acids (starting with Trp-479 of the INT1 sequence) that was fused to the C terminus of the maltose binding protein (MBP) from Escherichia coli. The resulting MBP-INT1 fusion protein was used to generate polyclonal antisera.
A protein gel blot with anti-INT1 antiserum (αINT1) and with protein extracts from total membrane preparations of INT1-expressing SSY36 yeast cells and of SSY37 control cells is shown in Figure 5. αINT1 labeled a protein band with an apparent molecular mass of ∼48 kD exclusively in the membrane protein fraction of INT1-expressing cells. No band was detected at this molecular mass in extracts of control membranes (SSY37), demonstrating that the identified protein band corresponds to recombinant INT1. The apparent molecular mass of this protein is slightly lower than the molecular mass predicted from the DNA sequence (54.2 kD). This is a typical observation for highly lipophilic membrane proteins that bind an excess of negatively charged SDS molecules, which causes faster movement on SDS gels (Beyreuther et al., 1980; Gahrtz et al., 1994; Barth et al., 2003; Schneider et al., 2006).
Figure 5.
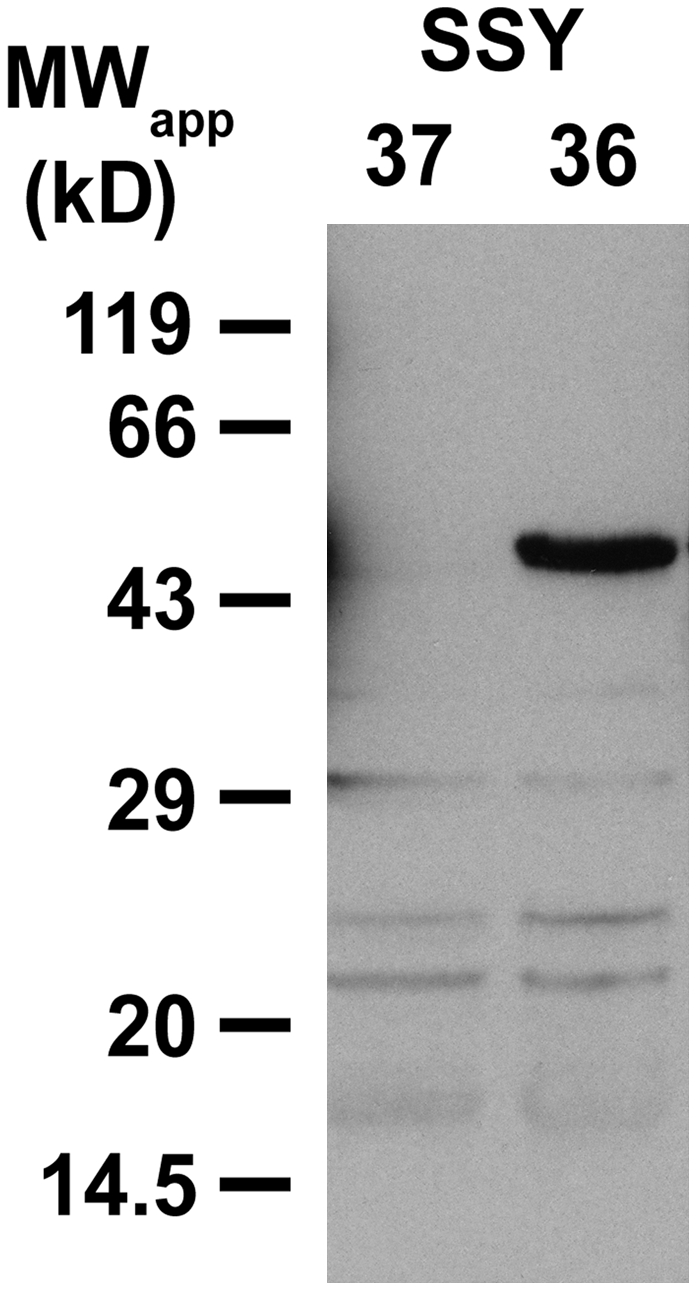
Protein Gel Blot of Total Membrane Proteins from SSY36 and SSY37 Yeast Cells.
Identification of recombinant INT1 protein with affinity-purified αINT1 (dilution, 1:10) in SDS extracts from yeast total membranes (10 μg/lane) after separation on a polyacrylamide gel and transfer to nitrocellulose.
αINT1 was used to immunolocalize INT1 protein in sections from different tissues and organs of Arabidopsis plants; however, we were not able to detect αINT1-specific fluorescence. We also tested the quality of the αINT1 on thin sections of SSY36 and SSY37 cells. These sections were prepared using the identical fixation and embedding protocol that had also been used for the plant material. Again, the αINT1 treatment did not yield any immunolabeling in the INT1-expressing SSY36 cells. These results demonstrate that either the antigenic epitope is destroyed during the fixation procedure or the amount of INT1 is too low for immunodetection in tissue sections.
Characterization of Mutant Lines Carrying T-DNA Insertions in the INT1 Gene
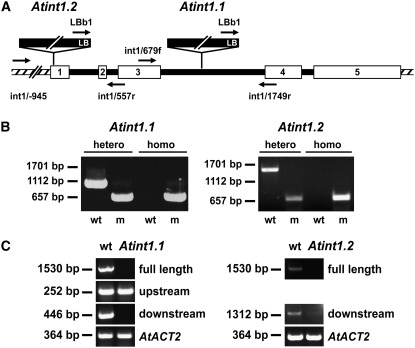
Analyses of publicly accessible collections of T-DNA insertion lines (http://signal.salk.edu/cgi-bin/tdnaexpress) identified two lines (SALK_085400 = int1.1 and SALK_018591 = int1.2) with insertions in the INT1 gene. For both mutant alleles, the position of the T-DNA left border was determined (Figure 6A). In the int1.2 allele, the left border sequence starts upstream from nucleotide 47, and in the int1.1 allele, it starts upstream from nucleotide 1218 (numbers refer to the genomic sequence after the start ATG). Thus, the int1.2 insertion is in the first exon and the int1.1 insertion in the third intron of the INT1 gene.
Figure 6.
Characterization of int1.1 and int1.2 Mutant Plants on the Genomic and mRNA Levels.
(A) Schematic presentation of the INT1 gene (white boxes, exons; black lines, introns; hatched areas, 5′ and 3′ flanking regions). Positions of T-DNA insertions, left borders (LB), and binding sites for the primers (arrows) used in (B) are indicated.
(B) PCR analyses of genomic int1.1 and int1.2 DNA. Heterozygous and homozygous plants were discriminated by PCR performed with wild-type-specific or mutant (m)–specific pairs of primers (int1.1 plants, int1/679f and int1/1749r for the wild type, LBb1 and int1-2/1749r for the mutant; int1.2 plants, int1/-945 and int1/557r for the wild type, LBb1 and int1/557r for the mutant).
(C) Mutant analyses on the mRNA level. Fragments were amplified from wild-type and mutant cDNAs that corresponded to the full INT1 mRNA or to mRNA fragments upstream or downstream from the T-DNA insertions. Control reactions were performed with primers for the Arabidopsis ACT2 cDNA.
For both insertion lines, homozygous plants could be isolated (Figure 6B). PCR analyses performed with cDNA that had been synthesized from total RNA of int1.1 plants showed that the T-DNA insertion in these plants resulted in a complete loss of full-length INT1 mRNA and of RNA sequences downstream from the insertion site (Figure 6C). However, a truncated mRNA corresponding to the region upstream from the insertion site could be identified (Figure 6C). Similarly, a full-length INT1 mRNA was no longer detected in int1.2 plants. In contrast with int1.1 plants, however, a weak band for a truncated downstream mRNA was identified in int1.2 plants (Figure 6C). This may result from a promoter activity in the left border region of the T-DNA. No PCR was performed to identify a truncated mRNA from the upstream region, as the insertion was too close to the 5′ end of the gene in the int1.2 allele.
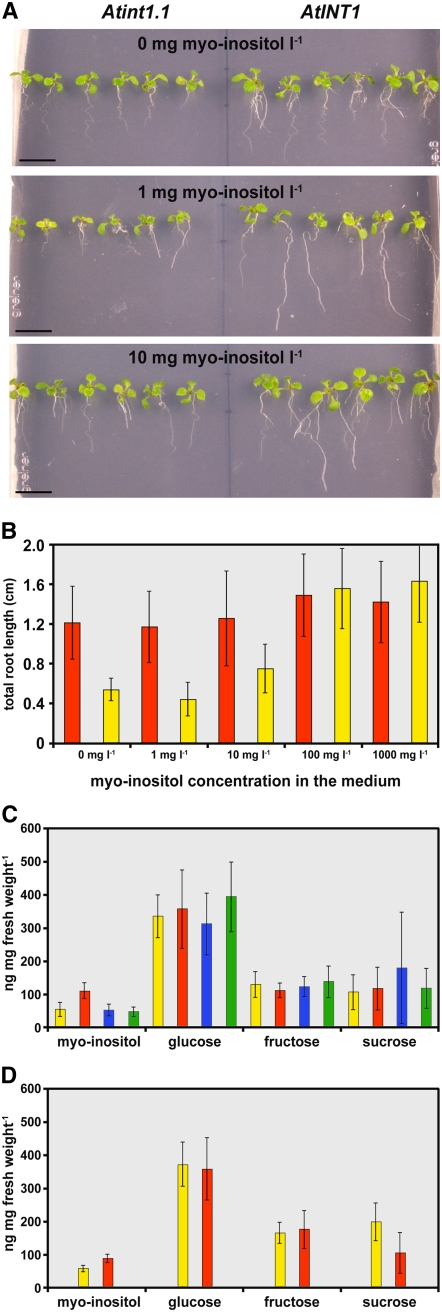
Homozygous int1.1 and int1.2 plants were used for analyses of potential phenotypic differences between mutant and wild-type plants. Germination analyses of seeds on agar medium, comparisons of plant growth on soil, and analyses of soil-grown plants after NaCl stress or cold treatment showed no differences. However, for int1.1 and int1.2 plants, reduced root lengths were detected on agar medium. Figure 7A shows 14-d-old seedlings that were grown on Murashige and Skoog (MS) medium (Murashige and Skoog, 1962) with the indicated concentrations of myo-inositol. At all three concentrations, wild-type plants developed longer roots than int1.1 plants. For the experiment shown in Figure 7B, the total root length (main and lateral roots) of 12 int1.1 plants and of 12 wild-type plants was determined. The data demonstrate that at low concentrations of myo-inositol, the total root length of int1.1 plants is only ∼50% of the total root length of wild-type plants. With increasing concentrations, however, this difference became smaller and finally disappeared at myo-inositol concentrations of 100 mg/L or higher (Figure 7B). Root lengths in wild-type plants were not affected by the different myo-inositol concentrations.
Figure 7.
Phenotypic Characterization of int1.1 and int1.2 Mutant Plants.
(A) Comparison of the root growth of int1.1 and wild-type seedlings (14 d old) on synthetic medium supplemented with the indicated concentrations of myo-inositol (mg/L).
(B) Quantitative analysis of root lengths of int1.1 (yellow) and wild-type (red) seedlings (10 d old) on synthetic medium supplemented with the indicated concentrations of myo-inositol (n = 12; ±sd).
(C) Comparison of myo-inositol, glucose, fructose, and sucrose concentrations in ethanol extracts from rosette leaves of int1.1 plants (red), of wild-type plants (yellow), and of int2.1 (blue) and int4.2 (green) mutant plants (n = 9; ±sd) that were grown, harvested, and analyzed in parallel.
(D) A similar data set for extracts (n = 10) of independently grown and analyzed int1.2 (red) and wild-type (yellow) plants. The significance of the difference between the inositol concentrations in int1.2 and wild-type extracts was tested with Student's t test (P = 4 × 10−6).
These data suggest that int1.1 plants suffer from a reduced availability of myo-inositol. Therefore, we determined the myo-inositol concentrations in ethanol extracts from rosette leaves of 75-d-old int1.1 and wild-type plants. Surprisingly, the myo-inositol concentration in these leaves was ∼2-fold higher than in wild-type leaves (Figure 7C). For comparison, we also determined the myo-inositol concentrations of rosette leaves of int2.1 (Schneider et al., 2007) and int4.2 (Schneider et al., 2006) mutant plants of the same age. These mutants carry T-DNA insertions that cause null mutations in the INT2 and INT4 genes, respectively. The analyzed plants showed the identical myo-inositol concentrations as wild-type plants (Figure 7C). In contrast with the myo-inositol concentrations, the concentrations of glucose, fructose, and sucrose were almost identical in all mutant lines and in wild-type plants (Figure 7C). The difference in total myo-inositol concentrations observed between int1.1 and wild-type plants was also seen when younger plants were analyzed or with plants after cold treatment or NaCl stress (data not shown). Moreover, this difference was observed in all double mutants that carried the int1.1 mutation (e.g., in int1.1 int2.1) but never in double mutants in which the INT1 gene was intact (e.g., in int2.1 int4.2; data not shown).
Finally, analyses of the total myo-inositol concentrations in rosette leaves of int1.2 plants showed a similar, although less pronounced, increase (Figure 7D). This smaller increase in the myo-inositol concentration in int1.2 plants might be explained by an observation already mentioned above. In contrast with int1.1, which seems to be a real knockout allele (Figure 6C), int1.2 plants possess a truncated mRNA corresponding to sequences downstream from the T-DNA insertion site (Figure 6C). Therefore, they may still have some residual INT1 transport activity.
The observed reduced availability of myo-inositol, the inositol-dependent root growth, and the simultaneous increase in cellular inositol can only be explained by the sequestration of inositol in an intracellular compartment, most likely within the vacuoles.
Patch-Clamp Analyses of Isolated Vacuoles from Arabidopsis Plants
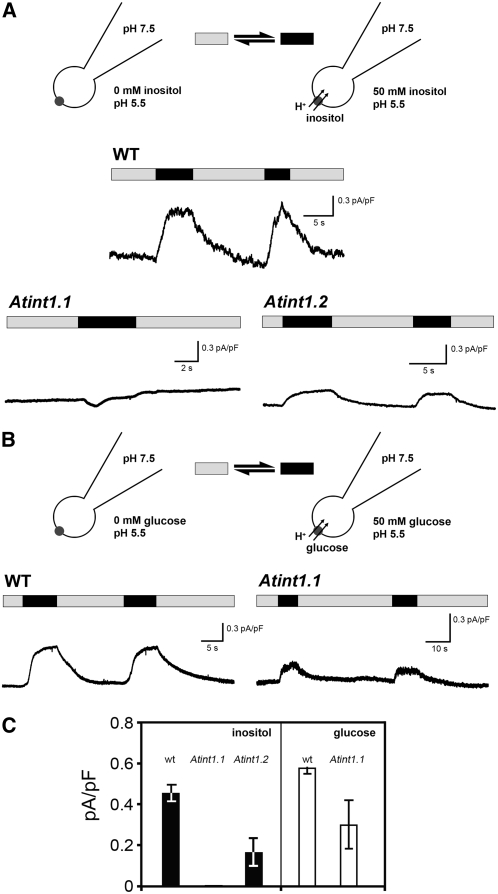
The data presented so far show a tonoplast localization of INT1 in yeast (Figure 2) and different plant species (Figure 3). Moreover, the results obtained from T-DNA insertion mutants (Figure 7) suggest that the role of INT1 might be the release of myo-inositol from vacuoles. To further elucidate the mechanism of INT1-dependent inositol transport across the vacuolar membrane, the patch-clamp technique was applied to isolated mesophyll vacuoles of different Arabidopsis lines. As only low currents were expected from a putatively H+-driven transporter, all analyses were performed in the whole vacuole configuration. Under these conditions, neither the pH nor the substrate concentrations in the vacuolar lumen can be changed after the tonoplast is attached to the patch pipette. Therefore, we established an inverse system with pH 7.5 in the patch pipette (vacuolar lumen) and pH 5.5 in the bath (cytoplasm) (Figure 8A). This approach allows changes in the extravacuolar inositol concentrations. In previous studies (Carpaneto et al., 2005), we could demonstrate that H+-driven symporters of the major facilitator superfamily (Marger and Saier, 1993) can work in this inversed mode, as expected from an ideal thermodynamic machine. In other words, H+ symporters can switch from a H+-driven substrate flux to a substrate-driven H+ flux if appropriate conditions are applied. A similar behavior was predicted for INT1; therefore, high extravacuolar inositol concentrations were expected to elicit H+ currents into the vacuolar lumen.
Figure 8.
Whole-Vacuole Recordings of Currents Triggered by myo-Inositol in Mesophyll Vacuoles of INT1 Wild-Type and int1 Mutant Plants.
(A) The stylized presentation of vacuoles with attached patch pipettes shows the experimental conditions in the absence (gray bars) or presence (black bars) of myo-inositol in the bath medium. Cytosolic application of 50 mM myo-inositol (black bars) resulted in INT1-mediated H+ currents into the vacuoles of wild-type plants, as represented by the upward deflection of the current traces. Under the same experimental conditions, no or significantly smaller myo-inositol–induced H+ currents were observed in int1.1 or int1.2 mutant plants, respectively. Note that INT1-dependent currents were determined in the inverse mode (i.e., pH values in the extravacuolar bath and the patch pipette solution were adjusted to 5.5 and 7.5, respectively).
(B) Similar measurement as in (A), but with 50 mM glucose on vacuoles from wild-type and int1.1 mutant plants.
(C) Average currents calculated from several experiments (n = 3; ±sd) performed under identical conditions as the original recordings shown in (A) and (B). Reproducibly, inositol induced no current (= 0 pA/pF) in vacuoles of int1.1 plants.
In fact, when the inositol concentration in the bath medium of vacuoles from Arabidopsis wild-type plants was raised from 0 to 50 mM (i.e., to a most likely saturating concentration), immediate positive currents were observed (Figures 8A and 8C). By convention (Bertl et al., 1992), currents in this direction are defined as currents out of the cytoplasm (i.e., in this case into the vacuole). This result suggests that INT1 transports myo-inositol and H+ out of the vacuole.
When identical analyses were performed on vacuoles from int1.1 mutant plants, this current could not be resolved (Figures 8A and 8C). As expected from the data presented in Figure 6C (which shows truncated int1.2 mRNA) and in Figure 7D (which shows a less pronounced change in the cellular inositol concentration of int1.2 mutant leaves), analyses of vacuoles from the int1.2 mutant yielded detectable currents that were significantly lower than those measured in the wild type (Figures 8A and 8C). These data confirm that int1.1 represents a knockout mutant, whereas int1.2 is a knockdown mutant.
The comparative analyses of cellular sugar and inositol concentrations in wild-type and different knockout plants (Figure 7C) revealed changes only for myo-inositol. Cellular glucose and fructose concentrations were not affected, suggesting that these hexoses are not substrates for INT1. This suggested that INT1 might be myo-inositol–specific, just like the related plasma membrane transporters INT2 and INT4. Nevertheless, we compared the capacity of vacuoles from wild-type and int1.1 mutant plants to mediate the efflux of glucose (Figures 8B and 8C). Obviously, glucose transport activities could be determined in vacuoles from both plants.
The presented patch-clamp data independently confirm that INT1 is a tonoplast-localized inositol transporter. Most importantly, however, they demonstrate that INT1 mediates the symport of myo-inositol and protons across the tonoplast. Na+ can be excluded as a potentially cotransported ion, as sodium salts were absent from both the bath solution and the buffer in the patch pipette.
DISCUSSION
Higher plant vacuoles occupy up to 95% of the volume of mature cells, represent the main storage compartment for solutes, are involved in the regulation of cell volume and turgor, mediate essential steps in the degradation of cellular components, and play a key role in cellular ion homeostasis. Moreover, vacuoles are indispensable for the deposition of secondary metabolites (Marinova et al., 2007) and for multiple detoxification processes, including heavy metals and xenobiotics (Wink, 1993). For all of these different tasks, vacuoles depend on specific and efficient transport proteins that are embedded into their membrane, the tonoplast (Martinoia et al., 2007). Despite these numerous essential functions of the tonoplast for plant growth and development, few tonoplast proteins have been functionally characterized. To date, only four solute transporters for amino acids or sugars were described in Arabidopsis: one amino acid transporter (CAT2; Su et al., 2004), two different monosaccharide transporters (TMT1; Wormit et al., 2006; VGT1; Aluri and Büttner, 2007), and one transporter for the disaccharide sucrose (SUC4; Endler et al., 2006). In sugar beet (Beta vulgaris), another vacuolar solute transporter has been identified (Chiou and Bush, 1996) that is similar to proteins (At1g75220 and At1g19450) of a so far uncharacterized subgroup of the Arabidopsis monosaccharide-transporter(-like) gene family (Büttner, 2007).
During the last few years, several groups performed analyses of the higher plant vacuolar proteome (Carter et al., 2004; Shimaoka et al., 2004; Szponarski et al., 2004; Endler et al., 2006; Jaquinod et al., 2007; Schmidt et al., 2007) that resulted in the identification of new, potentially tonoplast-localized solute transporters. However, detailed analyses will be necessary for each of these newly identified proteins to unequivocally confirm or disprove its vacuolar localization and its physiological function.
INT1 Is a Tonoplast-Localized, Energy-Dependent Transporter for myo-Inositol
Here, we present the characterization of INT1, a novel, tonoplast-localized transporter for myo-inositol from Arabidopsis. Intracellular localization of the protein was suggested by expression analyses in baker's yeast, Arabidopsis, and tobacco, where an INT1-GFP fusion is targeted to tonoplast membranes (Figures 2B and 3). Moreover, the presence of INT1 or INT1-GFP resulted in intracellular accumulation of myo-inositol in yeast (Figure 2C). These increased myo-inositol concentrations might result from the activity of recombinant, tonoplast-localized INT1 transporters that catalyze the accumulation of myo-inositol inside the yeast vacuoles. Alternatively, however, they might be mediated by a small amount of INT1 transporters that are mistargeted to the yeast plasma membrane. Previous publications repeatedly showed that plant tonoplast proteins can be targeted incorrectly to the yeast plasma membrane. Examples are the Arabidopsis tonoplast sucrose transporter SUC4 (Weise et al., 2000; Endler et al., 2006) and the Arabidopsis tonoplast water channels δ-TIP and γ-TIP (Klebl et al., 2003). However, because all attempts to study INT1 transport activities in intact yeast cells or in isolated yeast vacuoles failed, these analyses can only be used as a first direct evidence for an inositol transport activity of INT1 and INT1-GFP.
Attempts to express INT1 in Xenopus laevis oocytes for more detailed analyses of substrate specificity, kinetic properties, or energy dependence of INT1 were not successful, and expression of an INT1-GFP fusion construct in Xenopus oocytes demonstrated that the fusion protein did not localize to the plasma membrane (U. Hammes, P. Udvardi, and N. Sauer, unpublished data). Therefore, it was not possible to study the transport properties of INT1 in this expression system. However, the results obtained from Xenopus oocytes with the closely related plasma membrane–localized transporters INT2 and INT4 (Schneider et al., 2007) suggest that INT1 may also accept other naturally occurring inositols, such as scyllo-, chiro-, and muco-inositol.
Due to the lack of detectable INT1 transport activity in these well established expression systems, we aimed to characterize the function of INT1 by comparing mesophyll vacuoles isolated from wild-type plants or from transgenic Arabidopsis lines that carried a T-DNA insertion in the INT1 gene (int1.1 plants).
The results obtained from these analyses identified a myo-inositol transport activity that was present in wild-type plants but absent from int1.1 plants (Figures 8A and 8C). This demonstrates (1) that this activity can be attributed to the INT1 protein and (2) that INT1 is targeted to the tonoplast with (Figure 3) or without a C-terminal GFP fusion. Second, these analyses characterize INT1 as an energy-dependent transporter that generates inositol-dependent currents into the vacuoles of wild-type plants. However, as already pointed out above, for experimental reasons the patch-clamp analyses shown in Figure 8 were performed in the inverse mode (i.e., at high vacuolar and low cytoplasmic pH values). This allows changes in the myo-inositol concentrations during studies of a possible symporter activity of INT1. Previous characterizations of the closely related INT2 and INT4 proteins as H+/inositol symporters of the plasma membrane (Schneider et al., 2006, 2007) suggested an H+ symporter activity also for INT1. Under physiological conditions (i.e., low pH inside the vacuole, neutral pH in the cytoplasm, and a membrane potential that is negative on the cytoplasmic side), H+/inositol symport is expected to occur only from the vacuolar lumen into the cytoplasm. In contrast with the complete absence of recordable currents from int1.1 vacuoles, patch-clamp analyses on int1.2 vacuoles revealed low inositol-dependent currents that were ∼30% of the current detected in wild-type vacuoles (Figure 8C). Such low, residual currents were expected, as other data (Figures 6C and 7D) had suggested that int1.2 mutants carry a knockdown rather than a knockout allele.
The complete absence of inositol currents in int1.1 vacuoles (Figures 8A and 8C) demonstrates that INT1 is the only inositol exporter at the tonoplast, a fact that was most likely indispensable for the identification of the clear differences between vacuoles from wild-type and knockout plants (Figure 8C). Surprisingly enough, the comparison of glucose-induced currents in wild-type and int1.1 vacuoles (Figures 8A and 8C) showed reduced currents for this substrate in mutant vacuoles (50% of the wild type; Figure 8C). However, whether this reduction is due to the absence of INT1 that may have some marginal glucose transport activity, or whether INT1 is highly specific for inositol and the observed reduction is an indirect effect (e.g., of the altered inositol availability in the cytoplasm; see analyses in Figure 7), cannot be said. This question can only be answered in the absence of interfering transport activities of other tonoplast transporters. Crosses with knockout lines of already characterized and possibly yet to be identified tonoplast transporter genes will be necessary for detailed analyses of substrate specificities in planta.
Nevertheless, two lines of evidence suggest that INT1 is most likely a specific transporter for inositols: (1) the previously characterized transporters INT2 and INT4 are highly specific for different inositol epimers (Schneider et al., 2006, 2007); and (2) the int1.1 knockout line shows altered concentrations of inositol but not of glucose or fructose (Figure 7).
Our patch-clamp data obtained from reverse mode analyses are in agreement with the predicted physiological function of INT1 as a H+/inositol exporter from the vacuolar lumen into the cytoplasm.
INT1 Drives myo-Inositol Export from Arabidopsis Vacuoles
In contrast to INT2 and INT4, which are both expressed in specific regions of the vascular tissue, in pollen grains or in the anther tapetum (Schneider et al., 2006, 2007), INT1 is expressed in almost all cells and tissues of Arabidopsis (Figure 4). This suggests that INT1 has a more general function for the cellular myo-inositol metabolism. The phenotypic differences between int1.1 and int1.2 mutant lines on the one hand and wild-type plants on the other can only be explained by INT1-mediated myo-inositol export from the vacuoles into the cytoplasm, an interpretation that fully agrees with the H+/myo-inositol exporter activity of INT1 observed in plant vacuoles (Figure 8). A defect in a tonoplast-localized importer would be unlikely to cause the observed increase in total myo-inositol concentrations found for both int1 mutants (Figure 7C), nor would it be expected to result in reduced root growth (Figures 7A and 7B), as the mutation would not reduce the amount of myo-inositol available for biosynthetic processes in the cytoplasm. However, the inositol-dependent root growth in int1 mutant plants and the observation that wild-type roots grow normally at all myo-inositol concentrations (Figure 7B) clearly suggest that the lack of INT1 transport activity causes a reduction in the availability of myo-inositol.
What Is the Origin of Vacuolar myo-Inositol?
In higher plants, myo-inositol and its derivatives play multiple essential roles in numerous metabolic pathways under all physiological conditions (Brinch-Pedersen et al., 2006). For example, myo-inositol represents a precursor in the synthesis of the nucleotide sugar uridine diphosphoglucuronic acid, which in turn is used for the synthesis of galacturonic acid, xylose, apiose, and arabinose, important residues of plant cell wall polymers (Loewus and Murthy, 2000; Kanter et al., 2005). Moreover, myo-inositol acts as a carrier of activated galactose that is eventually transferred to sucrose, yielding raffinose and its longer derivatives (Kandler and Hopf, 1982). myo-Inositol has also been discussed as an initial substrate in a newly described biosynthetic route for l-ascorbic acid biosynthesis (Lorence et al., 2004), and it may be conjugated to auxins to prevent their biological activity during long-distance transport (Cohen and Bandurski, 1982). In glycosylphosphatidylinositol membrane anchors (Schultz et al., 1998) and in phospholipids (Lehle, 1990), myo-inositol provides the structural basis for membranes and membrane-attached proteins, and after hydrolysis by phospholipase C, inositol-1,4,5-triphosphate plays an important role as a second messenger. Moreover, the salts of phytate (myo-inositol-1,2,3,4,5,6-hexakisphosphate [IP6]), the product of inositol phosphorylation (Shi et al., 2005), represent storage forms for myo-inositol, phosphorus, and cations in seeds and embryos. Only recently was IP6 identified as a structural component of enzymes or receptors (Macbeth et al., 2005; Tan et al., 2007). Last but not least, IP6 may even represent a form of cellular energy currency (Raboy, 2003).
However, to our knowledge, the uptake of cytoplasmic myo-inositol into plant vacuoles or even accumulation to high concentrations within the vacuoles has not been described. Even in roots of ice plant, where myo-inositol is delivered from the phloem, myo-inositol accumulates in the cytoplasm and not in vacuoles (Nelson et al., 1999). Only one of the myo-inositol–dependent reactions described above may include vacuoles (i.e., the biosynthesis and degradation of IP6). IP6 is thought to be synthesized in the cytoplasm, to be transported into the endoplasmic reticulum lumen, and to move via endoplasmic reticulum–derived vesicles to specialized vacuoles (Greenwood and Bewley, 1984; Otegui et al., 2002). Only recently, an ABC transporter from maize (Zea mays) was described that is potentially involved in the import of IP6 into the endoplasmic reticulum or into the transport vesicles (Shi et al., 2007).
However, IP6 biosynthesis occurs mainly in seeds, where it is deposited as an organic storage form for phosphorus and minerals (Brinch-Pedersen et al., 2002; Raboy, 2003). During the early stages of germination, IP6 is degraded by vacuolar phytases (Nishimura and Beevers, 1978; Brinch-Pedersen et al., 2006) and eventually provides myo-inositol and phosphorus to the rapidly growing seedlings. At this stage, a tonoplast-localized myo-inositol exporter might be needed; in fact, in microarray analyses (http://www.bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi), the highest levels of INT1 expression were detected in Arabidopsis seeds that had been soaked in water for 24 h.
Nevertheless, IP6 or enzymes involved in IP6 biosynthesis were also found in leaves, roots, and tubers of different plant species (Brearley and Hanke, 2000; Bentsink et al., 2003; Phillippy et al., 2003). Comparisons of IP6 concentrations in leaves of different Arabidopsis cultivars revealed maximum concentrations of 25 μg/g (Bentsink et al., 2003).
Even a complete hydrolysis of this IP6 would result in an increase in free myo-inositol concentrations of only 4 μg/g, a value that is too small to account for the observed increase in myo-inositol in int1.1 plants (50 μg/g in Figure 7C). Moreover, it is not entirely clear whether all of the IP6 found in leaves is in vacuoles (Bentsink et al., 2003). However, other hydrolytic activities may also contribute to the production of vacuolar myo-inositol. Phosphatidylinositol 3-phosphate, for example, a phospholipid molecule that is involved in vesicle trafficking also in plant cells, seems to be synthesized in the trans-Golgi network and to move into the central vacuole of Arabidopsis for degradation (Kim et al., 2001). In fact, large amounts of soluble phospholipase D were found in vacuoles of young leaves in Robinia pseudoacacia (Yoshida, 1979) or castor bean (Ricinus communis) (Xu et al., 1996), but the physiological role of this vacuolar phospholipase D is not quite clear. Moreover, vacuolar phosphatases were identified in barley (Hordeum vulgare) mesophyll vacuoles that hydrolyzed inositol 1,4,5-trisphosphate and produced myo-inositol (Martinoia et al., 1993).
The sum of these and other reactions might result in the production of significant amounts of myo-inositol in the vacuolar lumen, and INT1 seems to mediate the release of this recycled myo-inositol into the cytoplasm. A lack of this transporter would then result in the observed phenotype. Similar reactions are likely to occur in all other plant species as well, and the identification of one or several transporters of the INT1 type in all plant species analyzed so far (Figure 1) suggests similar functions for these proteins.
METHODS
Strains and Growth Conditions
Arabidopsis thaliana (Columbia wild type) plants were grown in growth chambers on potting soil or on MS plates under long-day conditions (16 h of light/8 h of dark) at 22°C and 60% relative humidity for most analyses. For ion chromatographic analyses, plants were grown on soil under short-day conditions (8 h of light/16 h of dark). The Saccharomyces cerevisiae strain D458-1B (Nikawa et al., 1991) was used for heterologous expression of INT1 cDNA. The Escherichia coli strain DH5α (Hanahan, 1983) was used for basic cloning steps, and the MBP-INT1 fusion protein for antisera was expressed in E. coli strain Rosetta (Novagen). Transformation of Arabidopsis was performed using Agrobacterium tumefaciens strain GV3101 (Holsters et al., 1980).
cDNA Cloning and Constructs for Yeast Expression
The PCR-based cloning of the INT1 cDNA was described by Schneider et al. (2006). For cloning into the yeast expression vector NEV-N-Leu, NotI restriction sites had been added to both ends of the cDNA. NEV-N-Leu is a modification of the expression vector NEV-N (Sauer and Stolz, 1994), in which the Ura3 selection marker has been replaced by Leu2. D458-1B yeast cells were transformed (Gietz et al., 1992) with NEV-N Leu plasmids containing INT1 cDNA in the sense (pSS50s) and antisense (pSS50as) orientations. The obtained transformants were named SSY36 (sense) and SSY37 (antisense).
INT1 Promotor/GUS and INT1 Promotor/GFP Constructs
A 1210-bp INT1 promoter fragment was PCR-amplified from Arabidopsis genomic DNA using the oligonucleotides INT1-p5 (5′-AAAAATCGAAGCTTCGGTTCAGACC-3′) and INT1-p3 (5′-CGTCAATGCCATGGTTCTTATTAAGCTCTTCTCAATCTG-3′), which introduced N-terminal HindIII and C-terminal NcoI sites. Using these restriction sites, the sequenced promoter fragment was cloned into pAF6 (Klepek et al., 2005). From the resulting plasmid, the INT1 promoter/GUS cassette was excised with HindIII and EcoRI and cloned into the plant transformation vector pGPTVbar (Becker et al., 1992), yielding plasmid pSS14, which was used for Agrobacterium-mediated Arabidopsis transformation (Clough and Bent, 1998). For INT1 promoter/GFP plants, the same promoter fragment was used and cloned into the plasmid pMH4, a pUC19-based plasmid containing the ORF of GFP. The INT1 promoter/GFP cassette was cloned into pGPTVbar as described above, yielding plasmid pSS29.
Transient Expression of INT1-GFP
For transient expression of an INT1-GFP fusion, we used the plasmid pSO35e (Klepek et al., 2005). The INT1 coding sequence was amplified using the primers INT1-5-Pci (5′-ACATGTCATTGACGATCCCAAACG-3′) and INT1-3-Pci (5′-ACATGTTGGCAGATTGAGATCCCTGCTCGAG-3′), which introduced PciI sites at both ends of the INT1 ORF and replaced the stop codon of the original INT1 ORF. This modified INT1 sequence was cloned in-frame to the 5′ end of GFP into the unique NcoI cloning site of pSO35e. The continuous INT1-GFP ORF in the resulting plasmid pSS21 was confirmed by sequencing. pSS21 was used for transient expression of INT1-GFP in Arabidopsis protoplasts (polyethylene glycol transformation; Abel and Theologis, 1994) or in tobacco (Nicotiana tabacum) epidermis cells (particle bombardment; Klepek et al., 2005).
Complementation Analyses in Yeast
Overnight cultures of yeast strains SSY36 and SSY37 were grown to the same OD (OD600), washed with water, and adjusted to a concentration of 5000 cells/μL. Droplets of this cell suspension and of 1:8, 1:64, and 1:512 dilutions were applied to agar plates containing 0.67% yeast nitrogen base, 0.5% glucose, and the indicated concentrations of myo-inositol.
Preparation of Yeast and Plant Material for Ion Chromatographic Analyses
Yeast cells were grown in liquid culture (0.67% yeast nitrogen base, 0.5% glucose, 10 μg/mL myo-inositol) to an OD600 of 2.0 or 4.0, harvested, and washed with water. Per sample, 50 mg fresh weight was used for ethanol extraction. Analyses of sugar concentrations in Arabidopsis tissue were also performed from ethanol extracts. To this end, freshly harvested and immediately frozen plant or yeast material was incubated for 1 h at 80°C with 80% ethanol. After centrifugation, the supernatant was vacuum-dried and resolved in water. This solution was used for ion chromatography.
Ion Chromatography
Sugar and sugar alcohol concentrations were determined in an ICS-3000 system (Dionex), including a gradient pump (ICS-3000 SP), a degasser module, an autosampler (ICS-3000 AS), and a detector compartment (ICS-3000 DC). Anionic compounds were separated with a CarboPac MA1 column (4 × 250 mm) that was connected to a guard column of the same material (4 × 10 mm) and an ATC-1 anion trap column that was placed between the eluent and separation columns to remove the anionic contaminants. The eluent (612 mM NaOH) was made from purest water (Millipore) and 50% NaOH (Fluka). The column was equilibrated at a flow rate of 0.4 mL/min, and the run was 60 min. The calibration and quantitative calculation of carbohydrates was performed using Chromeleon software 6.7 (Dionex).
Analysis of the T-DNA Insertion Lines int1.1 and int1.2
T-DNA insertion lines int1.1 (Salk_085400) and int1.2 (Salk_018591) were identified using the Salk Institute T-DNA express gene-mapping tool (Alonso et al., 2003). Homozygous plants were identified by PCR with genomic DNA using the primers int1/679f (5′-TTTTGATATCGGGTCGTCTCTT-3′) and int1/1749r (5′-AGGGGATTCTGGCATGAAA-3′) for int1.1 and the primers int1/-945 (5′-CGGATTTGTTAAATTTAGGCCTTT-3′) and int1/557r (5′-ATGGCACCAACTAAAGCCATAC-3′) for int1.2, plus the T-DNA left border primer LBb1 (5′-GCGTGGACCGCTTGCTGCAACT-3′).
The primers INT1-5-Pci (5′-ACATGTCATTGACGATCCCAAACG-3′), INT1-3-Pci (5′-ACATGTTGGCAGATTGAGATCCCTGCTCGAG-3′), Int1cs218 (5′-CAGGAAACTATTGTAAGTATGGCTTT-3′), Int1g557r (5′-ATGGCACCAACTAAAGCCATAC-3′), Int1cs774 (5′-TCTTTCAGCGGCTGAAGAGG-3′), and Int1cs1220r (5′-GGGTATATCTCTGAGTTCACCG-3′) were used for RT-PCRs with total RNA from wild-type and mutant Arabidopsis plants. The primers AtACT2g+846f (5′-ATTCAGATGCCCAGAAGTCTTGTT-3′) and AtACT2g+1295r (5′-GAAACATTTTCTGTGAACGATTCCT-3′) were used to amplify the ACT2 mRNA.
Analyses of Arabidopsis Root Lengths
For analysis of root growth, wild-type, int1.1, and int1.2 plants were grown on plates with MS medium (Murashige and Skoog, 1962), and the concentrations of myo-inositol were adjusted as indicated. After 8 d, seedlings were transferred to larger, rectangular plates with identical media and were grown in an upright position. Photographs were taken from plants at 10, 12, and 14 d after sowing. Root length was measured with the ImageJ software package (http://rsb.info.nih.gov/ij/).
Immunohistochemical Techniques and Protein Gel Blot Analyses
A DNA fragment that encoded the 31 C-terminal amino acids of INT1 was generated by annealing two oligonucleotides and cloned into the EcoRI/HindIII-digested vector pMAL-c2 (New England Biolabs), yielding the plasmid pSS11-AK1, which was used to make a MBP/INT1-peptide fusion in E. coli (strain Rosetta; Novagen). Expression of the fusion construct was induced with isopropyl thiogalactoside, and proteins were solubilized with SDS and separated on polyacrylamide gels (Laemmli, 1970) and blotted (Dunn, 1986). Bands were excised, and the protein was extracted and lyophilized. Antisera were generated by Pineda-Antikörper-Service. Purification of anti-INT1 antibodies was performed as described (Sauer and Stadler, 1993).
Protein extracts of total membrane fractions from baker's yeast were prepared as described (Sauer and Stolz, 2000). αINT1-labeled bands were detected with anti-guinea pig IgG-peroxidase conjugate (diluted 1:4000) and the Lumi-Light kit (Roche Diagnostics).
Microscopy and Detection of GFP Fluorescence
Images of GFP fluorescence were made on a confocal laser-scanning microscope (Leica TCS SPII; Leica Microsystems) and processed with the Leica Confocal Software 2.5 (Leica Microsystems). Emitted fluorescence was monitored at detection wavelengths longer than 510 nm.
GUS plants were analyzed with a stereomicroscope (Leica MZFLIII; Leica Microsystems) or a microscope (Zeiss Axioskop; Carl Zeiss). Images were processed using the analySIS Doku 3.2 software (Soft Imaging System).
Patch-Clamp Analyses of Isolated Plant Vacuoles
For patch-clamp experiments in isolated mesophyll vacuoles, protoplasts were isolated by incubation in 0.5% (w/v) cellulase Onozuka R-10 (Serva), 0.05% (w/v) pectolyase Y23 (Seishin), 0.5% (w/v) macerozyme R10 (Serva), 1% (w/v) BSA (Sigma-Aldrich), 1 mM CaCl2, and 10 mM HEPES-Tris, pH 7.4, for 45 min at 23°C and 80 rpm on a rotary shaker. The enzyme solution was adjusted to an osmolality of 400 mosmol/kg with sorbitol. Released protoplasts were filtered through a 50-μm nylon mesh and washed with 400 mM sorbitol and 1 mM CaCl2 (centrifugation at 60g and 4°C for 10 min). Protoplast samples were kept on ice, and aliquots were used for vacuole isolation and patch-clamp experiments. Vacuoles were extruded spontaneously by the protoplasts upon exposure to hypotonic medium (10 mM EGTA, 10 mM HEPES-Tris, pH 7.4, adjusted to 200 mosmol/kg with d-sorbitol).
Patch-clamp experiments on mesophyll vacuoles were performed in the whole-vacuole configuration essentially as described by Schulz-Lessdorf and Hedrich (1995) and Ivashikina and Hedrich (2005). The data acquisition rate was set to 500 μs. Currents were low-pass-filtered at 40 Hz. The vacuolar membrane was clamped to 0 mV. The bath as well as the pipette medium contained 190 mM KCl, 1 mM CaCl2, and 2 mM MgCl2. pH values were adjusted with 10 mM HEPES-Tris, pH 7.5 (vacuolar lumen), or 10 mM MES-Tris, pH 5.5 (cytoplasmic side of the tonoplast membrane).
Phylogenetic Analyses
The sequences used to calculate the phylogenetic tree shown in Figure 1 were aligned with ClustalW2 (http://www.ebi.ac.uk/Tools/clustalw2/index.html) using the default settings (Gonnet matrix, gap open penalty = 10.0, gap extension penalty = 0.2, protein ENDGAP = −1, protein GAPDIST = 4). Results were stored in the PHYLIP output format and used to construct an unrooted phylognetic tree by the maximum-likelihood method using PhyML 2.4.4 (Guindon and Gascuel, 2003) with the JTT model for amino acid substitutions (1000 bootstrap samplings). TreeViewX 0.4.1 for Mac (http://darwin.zoology.gla.ac.uk; Page, 1996) was used for tree viewing. The full alignment used for tree calculation is shown in Supplemental Data Set 1 online.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: INT1, AJ973175; Mc ITR3, AAO74897; unnamed sequence from sugar beet (Chiou and Bush, 1996), U43629.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. GUS Reporter Expression in a Flower of an INT1 Promoter/GUS Plant.
Supplemental Data Set 1. Alignment of the 26 Sequences Used to Calculate the Phylogenetic Tree Shown in Figure 1.
Supplementary Material
Acknowledgments
We thank Enrico Martinoia (University of Zürich) for his help in the large-scale preparation of plant vacuoles. This work was supported by Grant SA 382/13 from the Deutsche Forschungsgemeinschaft to N.S.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Norbert Sauer (nsauer@biologie.uni-erlangen.de).
Online version contains Web-only data.
References
- Abel, S., and Theologis, A. (1994). Transient transformation of Arabidopsis leaf protoplasts: A versatile experimental system to study gene expression. Plant J. 5 421–427. [DOI] [PubMed] [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Aluri, S., and Büttner, M. (2007). Identification and functional expression of the Arabidopsis thaliana vacuolar glucose transporter 1 and its role in seed germination and flowering. Proc. Natl. Acad. Sci. USA 104 2537–2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth, I., Meyer, S., and Sauer, N. (2003). PmSUC3: Characterization of a SUT2/SUC3-type sucrose transporter from Plantago major. Plant Cell 15 1375–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker, D., Kemper, E., Schell, J., and Masterson, R. (1992). New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol. Biol. 20 8123–8128. [DOI] [PubMed] [Google Scholar]
- Bentsink, L., Yuan, K., Korneef, M., and Vreugdenhil, D. (2003). The genetics of phytate and phosphate accumulation in seeds and leaves of Arabidopsis thaliana, using natural variation. Theor. Appl. Genet. 106 1234–1243. [DOI] [PubMed] [Google Scholar]
- Bertl, A., et al. (1992). Electrical measurements on endomembranes. Science 258 873–874. [DOI] [PubMed] [Google Scholar]
- Beyreuther, K., Bieseler, B., Ehring, R., Griesser, H.-W., Mieschendahl, M., Müller-Hill, B., and Triesch, I. (1980). Investigation of structure and function of lactose permease of Escherichia coli. Biochem. Soc. Trans. 8 675–676. [DOI] [PubMed] [Google Scholar]
- Bohnert, H.J., Nelson, D.E., and Jensen, R.G. (1995). Adaptations to environmental stresses. Plant Cell 7 1099–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brearley, C.A., and Hanke, D.E. (2000). Metabolic relations of inositol 3,4,5,6-tetrakisphosphate revealed by cell permeabilization. Identification of inositol 3,4,5,6-tetrakisphosphate 1-kinase and inositol 3,4,5,6-tetrakisphosphate phosphatase activities in mesophyll cells. Plant Physiol. 122 1209–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinch-Pedersen, H., Hatzack, F., Stöger, E., Arcalis, E., Pontopidan, K., and Holm, P.B. (2006). Heat-stable phytases in transgenic wheat (Triticum aestivum L.): Deposition pattern, thermostability, and phytate hydrolysis. J. Agric. Food Chem. 54 4624–4632. [DOI] [PubMed] [Google Scholar]
- Brinch-Pedersen, H., Sørensen, L.D., and Holm, P.B. (2002). Engineering crop plants: Getting a handle on phosphate. Trends Plant Sci. 7 118–125. [DOI] [PubMed] [Google Scholar]
- Büttner, M. (2007). The monosaccharide transporter(-like) gene family in Arabidopsis. FEBS Lett. 581 2318–2324. [DOI] [PubMed] [Google Scholar]
- Carpaneto, A., Geiger, D., Bamberg, E., Sauer, N., Fromm, J., and Hedrich, R. (2005). Phloem-localized, proton-coupled sucrose carrier ZmSUT1 mediates sucrose efflux under control of sucrose gradient and pmf. J. Biol. Chem. 280 21437–21443. [DOI] [PubMed] [Google Scholar]
- Carter, C., Pan, S., Zouhar, J., Avila, E.L., Girke, T., and Raikhel, N.V. (2004). The vegetative vacuole proteome of Arabidopsis thaliana reveals predicted and unexpected proteins. Plant Cell 16 3285–3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan, S., Forsthoefel, N., Ran, Y., Quigley, F., Nelson, D.E., and Bohnert, H.J. (2000). Na+/myo-inositol symporters and Na+/H+-antiport in Mesembryanthemum crystallinum. Plant J. 24 511–522. [DOI] [PubMed] [Google Scholar]
- Chiou, T.J., and Bush, D.R. (1996). Molecular cloning, immunochemical localization to the vacuole, and expression in transgenic yeast and tobacco of a putative sugar transporter from sugar beet. Plant Physiol. 110 511–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Cohen, J.D., and Bandurski, R.S. (1982). Chemistry and physiology of the bound auxins. Annu. Rev. Plant Physiol. 33 403–430. [Google Scholar]
- Dunn, S.D. (1986). Effects of the modification of transfer buffer composition on the renaturation of proteins in gels on the recognition of proteins on Western blots by monoclonal antibodies. Anal. Biochem. 157 144–153. [DOI] [PubMed] [Google Scholar]
- Emr, S.D., Schekman, R., Flessel, M.C., and Thorner, J. (1983). An MF alpha 1–SUC2 (alpha-factor-invertase) gene fusion for study of protein localization and gene expression in yeast. Proc. Natl. Acad. Sci. USA 80 7080–7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endler, A., Meyer, S., Schelbert, S., Schneider, T., Weschke, W., Peters, S.W., Keller, F., Baginsky, S., Martinoia, E., and Schmidt, U.G. (2006). Identification of a vacuolar sucrose transporter in barley and Arabidopsis mesophyll cells by a tonoplast proteomic approach. Plant Physiol. 141 196–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahrtz, M., Stolz, J., and Sauer, N. (1994). A phloem specific sucrose-H+ symporter from Plantago major L. supports the model of apoplastic phloem loading. Plant J. 6 697–706. [DOI] [PubMed] [Google Scholar]
- Gao, Z., Maurousset, L., Lemoine, R., Yoo, S.D., Van Nocker, S., and Loescher, W. (2003). Cloning, expression, and characterization of sorbitol transporters from developing sour cherry fruit and leaf sink tissues. Plant Physiol. 131 1566–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Mata, R., Bebök, Z., Sorscher, E.J., and Sztul, E.S. (1999). Characterization and dynamics of aggresome formation by a cytosolic GFP-chimera. J. Cell Biol. 146 1239–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz, D., Jean, W.S., Woods, R.A., and Schiestl, R.H. (1992). Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granot, D., and Snyder, M. (1991). Glucose induces cAMP-independent growth-related changes in stationary phase cells of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 88 5724–5728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood, J.S., and Bewley, J.D. (1984). Subcellular distribution of phytin in the endosperm of developing castor bean: A possibility for its synthesis in the cytoplasm prior to deposition within protein bodies. Planta 160 113–120. [DOI] [PubMed] [Google Scholar]
- Guindon, S., and Gascuel, O. (2003). Systematic biology. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52 696–704. [DOI] [PubMed] [Google Scholar]
- Hanahan, D. (1983). Studies on transformation of E. coli with plasmids. J. Mol. Biol. 166 557–580. [DOI] [PubMed] [Google Scholar]
- Holsters, M., Silva, B., Van Vliet, F., Genetello, C., De Block, M., Dhaese, P., Depicker, A., Inze, D., Engler, G., Villarroel, R., Van Montagu, M., and Schell, J. (1980). The functional organization of the nopaline Agrobacterium tumefaciens plasmid pTiC58. Plasmid 3 212–230. [DOI] [PubMed] [Google Scholar]
- Ivashikina, N., and Hedrich, R. (2005). K+ currents through SV-type vacuolar channels are sensitive to elevated luminal sodium levels. Plant J. 41 606–614. [DOI] [PubMed] [Google Scholar]
- Jaquinod, M., Villiers, F., Kieffer-Jaquinod, S., Hugouvieux, V., Bruley, C., Garin, J., and Bourguignon, J. (2007). A proteomic dissection of Arabidopsis thaliana vacuoles isolated from cell culture. Mol. Cell. Proteomics 6 394–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandler, O., and Hopf, H. (1982). Oligosaccharides based on sucrose (sucrosyl oligosaccharides). In Plant Carbohydrates 1. Encyclopedia of Plant Physiology: Plant Carbohydrates I, Intracellular Carbohydrates, New Series, Vol. 13A, F.A. Loewus and W. Tanner, eds (Berlin: Springer-Verlag), pp. 348–383.
- Kanter, U., Usadel, B., Guerineau, F., Li, Y., Pauly, M., and Tenhaken, R. (2005). The inositol oxygenase gene family of Arabidopsis is involved in the biosynthesis of nucleotide sugar precursors for cell-wall matrix polysaccharides. Planta 221 243–254. [DOI] [PubMed] [Google Scholar]
- Kim, D.H., Eu, Y.-J., Yoo, C.M., Kim, Y.-W., Pih, K.T., Jin, J.B., Kim, S.J., Stenmark, H., and Inhwan Hwang, I. (2001). Trafficking of phosphatidylinositol 3-phosphate from the trans-Golgi network to the lumen of the central vacuole in plant cells. Plant Cell 13 287–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebl, F., Wolf, M., and Sauer, N. (2003). A defect in the yeast plasma membrane urea transporter Dur3p is complemented by CpNIP1, a Nod26-like protein from zucchini (Cucurbita pepo L.), and by Arabidopsis thaliana δ-TIP or γ-TIP. FEBS Lett. 547 69–74. [DOI] [PubMed] [Google Scholar]
- Klepek, Y.S., Geiger, D., Stadler, R., Klebl, F., Landouar-Arsivaud, L., Lemoine, R., Hedrich, R., and Sauer, N. (2005). Arabidopsis POLYOL TRANSPORTER5, a new member of the monosaccharide transporter-like superfamily, mediates H+-symport of numerous substrates, including myo-inositol, glycerol, and ribose. Plant Cell 17 204–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli, U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 680–685. [DOI] [PubMed] [Google Scholar]
- Lehle, L. (1990). Phosphatidyl inositol metabolism and its role in signal transduction in growing plants. Plant Mol. Biol. 15 647–658. [DOI] [PubMed] [Google Scholar]
- Loewus, F.A., and Murthy, P.P.N. (2000). myo-Inositol metabolism in plants. Plant Sci. 150 1–19. [Google Scholar]
- Lorence, A., Chevone, B.I., Mendes, P., and Nessler, C.L. (2004). myo-Inositol oxygenase offers a possible entry point into plant ascorbate biosynthesis. Plant Physiol. 134 1200–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macbeth, M.R., Schubert, H.L., VanDemark, A.P., Lingam, A.T., Hill, C.P., and Bass, B.L. (2005). Inositol hexakisphosphate is bound in the ADAR2 core and required for RNA editing. Science 309 1534–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marger, M.D., and Saier, M.H., Jr. (1993). A major superfamily of transmembrane facilitators that catalyse uniport, symport and antiport. Trends Biochem. Sci. 18 13–20. [DOI] [PubMed] [Google Scholar]
- Marinova, K., Pourcel, L., Weder, B., Schwarz, M., Barron, D., Routaboul, J.-M., Debeaujon, I., and Klein, M. (2007). The Arabidopsis MATE transporter TT12 acts as a vacuolar flavonoid/H+-antiporter active in proanthocyanidin-accumulating cells of the seed coat. Plant Cell 19 2023–2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinoia, E., Locher, R., and Vogt, E. (1993). Inositol triphosphate metabolism in subcellular fractions of barley (Hordeum vulgare L.) mesophyll cells. Plant Physiol. 102 101–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinoia, E., Maeshima, M., and Neuhaus, E. (2007). Vacuolar transporters and their essential role in plant metabolism. J. Exp. Bot. 58 83–102. [DOI] [PubMed] [Google Scholar]
- Miyashita, M., Shugyo, M., and Nikawa, J. (2003). Mutational analysis and localization of the inositol transporters of Saccharomyces cerevisiae. J. Biosci. Bioeng. 96 291–297. [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15 473–497. [Google Scholar]
- Nelson, D.E., Koukoumanos, M., and Bohnert, H.J. (1999). Myo-inositol-dependent sodium uptake in ice plant. Plant Physiol. 119 165–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, D.E., Rammesmayer, G., and Bohnert, H.J. (1998). Regulation of cell-specific inositol metabolism and transport in plant salinity tolerance. Plant Cell 10 753–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikawa, J., Hosaka, K., and Yamashita, S. (1993). Differential regulation of two myo-inositol transporter genes of Saccharomyces cerevisiae. Mol. Microbiol. 10 955–961. [DOI] [PubMed] [Google Scholar]
- Nikawa, J., Tskugoshi, Y., and Yamashita, S. (1991). Isolation and characterization of two distinct myo-inositol transporter genes of Saccharomyces cerevisiae. J. Biol. Chem. 266 11184–11191. [PubMed] [Google Scholar]
- Nishimura, M., and Beevers, H. (1978). Hydrolases in vacuoles from castor bean endosperm. Plant Physiol. 62 44–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noiraud, N., Maurousset, L., and Lemoine, R. (2001). Identification of a mannitol transporter, AgMaT1, in celery phloem. Plant Cell 13 695–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otegui, M.S., Capp, R., and Staehelin, L.A. (2002). Developing seeds of Arabidopsis store different minerals in two types of vacuoles and in the endoplasmic reticulum. Plant Cell 14 1311–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page, R.D.M. (1996). TREEVIEW: An application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12 357–358. [DOI] [PubMed] [Google Scholar]
- Phillippy, B.Q., Bland, J.M., and Evens, T.J. (2003). Ion chromatography of phytate in roots and tubers. J. Agric. Food Chem. 51 350–353. [DOI] [PubMed] [Google Scholar]
- Raboy, V. (2003). myo-Inositol-1,2,3,4,5,6-hexakisphosphate. Phytochemistry 64 1033–1043. [DOI] [PubMed] [Google Scholar]
- Ramsperger-Gleixner, M., Geiger, D., Hedrich, R., and Sauer, N. (2004). Differential expression of sucrose transporter and polyol transporter genes during maturation of common plantain companion cells. Plant Physiol. 134 147–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinders, A., Panshyshyn, J.A., and Ward, J.M. (2005). Analysis of transport activity of Arabidopsis sugar alcohol permease homolog AtPLT5. J. Biol. Chem. 280 1594–1602. [DOI] [PubMed] [Google Scholar]
- Sato, T., Ohsumi, Y., and Anraku, Y. (1984). Substrate specificities of active transport systems for amino acids in vacuolar-membrane vesicles of Saccharomyces cerevisiae. J. Biol. Chem. 259 11505–11508. [PubMed] [Google Scholar]
- Sauer, N. (2007). Molecular physiology of higher plant sucrose transporters. FEBS Lett. 581 2309–2317. [DOI] [PubMed] [Google Scholar]
- Sauer, N., Friedlander, K., and Gräml-Wicke, U. (1990). Primary structure, genomic organization and heterologous expression of a glucose transporter from Arabidopsis thaliana. EMBO J. 9 3045–3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer, N., and Stadler, R. (1993). A sink-specific H+/monosaccharide co-transporter from Nicotiana tabacum: Cloning and heterologous expression in baker's yeast. Plant J. 4 601–610. [DOI] [PubMed] [Google Scholar]
- Sauer, N., and Stolz, J. (1994). SUC1 and SUC2: Two sucrose transporters from Arabidopsis thaliana. Expression and characterization in baker's yeast and identification of the histidine-tagged protein. Plant J. 6 67–77. [DOI] [PubMed] [Google Scholar]
- Sauer, N., and Stolz, J. (2000). Expression of foreign transport proteins in yeast. In Practical Approach Series, S.A. Baldwin, ed (Oxford, UK: Oxford University Press), pp. 79–105.
- Schmidt, U.G., Endler, A., Schelbert, S., Brunner, A., Schnell, M., Neuhaus, H.E., Marty-Mazars, D., Marty, F., Baginsky, S., and Martinoia, E. (2007). Novel tonoplast transporters identified using a proteomic approach with vacuoles isolated from cauliflower buds. Plant Physiol. 145 216–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, S., Schneidereit, A., Konrad, K.R., Hajirezaei, M.-R., Gramann, M., Hedrich, R., and Sauer, N. (2006). Arabidopsis thaliana INOSITOL TRANSPORTER 4 mediates high affinity H+-transport of myo-inositol across the plasma membrane. Plant Physiol. 141 565–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, S., Schneidereit, A., Udvardi, P., Hammes, U., Gramann, M., Dietrich, P., and Sauer, N. (2007). Arabidopsis thaliana INOSITOL TRANSPORTER2 mediates high affinity H+-symport of different inositols across the plasma membrane. Plant Physiol. 145 1395–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz, C., Gilson, P., Oxley, D., Youl, J., and Bacic, A. (1998). GPI-anchors on arabinogalactan-proteins: Implications for signalling in plants. Trends Plant Sci. 3 426–431. [Google Scholar]
- Schulz-Lessdorf, B., and Hedrich, R. (1995). Protons and calcium modulate SV-type channels in the vacuolar-lysosomal compartment—Channel interaction with calmodulin inhibitors. Planta 197 655–671. [Google Scholar]
- Sherson, S.M., Hemmann, G., Wallace, G., Forbes, S., Germain, V., Stadler, R., Bechtold, N., Sauer, N., and Smith, S.M. (2000). Monosaccharide/proton symporter AtSTP1 plays a major role in uptake and response of Arabidopsis seeds and seedlings to sugars. Plant J. 24 849–857. [DOI] [PubMed] [Google Scholar]
- Shi, J., Wang, H., Hazebroek, J., Ertl, D.S., and Harp, T. (2005). The maize low-phytic acid 3 encodes a myo-inositol kinase that plays a role in phytic acid biosynthesis in developing seeds. Plant J. 42 708–719. [DOI] [PubMed] [Google Scholar]
- Shi, J., Wang, H., Schellin, K., Li, B., Faller, F., Stoop, J.M., Meeley, R.B., Ertl, D.S., Ranch, J.P., and Glassman, K. (2007). Embryo-specific silencing of a transporter reduces phytic acid content of maize and soybean seeds. Nat. Biotechnol. 25 930–937. [DOI] [PubMed] [Google Scholar]
- Shimaoka, T., Ohnishi, M., Sazuka, T., Mitsuhashi, N., Hara-Nishimura, I., Shimazaki, K., Maeshima, M., Yokota, A., Tomizawa, K., and Mimura, T. (2004). Isolation of intact vacuoles and proteomic analysis of tonoplast from suspension-cultured cells of Arabidopsis thaliana. Plant Cell Physiol. 45 672–683. [DOI] [PubMed] [Google Scholar]
- Su, Y.-H., Frommer, W.B., and Ludewig, U. (2004). Molecular and functional characterization of a family of amino acid transporters from Arabidopsis. Plant Physiol. 136 3104–3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szponarski, W., Sommerer, N., Boyer, J.C., Rossignol, M., and Gibart, R. (2004). Large-scale characterization of integral proteins from Arabidopsis vacuolar membrane by two-dimensional liquid chromatography. Proteomics 4 397–406. [DOI] [PubMed] [Google Scholar]
- Tan, X., Calderon-Villalobos, L.I.A., Sharon, M., Zheng, C., Robinson, C.V., Estelle, M., and Zheng, N. (2007). Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446 640–645. [DOI] [PubMed] [Google Scholar]
- Wang, H.X., Weerasinghe, R.R., Perdue, T.D., Cakmakci, N.G., Taylor, J.P., Marzluff, W.F., and Jones, A.M. (2006). A Golgi-localized hexose transporter is involved in heterotrimeric G protein-mediated early development in Arabidopsis. Plant Cell 17 4257–4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, A., Servaites, J.C., Geiger, D.R., Kofler, H., Hille, D., Groner, F., Hebbeker, U., and Flügge, U.I. (2000). Identification, purification, and molecular cloning of a putative plastidic glucose translocator. Plant Cell 12 787–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weise, A., Barker, L., Kühn, C., Lalonde, S., Buschmann, H., Frommer, W.B., and Ward, J.M. (2000). A new subfamily of sucrose transporters, SUT4, with low affinity/high capacity localized in enucleate sieve elements of plants. Plant Cell 12 1345–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wink, M. (1993). The plant vacuole: A multifunctional compartment. J. Exp. Bot. 44 231–246. [Google Scholar]
- Wormit, A., Trentmann, O., Feifer, I., Lohr, C., Tjaden, J., Meyer, S., Schmidt, U., Martinoia, E., and Neuhaus, H.E. (2006). Molecular identification and physiological characterization of a novel monosaccharide transporter from Arabidopsis involved in vacuolar sugar transport. Plant Cell 18 3476–3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, S. (1979). Freezing injury and phospholipid degradation in vivo in woody plant cells. I. Subcellular localization of phospholipase D in living bark tissue of the black locust tree (Robinia pseudoacacia L.). Plant Physiol. 64 241–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, L., Paulsen, A.Q., Ryu, S.B., and Wang, X. (1996). lntracellular localization of phospholipase D in leaves and seedling tissues of castor bean. Plant Physiol. 111 101–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.