Abstract
Cortical microtubule arrays are highly organized networks involved in directing cellulose microfibril deposition within the cell wall. Their organization results from complex interactions between individual microtubules and microtubule-associated proteins. The precise details of these interactions are often not evident using optical microscopy. Using high-resolution scanning electron microscopy, we analyzed extensive regions of cortical arrays and identified two spatially discrete microtubule subpopulations that exhibited different stabilities. Microtubules that lay adjacent to the plasma membrane were often bundled and more stable than the randomly aligned, discordant microtubules that lay deeper in the cytoplasm. Immunolabeling revealed katanin at microtubule ends, on curves, or at sites along microtubules in line with neighboring microtubule ends. End binding 1 protein also localized along microtubules, at microtubule ends or junctions between microtubules, and on the plasma membrane in direct line with microtubule ends. We show fine bands in vivo that traverse and may encircle microtubules. Comparing confocal and electron microscope images of fluorescently tagged arrays, we demonstrate that optical images are misleading, highlighting the fundamental importance of studying cortical microtubule arrays at high resolution.
INTRODUCTION
Microtubules form highly organized arrays within the plant cell cortex. These cortical arrays are crucial in directing normal cell morphogenesis and in elongating regions are perpendicular to the direction of organ growth. This directed organization is reflected in cellulose microfibril deposition within the cell wall, demonstrating a functional interrelationship between the two networks. Disrupting cellulose microfibril deposition within the cell wall alters the orientation of cortical microtubule arrays (Chu et al., 2007), and cortical microtubules act as guides for the movement of the cellulose synthase complexes within the plasma membrane (Paredez et al., 2006).
Plant cortical microtubule arrays lack defined organizing centers such as centrosomes. Instead, array self-organization results from interactions between individual microtubules (Dixit and Cyr, 2004). Microtubules are highly dynamic filaments and grow or shrink through a hybrid treadmilling process (Shaw et al., 2003). When a growing microtubule end encounters a second microtubule at a steep angle, it either passes over the microtubule or initiates depolymerization (Dixit and Cyr, 2004; Wightman and Turner, 2007). However, if the angle of encounter is shallow, the growing end aligns with the second microtubule and forms a bundle. Microtubule bundles were recently described as the basic unit of array behavior (Chan et al., 2007), suggesting that they are the foundations upon which an array is organized. So, by aligning divergent microtubules into bundles, an array becomes ordered (Dixit et al., 2006).
Cortical microtubule organization and dynamics are regulated in part by the activities of microtubule-associated proteins (MAPs). Structural MAPs, such as those of the MAP65 family, form cross-bridges between bundled microtubules (Chan et al., 1999; Van Damme et al., 2004b). Plus end tracking proteins, including the end binding 1 (EB1) proteins, are located at microtubule plus ends, where they facilitate end growth (Bisgrove et al., 2004). The activities of other MAPs, such as γ-tubulin, which nucleates microtubules (Murata et al., 2005), and katanin, which severs microtubules (Stoppin-Mellet et al., 2002, 2006), are fundamental to microtubule array self-organization (Wasteneys, 2002). To date, γ-tubulin is the only MAP localized to microtubules within plant cortical arrays using electron microscopy (Hoffman et al., 1994; Murata et al., 2005). It is located along microtubules, specifically at sites of microtubule nucleation. The new microtubules branch out at 40° (Murata et al., 2005) and extend into the cytoplasm. Katanin severs microtubules from these nucleation sites, releasing them to encounter other microtubules and become incorporated into bundles, and so into the main axis of an array (Wasteneys, 2002; Stoppin-Mellet et al., 2006).
There are three EB1 homologs in Arabidopsis thaliana, EB1a, EB1b, and EB1c (Chan et al., 2003; Van Damme et al., 2004a; Bisgrove et al., 2008). Fluorescently tagged EB1a and EB1b localize to growing microtubule plus ends (Van Damme et al., 2004a; Dixit et al., 2006) and, when highly expressed, to microtubule minus ends and along the microtubules themselves (Chan et al., 2003; Mathur et al., 2003). By contrast, EB1c localizes to the nucleus during interphase (Van Damme et al., 2004a) but also to microtubule plus ends (Bisgrove et al., 2008). MAPs, such as EB1 or katanin, are smaller than microtubules, and when fluorescently tagged their precise positions are not resolved using optical microscopy. Given the growing number of publications investigating the construction of cortical arrays or the interaction between cortical microtubules and MAPs using fluorescently tagged tubulin or MAPs, this raises questions about the nature of the conclusions drawn.
Here, we explored the complex organization of cortical arrays as well as the localization of MAPs to individual microtubules using high-resolution scanning electron microscopy (HRSEM). We show that the arrays are composed of two spatially discrete subpopulations with different stabilities. Microtubules lying directly adjacent to the plasma membrane, the majority of which were coaligned and bundled, are more stable than both bundled and solitary discordant microtubules that are randomly aligned throughout an array and lie deeper into the cytoplasm. At high resolution, EB1 and katanin localized to microtubules. Finally, we demonstrate using correlative confocal microscopy and HRSEM that optical images of fluorescently tagged microtubule arrays may be misinterpreted.
RESULTS
Cortical Microtubule Arrays Imaged with HRSEM
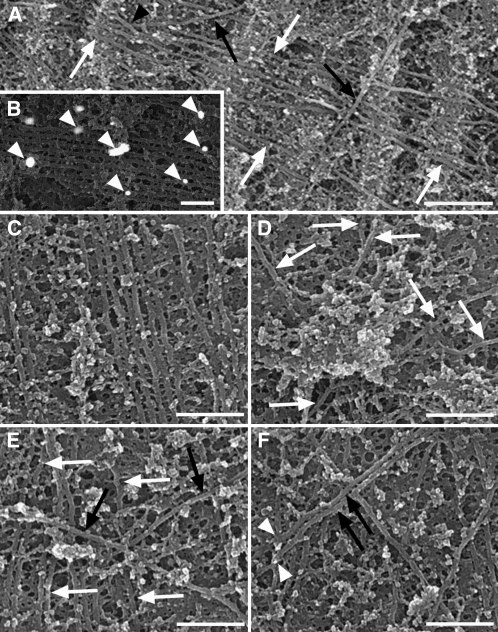
Cortical microtubule arrays are a network of filaments positioned above the plasma membrane (Figure 1; see Supplemental Figure 1A online). Individual microtubules, ∼25 nm in diameter, were decorated with gold particles after labeling with an antibody against α-tubulin (Figure 1B). Two spatially discrete subpopulations were identified within the arrays (Figure 1A). Randomly aligned discordant microtubules, lying deeper in the cytoplasm, crossed over microtubules positioned directly above the plasma membrane. Both of these subpopulations contained solitary microtubules as well as coaligned microtubules linked by cross-bridges into bundles. Within the arrays (an extensive region is included as Supplemental Figure 1A online), there were localized regions of high coalignment between microtubules (Figure 1C; see box 1 of Supplemental Figure 1A online), regions of complete microtubule disorder (Figure 1D; see box 4 of Supplemental Figure 1A online), and regions in which discordant microtubules, often bundled, crossed above microtubules on the plasma membrane (Figures 1E and 1F; see boxes 2 and 3 of Supplemental Figure 1A online). Occasionally, discordant microtubule ends were situated on microtubules of the subpopulation adjacent to the plasma membrane (Figure 1F; see box 3 of Supplemental Figure 1A online), suggesting that they were either nucleated or severed at these sites. Microtubule bundles were typically in two dimensions, lying parallel to the plasma membrane, but on rare occasions three-dimensional bundles were observed. Another rare observation was of discordant microtubules crossing beneath microtubules of the subpopulation on the plasma membrane (Figure 1A).
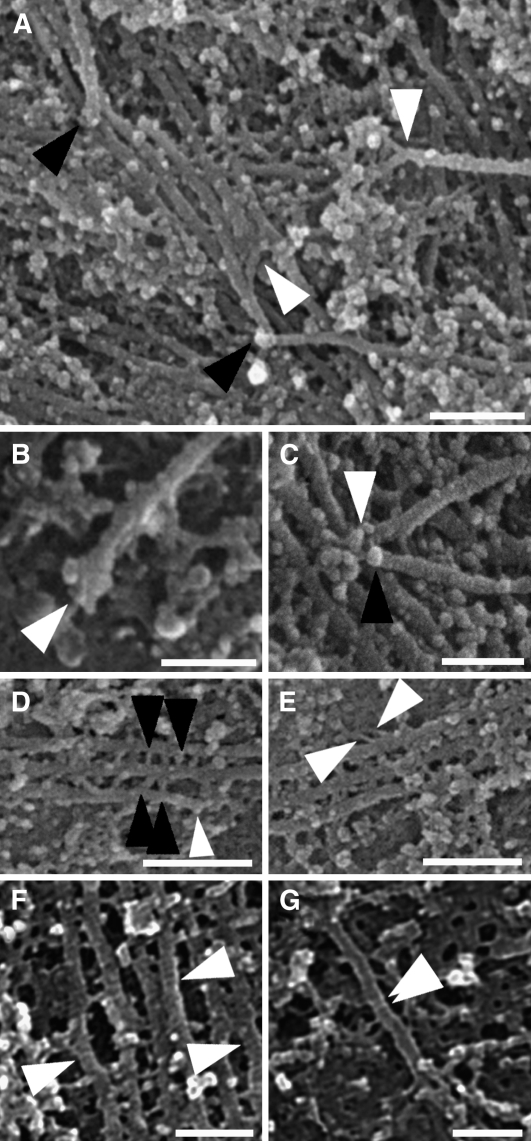
Figure 1.
Cortical Microtubule Arrays Imaged Using HRSEM.
(A) Two microtubule subpopulations were present within the arrays: microtubules that lay adjacent to the plasma membrane (white arrows) and discordant microtubules lying above them (black arrows). On rare occasions, discordant microtubules lay beneath microtubules adjacent to the membrane (black arrowhead).
(B) In cells treated with an anti-α-tubulin primary antibody and imaged using a back-scattered electron detector, gold particles (arrowheads) of the secondary antibody were scattered along microtubules.
(C) to (F) Regions of a microtubule array corresponding to boxes 1 to 4 of the montage in Supplemental Figure 1A online.
(C) A region of high coalignment between bundled microtubules lying adjacent to the plasma membrane.
(D) Microtubule disorganization exhibiting six different alignments (white arrows).
(E) Discordant microtubules of different alignments (black arrows) lay above microtubules on the membrane (white arrows).
(F) A bundle of two discordant microtubules (black arrows) with ends positioned on a microtubule lying adjacent to the plasma membrane (white arrowheads).
Bars = 500 nm in (A), 200 nm in (B), and 300 nm in (C) to (F).
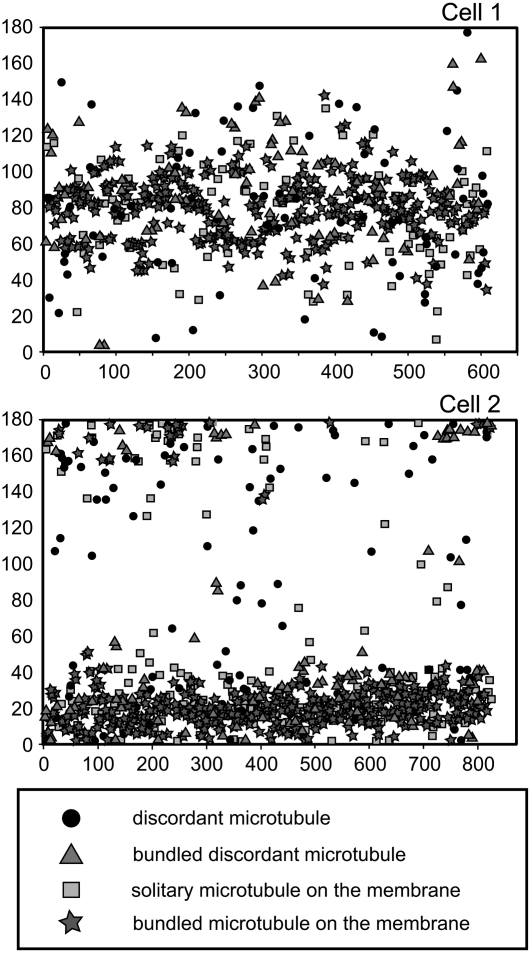
The position, length, angle, and surface area of every microtubule within two extensive regions of cortical arrays, each covering ∼190 μm2, were measured and analyzed (Table 1, Figure 2). Each cortical microtubule array covered 13% of the plasma membrane surface area. Discordant microtubules, both solitary and bundled, accounted for 25% of the microtubules within the arrays and were randomly aligned. Bundled microtubules in the subpopulation adjacent to the plasma membrane accounted for more than half of the microtubules within the arrays. In general, microtubules on the plasma membrane were aligned to the main axes of the arrays. In both subpopulations, bundled microtubules were longer than solitary microtubules. Bundled microtubules, particularly those lying adjacent to the membrane, were also more closely aligned to the main axis of the arrays. The average microtubule length of each cell was approximately one-eighth of the cell's circumference.
Table 1.
Analysis of Large Regions of Cortical Microtubule Arrays within Two Cells
| Features of Microtubule Montages | Cell 1 | Cell 2 | |
|---|---|---|---|
| Percentage of microtubules in different subpopulations | Solitary discordant | 12.7 | 12.8 |
| Bundled discordant | 13.2 | 12.1 | |
| Solitary on the membrane | 17.8 | 21.8 | |
| Bundled on the membrane | 56.3 | 53.3 | |
| Estimated average microtubule length of different subpopulations (μm) | Solitary discordant | 7.4 | 5.9 |
| Bundled discordant | 11.9 | 10.9 | |
| Solitary on the membrane | 8.4 | 5.1 | |
| Bundled on the membrane | 15.7 | 10.8 | |
| All microtubules | 12.4 | 8.6 | |
| Other features | One-eighth cell circumference (μm) | 13.0 | 9.4 |
| Longest microtubule measured (μm) | 17.7 | 16.9 | |
| Surface area of plasma membrane examined (μm2) | 189 | 187 | |
| Surface area covered by microtubules | 13.3% | 12.7% | |
| Total number of microtubules examined | 609 | 825 | |
Cell 1 corresponds to the cortical array presented in Supplemental Figure 1 online.
Figure 2.
Scatterplots Representing the Angle (y Axis) of Every Microtubule (x Axis) within the Two Cortical Arrays of Cells 1 and 2 Corresponding to Table 1.
Microtubules of 0° and 180° were transversely aligned, and microtubules of 90° were longitudinally aligned with respect to the longitudinal axis of the leaf. Microtubules within the cortical array of cell 1 were loosely aligned in a longitudinal direction and those of cell 2 were more tightly aligned into an array that was almost transverse. Bundled microtubules were more aligned to the main axis of the arrays than solitary microtubules. Solitary discordant microtubules were randomly aligned throughout the arrays.
Interactions between Microtubules
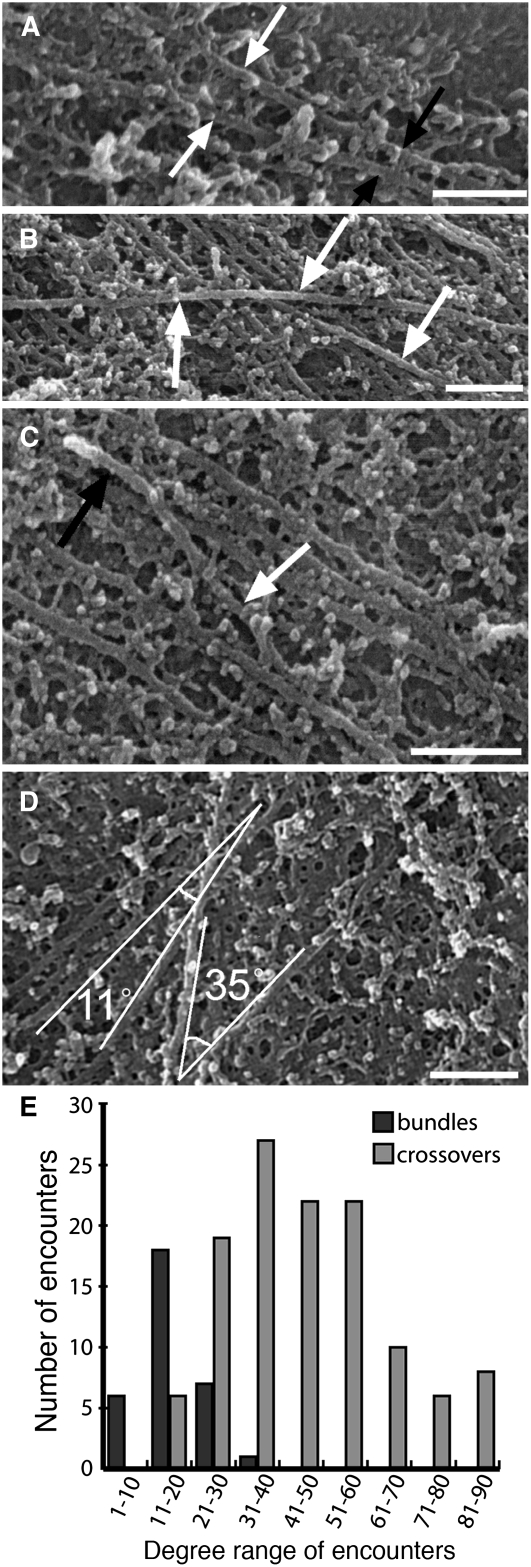
Microtubule interactions were identified either as steep angle encounters resulting in crossovers or shallow angle encounters resulting in bundling (Figure 3). This is consistent with observations of microtubule interactions in live cells, where microtubules converge into bundles rather than diverging from bundles into different alignments (Dixit and Cyr, 2004). Bundling predominantly occurred between microtubules lying adjacent to the plasma membrane that met at ∼20° or less (Figure 3A). Steep angle encounters, in which microtubules met at angles greater than ∼20°, occurred when discordant microtubules crossed above microtubules lying adjacent to the plasma membrane (Figure 3B). On one occasion, a discordant microtubule appeared to alter its trajectory and align to others on the plasma membrane, forming a three-dimensional bundle (Figure 3C). We measured the angles at which the microtubules in Supplemental Figure 1A online interacted (Figure 3D), and of the 153 encounters identified, 21.5% resulted in bundling and 78.5% resulted in crossovers (Figure 3E).
Figure 3.
Interactions between Individual Microtubules.
(A) and (B) Microtubules either coaligned (white arrows) to form bundles (black arrows) (A) or crossed other microtubules (arrows) (B).
(C) A discordant microtubule (white arrow) coaligned with the central microtubule of a bundle at the top of the image (black arrow).
(D) Measurement of encounters between microtubules within box 5 of Supplemental Figure 1A online. A solitary microtubule aligned to a bundle at 11°, and a discordant microtubule passed over other microtubules at 35°.
(E) Angles of encounter within Supplemental Figure 1A online. The number of shallow angle encounters peaked between 11° and 20° and rarely occurred at >30°. Steep angle encounters resulting in crossovers occurred more frequently than shallow angle encounters, with the majority between 21° and 60°.
Bars = 200 nm.
Effects of Microtubule-Stabilizing and -Destabilizing Drugs
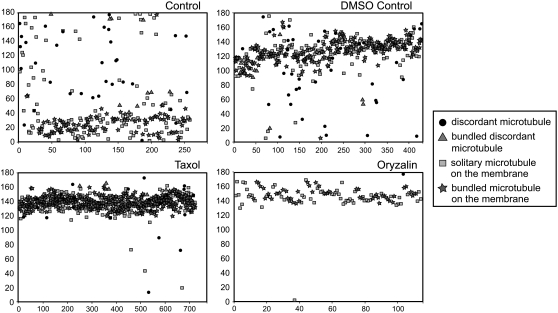
To determine the stability of the different microtubule subpopulations, cortical arrays were treated with the microtubule-stabilizing drug taxol (for 30 min) or the microtubule-depolymerizing drug oryzalin (for 15 min) and compared with arrays from a DMSO control (for 30 min) and a non-DMSO control (Table 2, Figure 4). The cortical microtubule surface area was less than in cells 1 and 2 (Table 1), which may be due to inherent variation between the different cells used for the experiments. Microtubule surface area increased following taxol treatment and decreased following oryzalin treatment, indicating increased microtubule polymerization and stabilization or microtubule depolymerization, respectively. After oryzalin treatment, the microtubules that remained were tightly coaligned (Figure 4), long, and either solitary or in small bundles. The bundling of microtubules on the plasma membrane increased after DMSO treatment and to a greater extent after taxol, coinciding with a decrease in the number of solitary microtubules. After oryzalin treatment, single microtubules on the plasma membrane were the most abundant. Discordant microtubules, both solitary and bundled, decreased after DMSO and taxol treatments and were almost absent following oryzalin treatment. This is reflected in the range of microtubule alignments present in the arrays (Figure 4). In the control cell, discordant microtubules were randomly aligned throughout the array, whereas microtubules on the plasma membrane were aligned in a wide band of angles defining the main axis of the array (Figure 4). This band narrowed and became more dense following DMSO, taxol, and oryzalin treatments, as the discordant microtubules depolymerized or were incorporated into the aligned bundles on the plasma membrane. Discordant microtubules, therefore, are less stable than bundled microtubules, and the most stable microtubules are those aligned to the main axis of a cortical array.
Table 2.
Analysis of Cortical Microtubule Arrays Treated with 0.1% DMSO (30 min), 10 μM Taxol (30 min), or 10 μM Oryzalin (15 min) Compared with a Control (30 min)
| Features of Microtubule Montages | Control | DMSO | Taxol | Oryzalin | |
|---|---|---|---|---|---|
| Percentage of microtubules in different subpopulations | Solitary discordant | 13.2 | 9.3 | 3.2 | 0.9 |
| Bundled discordant | 5.4 | 3.9 | 1.3 | 0 | |
| Solitary on the membrane | 44.0 | 34.8 | 35.1 | 56.1 | |
| Bundled on the membrane | 37.4 | 52.0 | 60.4 | 43.0 | |
| Estimated average microtubule length of different subpopulations (μm) | Solitary discordant | 6.0 | 13.0 | 6.3 | 1.8 |
| Bundled discordant | 14.0 | 23.1 | 23.7 | 0 | |
| Solitary on the membrane | 15.0 | 24.8 | 14.7 | 17.8 | |
| Bundled on the membrane | 23.2 | 28.5 | 18.9 | 29.5 | |
| All microtubules | 15.0 | 25.1 | 16.8 | 21.4 | |
| Other features | One-eighth cell circumference (μm) | 16.4 | 12.7 | 11.1 | 9.6 |
| Longest microtubule measured (μm) | 16.0 | 32.8 | 20.2 | 16.5 | |
| Surface area of plasma membrane examined (μm2) | 196 | 378 | 282 | 286 | |
| Surface area covered by microtubules | 6.2% | 6.4% | 10.7% | 3.5% | |
| Total number of microtubules examined | 257 | 431 | 721 | 114 | |
Figure 4.
Scatterplots of the Angle (y Axis) of Every Microtubule (x Axis) within Cells Treated with Microtubule-Stabilizing (Taxol) or Microtubule-Destabilizing (Oryzalin) Drugs.
Microtubules of the control cell were loosely aligned around 20°, and those within the array treated with 0.1% DMSO were aligned obliquely in relation to the longitudinal axis of the cell (0 and 180°). Most microtubules within the array treated with 10 μM taxol were tightly aligned in an oblique direction around 140°. Microtubules of the cell treated with 10 μM oryzalin were tightly aligned in an oblique direction (150°) relative to the longitudinal axis of the cell. In all treatments, bundled microtubules were generally aligned within the main axis of each array, whereas discordant microtubules, particularly solitary discordant microtubules, were randomly aligned throughout the arrays.
Morphology of Microtubule Ends and Cross-Bridges
Microtubule ends ranged from blunt, pointed, and slightly flared to more widely flared (Figures 5A to 5C). On occasion, discordant microtubule ends terminated on microtubules of the subpopulation on the plasma membrane (Figure 1C). In Figure 5C, two ends converged at a single point on a microtubule. The lower of these ends appears to be capped with a structure of lighter hue not unlike that proposed for a γ-tubulin ring complex (Moritz et al., 2000).
Figure 5.
Features of Microtubule Arrays Imaged Using HRSEM.
(A) Often, microtubule ends were blunt (black arrowheads), but occasionally they were splayed (white arrowheads).
(B) A flared microtubule end (arrowhead).
(C) Two microtubule ends terminating at the same point on another microtubule (arrowheads). One end is capped with a lighter structure (black arrowhead).
(D) Cross-bridges between neighboring microtubules (black arrowheads) occurred behind a microtubule end (white arrowhead).
(E) Cross-bridges linking a microtubule to the plasma membrane (white arrowheads).
(F) and (G) Fine bands that crossed microtubules (white arrowheads).
Bars = 200 nm in (A) and 100 nm in (B) to (G).
Cross-bridges, probably composed of MAP65 proteins (Chan et al., 1999), linked bundled microtubules (Figure 5D) but not necessarily along an entire bundle. In Figure 5D, cross-bridges only occurred directly behind a microtubule end, suggesting that they either stabilized the end or guided its growth parallel to other microtubules in the bundle. Cross-bridges were often aligned perpendicular to microtubules but were also at angles of up to 45°. The dimensions of the cross-bridges were not uniform and varied as the distances between neighboring microtubules changed. Cross-bridges, such as those that may be formed by phospholipase D (Gardiner et al., 2001), also linked microtubules to the plasma membrane (Figure 5E). Very fine bands that traversed microtubules (Figures 5F and 5G) were observed throughout the arrays. These bands, ∼4 to 5 nm in diameter, were light in hue and positioned perpendicularly across microtubules independent of their alignment.
Immunogold Localizations
FluoroNanogold secondary antibodies were used to identify antigen sites with HRSEM, as these label microtubules within whole mount cells more efficiently than larger colloidal gold antibody complexes (Robinson and Vandre, 1997; Robinson et al., 2000). Anti-tubulin labeling was punctate and considerably less dense than expected (Figure 1B). However, the gold particles were preferentially on microtubules when compared with a secondary antibody control and an anti-actin control (see Supplemental Figure 2 online). The sporadic, punctate labeling observed may be a result of the gold enhancement process employed to increase the size of the 1.4-nm Nanogold particles. Punctate FluoroNanogold labeling followed by gold or silver enhancement has also been observed in other tissues with electron microscopy (Robinson and Vandre, 1997).
EB1 Labeling
An antibody raised against Arabidopsis EB1c, which also recognizes EB1a and EB1b (Bisgrove et al., 2008), was localized along cortical microtubules (Figure 6; see Supplemental Figure 2 online). It was observed on occasion at microtubule ends (Figures 6C and 6D) and at junctions where microtubules crossed or aligned into bundles (Figures 6A and 6B). Occasionally, it localized to the plasma membrane just beyond, but in line with, a microtubule end (Figures 6C to 6F). In Figures 6E and 6F, four gold particles representing EB1 decorated the plasma membrane in a straight line beyond a microtubule end.
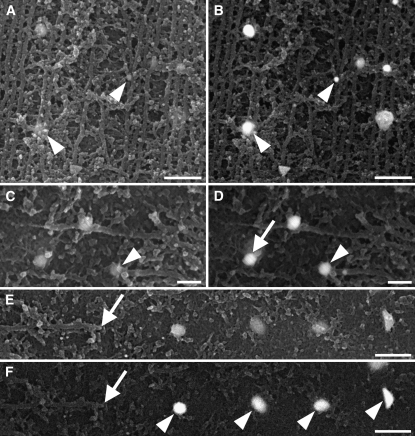
Figure 6.
EB1 Immunolabeling of Cortical Microtubule Arrays.
Secondary electron ([A], [C], and [E]) and corresponding back-scattered electron ([B], [D], and [F]) images.
(A) and (B) EB1 localized along microtubules and decorated junctions where microtubules aligned into bundles (arrowheads).
(C) and (D) EB1 at a microtubule end (arrowheads). EB1 was also on the plasma membrane (arrow in [D]) beyond the microtubule end but aligned to the axis of the microtubule.
(E) and (F) Four gold particles representing EB1 (arrowheads) were on the plasma membrane in line with the angle of the microtubule. The microtubule end is indicated by the arrows.
Bars = 200 nm in (A) and (B), 100 nm in (C) and (D), and 300 nm in (E) and (F).
Katanin Labeling
An antibody directed to the catalytic subunit of the microtubule-severing protein, katanin, also localized to cortical microtubules (Figure 7; see Supplemental Figure 2 online). On occasion, it labeled microtubule ends (Figures 7A to 7D) and curves (Figures 7A and 7B). In Figures 7A and 7B, katanin localized to a microtubule end, while a second end sharing the same alignment as the first was in close proximity, suggesting that katanin had severed the microtubule, initiating the depolymerization of either end or both ends. Katanin was also observed at sites along bundled microtubules adjacent to or in line with neighboring microtubule ends within the bundle (Figures 7E and 7F).
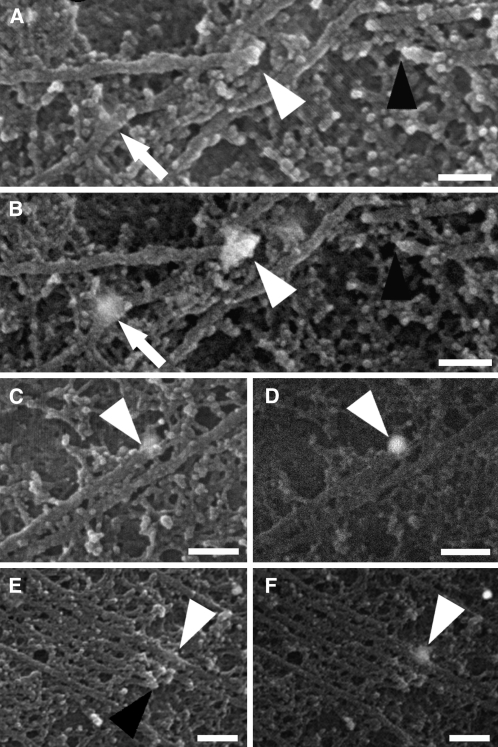
Figure 7.
Cortical Microtubules Labeled with an Antibody against Katanin.
Microtubules were imaged using a secondary electron detector ([A], [C], and [E]) or a back-scattered electron detector ([B], [D], and [F]).
(A) and (B) Katanin localized to a microtubule end (white arrowheads) with another end nearby (black arrowhead). Katanin also localized to a curve in a microtubule (arrows).
(C) and (D) Katanin (arrowheads) on the end of a bundled microtubule.
(E) and (F) Katanin (white arrowheads) on a bundled microtubule in line with other ends (black arrowhead) of the bundle.
Bars = 100 nm in (A) to (D) and 150 nm in (E) and (F).
Correlative Microscopy
To analyze the relative positions of cortical microtubules in corresponding optical and electron microscope images, a single array, labeled with a FluoroNanogold secondary antibody, was imaged with both confocal microscopy and HRSEM. The array was visible in the fluorescence image (Figure 8A) but not at the same magnification using HRSEM (Figure 8B). At higher magnification, individual microtubules were resolved (Figure 8C) and the position of each was traced. This tracing was positioned above the corresponding region of the fluorescence image (Figure 8D). Bundled microtubules corresponded to fluorescent lines, but solitary microtubules were often indistinguishable from background fluorescence. Regions of the tracing without any microtubules, but that corresponded to fluorescent lines, represented regions where microtubules were either lost during dry cleaving or were obscured beneath unextracted cytoplasmic material. The fluorescence intensity of every microtubule was measured (Figures 8E and 8F). Isolated individual microtubules (>150 nm from other microtubules) were dim, but as they approached other microtubules their fluorescence intensity increased. Isolated bundles of two or three microtubules were significantly brighter (P < 0.001) than isolated individual microtubules, but they were not significantly brighter than two single microtubules found <150 nm apart. As bundles of two or three approached another microtubule, their fluorescence also increased. Bundles of four microtubules were bright, but the brightness did not intensify significantly when they approached another microtubule. At regions where a number of microtubules of different alignments converged, the fluorescence intensity increased markedly (Figure 8E). Of the 46 discordant microtubules in the HRSEM image, 32 were not distinguishable as separate lines from nearby fluorescence. Nine bundled discordant microtubules were distinguished as fluorescent lines, but only five solitary discordant microtubules were visible as fluorescent lines.
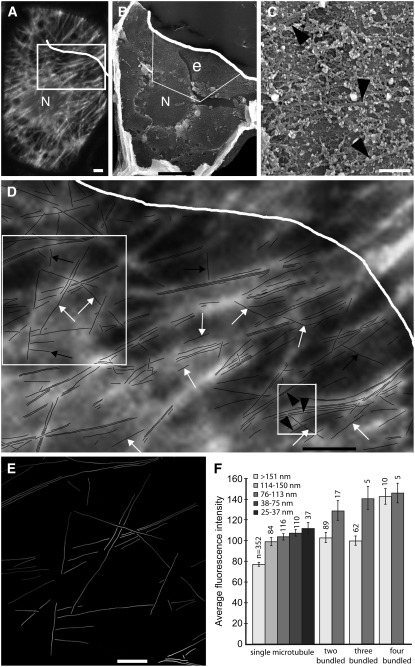
Figure 8.
Correlative Microscopy of FluoroNanogold-Labeled Microtubules within the Same Cell Using Confocal Microscopy and HRSEM.
(A) and (B) The position of the nucleus (N) in the confocal microscopy image in (A) corresponds to the hole in the cytoplasm made after the nucleus was dislodged during dry cleaving in the HRSEM image in (B). The white line marks the boundary of the cell as seen with HRSEM. A region of extraction e, in which microtubules were exposed, is outlined by the white box in (B).
(C) Individual microtubules within the region of extraction in (B) at higher magnification and corresponding to the traced lines in the small box in (D). Arrowheads indicate individual microtubules and correspond to the arrowheads in (D).
(D) A tracing of individual microtubules found in region e in (B). The tracing overlies the corresponding boxed region of the fluorescence image in (A). Individual microtubules sometimes corresponded to fluorescent lines (black arrows), but often they were indistinguishable from the fluorescence of other microtubules (white arrows). Black arrowheads within the small boxed region correspond to the individual microtubules of the corresponding HRSEM image in (C).
(E) Fluorescence intensity of each microtubule of the large boxed region in (D). As individual microtubules approached other microtubules of different orientations, their intensities generally increased.
(F) The average ± se fluorescence intensities of single microtubules and microtubules found in bundles of two, three, and four at a range of distances to their nearest neighbors. Numbers above each column represent the total number of measurements used to calculate average and se figures for each data set.
Bars = 3 μm in (A) and (D), 10 μm in (B), and 1 μm in (C) and (E).
DISCUSSION
Analysis of cortical microtubule arrays at high resolution revealed that they are not simply an ordered reticulum of overlapping individuals lying directly adjacent to the plasma membrane (Hardham and Gunning, 1977, 1978). Instead, up to one-quarter of the microtubules of an array are randomly aligned, more labile, and lie deeper into the cytoplasm than the subpopulation on the membrane. This raises questions regarding the function of these discordant microtubules within an array and how they interact with the microtubule subpopulation lying adjacent to the plasma membrane.
There is evidence that discordant microtubules are the most recently nucleated microtubules of an array. In movies of array dynamics, discordant microtubules nucleate and polymerize away from existing microtubules (Shaw et al., 2003), traversing microtubules of different alignments (Wightman and Turner, 2007), and supporting our findings that they are in a separate plane. Murata et al. (2005) also showed that newly formed microtubules grow away from existing microtubules at 40°. Here, we show that discordant microtubule ends lie on the cytoplasmic faces of microtubules that are adjacent to the plasma membrane. This is not consistent with the conclusions drawn by Murata et al. (2005), and it may be that the observed ends were severed at crossover sites (Wightman and Turner, 2007). However, we suggest that microtubules are nucleated above the ordered reticulum on the membrane, allowing them initially to grow unimpeded by other microtubules.
Dynamically active discordant microtubules detach from nucleation sites (Shaw et al., 2003) or are severed at crossover junctions (Wightman and Turner, 2007) and by interacting with other microtubules integrate within the main axis of the array. In tobacco (Nicotiana tabacum) BY-2 suspension culture cells, the definitive angle of encounter is 40°, below which microtubules form bundles and above which they cross paths or the growing ends initiate depolymerization (Dixit and Cyr, 2004). The definitive angle measured here was 20° and related to the planes in which microtubules interacted. Shallow angle encounters occurred on the plasma membrane, whereas steep angle encounters occurred as discordant microtubules crossed other microtubules. We rarely observed bundling at angles >30°, possibly because the growing microtubule ends had initiated depolymerization. Bundling at 20° also occurs in Arabidopsis cotyledon cells (Wightman and Turner, 2007). These differences in angles compared with those measured by Dixit and Cyr (2004) may be due to morphological differences between suspension culture cells and cells within whole tissue. On one occasion, we observed what appeared to be a discordant microtubule aligning to a microtubule bundle on the plasma membrane. Since the growing end did not directly encounter microtubules on the membrane, the coalignment may have been affected by a MAP. Wightman and Turner (2007) observed discordant microtubules pausing for several seconds as they crossed microtubules. MAPs on the underlying microtubules could be interacting with discordant microtubules at these junctions, either forming a stable link between the two or severing the attachment and initiating discordant microtubule depolymerization.
Cortical microtubules have been implicated in guiding cellulose synthase complexes within the plasma membrane, so directing cellulose synthesis parallel to microtubule alignment (Giddings and Staehelin, 1988; Paredez et al., 2006). It is highly unlikely that the labile, randomly oriented, discordant microtubule subpopulation lying deeper into the cytoplasm would interact with these complexes. Their role may be as sensors for environmental signals such as electrical fields or wounding responses, as they could respond more quickly than the stable microtubules on the plasma membrane. These stable, predominately bundled microtubules are coaligned and so are more likely to be directly involved in producing the ordered cellulose microfibril arrays within the cell wall (as seen in Sugimoto et al., 2000). The most stable microtubules observed in the cortical arrays were tightly coaligned, long, and solitary or in small bundles, suggesting that they had been selectively stabilized to reflect the main axis of the array. Selective stabilization of microtubules has been suggested previously to be important in cortical array self-organization (Dixit et al., 2006). We propose that these individual, selectively stabilized microtubules, rather than the bundles proposed by Chan et al. (2007), are the framework upon which an array is constructed. Bundles would be constructed around these microtubules, thus promoting the alignment of other microtubules to the main axis of the array.
Remarkably, our data support the observation of Hardham and Gunning (1978) that a cell's average microtubule length is approximately equal to one-eighth of its circumference. This suggests that innate cell dimensions, rather than interactions between individual microtubules or microtubules and MAPs, influence microtubule lengths. Interestingly, Wasteneys and Fujita (2006) suggested that the length of cellulose microfibrils within the cell wall is affected by microtubule length and so, in this way, microtubules may affect cell size and shape.
The composition of the fine bands that traversed microtubules is unknown. Possible contenders are the proteins or protein complexes that form ring-like structures around microtubules in vitro. These include the Dam1 complex that forms rings at the depolymerizing ends of microtubules (Westermann et al., 2006) or kinesin-13s that form rings and spirals along microtubules (Tan et al., 2006). They may also be constructed from katanin oligomers, similar to those formed by subunits of animal (Hartman et al., 1998) and plant (Stoppin-Mellet et al., 2007) katanin in vitro. Further investigations using immunogold labeling are needed to identify the precise composition of these bands.
Our observation of katanin on small curves in microtubules supports previous findings that microtubules are severed at sites of lattice defects (Davis et al., 2002) or curves (Waterman-Storer and Salmon, 1997). We also provide evidence to show that katanin may act to sever bundles in vivo, as demonstrated previously in vitro (Wicker-Planquart et al., 2004).
An antibody that recognizes all three EB1 proteins localized not only along microtubules but also at microtubule ends and on the plasma membrane directly past microtubule ends. This is similar to the observations of Chan et al. (2003), in which the overexpression of fluorescently tagged At EB1a led to signal not only along microtubules but also at small, discrete foci attached to the plasma membrane. Two possible explanations for our observations are either that EB1 on the plasma membrane guides microtubule growth or that as a microtubule end depolymerizes, EB1 is exposed on the plasma membrane behind the shrinking end. The yeast EB1 homolog, Mal3p, binds along the microtubule seam and in doing so stabilizes the microtubule lattice (Sandblad et al., 2006). We suggest that EB1 could fulfill a similar role in plant cortical microtubule arrays, and positioned on the plasma membrane face of microtubules, it may be one of a number of proteins, including CLASP (Ambrose et al., 2007) or MOR1 (Kawamura et al., 2006), in a complex mechanism that links microtubules to the plasma membrane. Indeed, EB1 accumulation on microtubules of mammalian cells is thought to be due to a direct interaction between EB1 and CLASP (Mimori-Kiyosue et al., 2005). Such a complex also could be directly involved in cellular growth and expansion.
Imaging fluorescently tagged tubulin or MAPs is being used increasingly to identify specific interactions between microtubules and their associated proteins or to investigate the dynamics of cortical microtubules. However, due to the resolution limits of optical microscopes (∼250 nm), it is not possible to resolve either individual microtubules or MAPs. Here, we have shown that images of fluorescently tagged cortical microtubule arrays are not a realistic representation of the individual microtubules that make up the arrays. Bundled microtubules do correspond to fluorescent lines. However, while individual microtubules and, in particular, solitary discordant microtubules were sometimes resolved as single, fluorescent lines, they were often obscured by the fluorescence emitted from other microtubules, especially when in close proximity to them. In addition, bundles of two microtubules had the same fluorescence intensity as two individual microtubules 50 to 150 nm apart, highlighting the fact that it is impossible to distinguish whether a fluorescent line is a bundle or simply two individuals. Others have shown that when individual microtubules, grown in vitro or in mammalian cells, were at distances greater than the diffraction limit of the fluorescence tag (0.29 μm), they were distinguished as single microtubules (Osborn et al., 1978; Sammak and Borisy, 1988). If they were situated within that limit, neighboring microtubules were not resolved using fluorescence microscopy. So, as discussed by Williamson (1991), images of fluorescently labeled microtubule arrays can be misleading, and we suggest that fluorescence tags, used to highlight the positions of microtubules and MAPs within cells, in fact mask the intricate nature of microtubule arrays as well as disguise the complex interactions between individual microtubules and their associated proteins.
METHODS
Plant Material
Tradescantia virginiana plants were grown in a glasshouse at 23°C under natural light conditions. Epidermal peels were made from the outer surface of young leaves (Cleary, 1995), affixed to cover slips with 1% (w/v) agarose, and bathed in Tradescantia bathing medium (TBM; 2.5 mM HEPES, 2.5 mM KCl, and 0.1 mM CaCl2). Unless specified otherwise, all experimental treatments were conducted at room temperature and all chemicals were purchased from Sigma-Aldrich.
Drug Treatments
Randomly selected peels were treated with TBM (control), 0.1% DMSO (control; Research Organics), 10 μM taxol, or 10 μM oryzalin, all in TBM. Taxol and oryzalin solutions were diluted from 10 mM DMSO stock solutions. The treatments were for either 15 min (oryzalin) or 30 min (controls and taxol).
Fixation and Extraction
Peels were fixed in 0.75% paraformaldehyde and 0.5% glutaraldehyde (Proscitech) in 10 mM PIPES, 10 mM CaCl2, and 5 mM sucrose for 30 min. After rinsing in PME buffer (50 mM PIPES, 5 mM MgSO4, and 5 mM EGTA, pH 6.9), the peels, still attached to cover slips, were plunged into liquid nitrogen and then thawed into PBS (3.3 mM Na2HPO4, 1.6 mM NaH2PO4, and 145 mM NaCl, pH 6.9). They were extracted with 0.04% saponin for 20 min.
Immunolabeling and Imaging
Peels were blocked in 10% (v/v) normal goat serum (Institute of Medical and Veterinary Science) and 3% (w/v) BSA, then incubated for 2 h at 37°C with monoclonal anti-α-tubulin (clone B-5-1-2) raised in mouse and diluted 1:500 in PBS or with anti-actin (clone C4; MP Biomedicals) raised in mouse and diluted 1:200 in PBS. A primary polyclonal antibody raised in rabbit against the recombinant katanin catalytic subunit of Arabidopsis thaliana (At KSS for Arabidopsis katanin small subunit) and purified against the recombinant At KSS (Stoppin-Mellet et al., 2006) was applied to other peels at 1:10 (v/v) for 2 h at 37°C. A second primary polyclonal antibody raised in rabbit against Arabidopsis EB1c, which recognizes all three EB1 proteins (Bisgrove et al., 2008; a kind gift from Bo Liu and Sherryl Bisgrove), was applied to peels at 1:500 for 2 h at 37°C. After rinsing, the peels were incubated for 90 min at 37°C with the secondary antibodies fluorescein-FluoroNanogold anti-mouse or anti-rabbit (Nanoprobes), both diluted 1:25 in PBS. Control peels were treated with secondary antibodies only. Cover slips were mounted onto slides in 1:1 (v/v) PBS:Citifluor (Leica Microsystems) including 20 mM ascorbic acid. The cells were imaged with a LSM 5 Pascal (Carl Zeiss) confocal apparatus attached to a Zeiss Axiovert 200M microscope. Images were captured using a Plan-Neofluar 100X/0.30 oil-immersion lens (Carl Zeiss) and a 488-nm argon laser. Z-series of sequential images at different focal planes were captured for each array, and these were combined to produce projected images.
HRSEM
Cover slips were detached from slides and the peels rinsed in PBS, then 50 mM Gly in PBS, PBSTB (PBS with 0.1% [v/v] Tween 20 and 1% [w/v] BSA), and deionized water. Nanogold particles of the secondary antibodies were enlarged using a Goldenhance electron microscopy kit (Nanoprobes) in which the enhance solution was applied for 10 min. After rinsing in water, all peels were postfixed with 0.5% (v/v) OsO4 (Ted Pella) for 10 min at 4°C. The peels were dehydrated through an ethanol (CSR Distilleries) series and dried in a critical point drier (CPD030; Baltec). Dried peels were lifted off the cover slips and cleaved open between two pieces of single-sided sticky tape (3M). They were mounted onto copper stubs, coated with 2 to 3 nm of platinum using a planar magnetron sputter-coater, and examined using a JEOL 6000F in-lens field emission scanning electron microscope fitted with a solid-state back-scattered electron detector using accelerating voltages of 5, 10, or 12 kV. Secondary electron images were captured for all specimens, and back-scattered electron images were collected to determine the localization of the secondary antibody gold particles.
HRSEM Montages and Analysis
Six large regions of microtubule arrays, three control and one each of cells treated with taxol, oryzalin, or DMSO, were captured at high magnification in sequential HRSEM images. These were collated into montages using Adobe Photoshop (version 7.0), and the position of every microtubule was traced on a separate layer. Tracings of microtubules physically broken or partially hidden beneath unextracted cytoplasm were extended to join the fragments into a single line (see Supplemental Figure 1C online). Where microtubules crossed paths, the lines representing microtubules on the plasma membrane were broken and those of the overlying microtubules were continuous. To avoid problems with analysis, lines that crossed others, representing discordant microtubules or microtubules forming three-dimensional bundles, were cut from their original positions and transferred to an area free of lines. The tracings of the microtubules on the plasma membrane were then joined to form single lines (see Supplemental Figure 3A online). Four modified tracings of each montage (see Supplemental Figure 3 online) were prepared for analysis: one representing the entire microtubule population, the second representing the bundled microtubules on the membrane, the third representing solitary discordant microtubules, and the fourth representing bundled discordant microtubules. Each modified tracing was analyzed with NIH Image version 1.63 (U.S. National Institutes of Health; http://rsb.info.nih.gov/nih-image), and the surface area, length, and angle of each microtubule were recorded. The surface area measurements were calibrated by averaging 50 actual microtubule widths in HRSEM images compared with the average of 50 lines drawn using Photoshop. The surface area of the plasma membrane covered by microtubules was converted to a percentage of the total plasma membrane surface area, also measured using NIH Image. The angle of each microtubule was relative to the longitudinal axis of the cell and the leaf.
Microtubule Lengths
The average microtubule length for each montage was calculated by summing the lengths of each microtubule, dividing by the number of ends in the array, and multiplying by 2, following previous methods (Hardham and Gunning, 1977, 1978). Microtubule ends obscured under cytoplasmic material, or broken at the edge of a cell or region of overextraction, were not included.
Cell Dimensions
The widths of the cells from which the montages were collated were measured from low-magnification HRSEM images. The circumference of each cell was estimated as follows: 2 × width + 2 × depth. The depth of each cell was calculated according to the following formula defined for these cells (Lang et al., 2004): depth = 0.7473 × width + 6.0963.
Statistical Analyses and Correlative Microscopy
The positions of selected cells imaged with confocal microscopy were recorded for later identification using HRSEM. A HRSEM montage was created for one cell with a large region of microtubules exposed on the plasma membrane. For correlative microscopy analysis, the tracing of this array was superimposed on the same region in the corresponding confocal image. The fluorescence intensities of single microtubules located either far from or close to other microtubules, as well as the intensities of microtubules in bundles of two, three, or four, were calculated using the histogram function in Photoshop. Single-factor analysis of variance was used to determine whether the average intensities of each group were significantly different.
Statistical Analyses of Immunogold Labeling
In at least six regions of different cells treated with each antibody and captured with HRSEM at magnifications of less than ×40,000, the number of gold particles on or within 25 nm of microtubules was counted and presented as a percentage of the total number of gold particles visible (see Supplemental Figure 2 online). These data were compared with those of the gold-only controls using single-factor analysis of variance. For all antibody treatments except anti-actin, the percentage of gold particles found on microtubules was statistically significantly higher than that of the gold-only control (P < 0.05).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. A Montage Composed of 104 HRSEM Images Revealing the Intricate Construction of a Cortical Microtubule Array within a T. virginiana Leaf Epidermal Cell.
Supplemental Figure 2. The Average Percentage Distribution of Secondary Antibody Gold Particles within Cells Treated with Different Primary Antibodies.
Supplemental Figure 3. The Four Modified Tracings of Montage 1 Used for Analysis.
Supplementary Material
Acknowledgments
This work was supported by Australian Research Council Grant A19905246 to R.L.O. and by an Australian Postgraduate Research Award to D.A.B. We are grateful to Jérémie Gaillard for purification of the katanin antibody, to B. Liu and S. Bisgrove for the kind gift of the EB1 antibody, and to M. Vesk, T. Dibbayawan, and T. Romeo for assistance in developing the HRSEM preparation technique.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Robyn L. Overall (roverall@mail.usyd.edu.au).
Online version contains Web-only data.
References
- Ambrose, J.C., Shoji, T., Kotzer, A.M., Pighin, J.A., and Wasteneys, G.O. (2007). The Arabidopsis CLASP gene encodes a microtubule-associated protein involved in cell expansion and division. Plant Cell 19 2763–2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisgrove, S.R., Hable, W.E., and Kropf, D.L. (2004). +TIPs and microtubule regulation. The beginning of the plus end in plants. Plant Physiol. 136 3855–3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisgrove, S.R., Lee, Y.R., Liu, B., Peters, N.T., and Kropf, D.L. (2008). The microtubule plus-end binding protein EB1 functions in root responses to touch and gravity signals in Arabidopsis. Plant Cell 20 396–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, J., Calder, G., Fox, S., and Lloyd, C. (2007). Cortical microtubule arrays undergo rotary movements in Arabidopsis hypocotyl epidermal cells. Nat. Cell Biol. 9 171–175. [DOI] [PubMed] [Google Scholar]
- Chan, J., Calder, G.M., Doonan, J.H., and Lloyd, C.W. (2003). EB1 reveals mobile microtubule nucleation sites in Arabidopsis. Nat. Cell Biol. 5 967–971. [DOI] [PubMed] [Google Scholar]
- Chan, J., Jensen, C.G., Jensen, L.C.W., Bush, M., and Lloyd, C.W. (1999). The 65-kDa carrot microtubule-associated protein forms regularly arranged filamentous cross-bridges between microtubules. Proc. Natl. Acad. Sci. USA 96 14931–14936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu, Z., Chen, H., Zhang, Y., Zhang, Z., Zheng, N., Yin, B., Yan, H., Zhu, L., Zhao, X., Yuan, M., Zhang, X., and Xie, Q. (2007). Knockout of the AtCESA2 gene affects microtubule orientation and causes abnormal cell expansion in Arabidopsis. Plant Physiol. 143 213–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary, A.L. (1995). F-actin redistributions at the division site in living Tradescantia stomatal complexes as revealed by microinjection of rhodamine-phalloidin. Protoplasma 185 152–165. [Google Scholar]
- Davis, L.J., Odde, D.J., Block, S.M., and Gross, S.P. (2002). The importance of lattice defects in katanin-mediated microtubule severing in vitro. Biophys. J. 82 2916–2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit, R., Chang, E., and Cyr, R. (2006). Establishment of polarity during organization of the acentrosomal plant cortical microtubule array. Mol. Biol. Cell 17 1298–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit, R., and Cyr, R. (2004). Encounters between dynamic cortical microtubules promote ordering of the cortical array through angle-dependent modifications of microtubule behavior. Plant Cell 16 3274–3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner, J.C., Harper, J.D.I., Weerakoon, N.D., Collings, D.A., Ritchie, S., Gilroy, S., Cyr, R.J., and Marc, J. (2001). A 90-kD phospholipase D from tobacco binds to microtubules and the plasma membrane. Plant Cell 13 2143–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giddings, T.H., and Staehelin, L.A. (1988). Spatial relationship between microtubules and plasma-membrane rosettes during the deposition of primary wall microfibrils in Closterium sp. Planta 173 22–30. [DOI] [PubMed] [Google Scholar]
- Hardham, A.R., and Gunning, B.E.S. (1977). The length and disposition of cortical microtubules in plant cells fixed in glutaraldehyde-osmium tetroxide. Planta 134 201–203. [DOI] [PubMed] [Google Scholar]
- Hardham, A.R., and Gunning, B.E.S. (1978). Structure of cortical microtubule arrays in plant cells. J. Cell Biol. 77 14–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman, J.J., Mahr, J., McNally, K., Okawa, K., Iwamatsu, A., Thomas, S., Cheesman, S., Heuser, J., Vale, R.D., and McNally, F.J. (1998). Katanin, a microtubule-severing protein, is a novel AAA ATPase that targets to the centrosome using a WD40-containing subunit. Cell 93 277–287. [DOI] [PubMed] [Google Scholar]
- Hoffman, J.C., Vaughn, K.C., and Joshi, H.C. (1994). Structural and immunocytochemical characterization of microtubule organizing centers in pteridophyte spermatogenous cells. Protoplasma 179 46–60. [Google Scholar]
- Kawamura, E., Himmelspach, R., Rashbrooke, M.C., Whittington, A.T., Gale, K.R., Collings, D.A., and Wasteneys, G.O. (2006). MICROTUBULE ORGANIZATION 1 regulates structure and function of microtubule arrays during mitosis and cytokinesis in the Arabidopsis root. Plant Physiol. 140 102–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang, I., Barton, D.A., and Overall, R.L. (2004). Membrane-wall attachments in plasmolysed plant cells. Protoplasma 224 231–243. [DOI] [PubMed] [Google Scholar]
- Mathur, J., Mathur, N., Kernebeck, B., Shrinivas, B.P., and Hulskamp, M. (2003). A novel localization pattern for an EB1-like protein links microtubule dynamics to endomembrane organization. Curr. Biol. 13 1991–1997. [DOI] [PubMed] [Google Scholar]
- Mimori-Kiyosue, Y., Grigoriev, I., Lansbergen, G., Sasaki, H., Matsui, C., Severin, F., Galjart, N., Grosveld, F., Vorobjev, I., Tsukita, S., and Akhmanova, A. (2005). CLASP1 and CLASP2 bind to EB1 and regulate microtubule plus-end dynamics at the cell cortex. J. Cell Biol. 168 141–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz, M., Braunfeld, M.B., Guenebaut, V., Heuser, J., and Agard, D.A. (2000). Structure of the γ-tubulin ring complex: A template for microtubule nucleation. Nat. Cell Biol. 2 365–370. [DOI] [PubMed] [Google Scholar]
- Murata, T., Sonobe, S., Baskin, T.I., Hyodo, S., Hasezawa, S., Nagata, T., Horio, T., and Hasebe, M. (2005). Microtubule-dependent microtubule nucleation based on recruitment of γ-tubulin in higher plants. Nat. Cell Biol. 7 961–968. [DOI] [PubMed] [Google Scholar]
- Osborn, M., Webster, R.E., and Weber, K. (1978). Individual microtubules viewed by immunofluorescence and electron microscopy in the same PtK2 cell. J. Cell Biol. 77 R27–R34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredez, A.R., Somerville, C.R., and Ehrhardt, D.W. (2006). Visualization of cellulose synthase demonstrates functional association with microtubules. Science 312 1491–1495. [DOI] [PubMed] [Google Scholar]
- Robinson, J.M., Takizawa, T., and Vandre, D.D. (2000). Enhanced labeling efficiency using ultrasmall immunogold probes: Immunocytochemistry. J. Histochem. Cytochem. 48 487–492. [DOI] [PubMed] [Google Scholar]
- Robinson, J.M., and Vandre, D.D. (1997). Efficient immunocytochemical labeling of leukocyte microtubules with FluoroNanogold: An important tool for correlative microscopy. J. Histochem. Cytochem. 45 631–642. [DOI] [PubMed] [Google Scholar]
- Sammak, P.J., and Borisy, G.G. (1988). Detection of single fluorescent microtubules and methods for determining their dynamics in living cells. Cell Motil. Cytoskeleton 10 237–245. [DOI] [PubMed] [Google Scholar]
- Sandblad, L., Busch, K.E., Tittmann, P., Gross, H., Brunner, D., and Hoenger, A. (2006). The Schizosaccharomyces pombe EB1 homolog Mal3p binds and stabilizes the microtubule lattice seam. Cell 127 1415–1424. [DOI] [PubMed] [Google Scholar]
- Shaw, S.L., Kamyar, R., and Ehrhardt, D.W. (2003). Sustained microtubule treadmilling in Arabidopsis cortical arrays. Science 300 1715–1718. [DOI] [PubMed] [Google Scholar]
- Stoppin-Mellet, V., Gaillard, J., Timmers, T., Neumann, E., Conway, J., and Vantard, M. (2007). Arabidopsis katanin binds microtubules using a multimeric microtubule-binding domain. Plant Physiol. Biochem. 45 867–877. [DOI] [PubMed] [Google Scholar]
- Stoppin-Mellet, V., Gaillard, J., and Vantard, M. (2002). Functional evidence for in vitro microtubule severing by the plant katanin homologue. Biochem. J. 365 337–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoppin-Mellet, V., Gaillard, J., and Vantard, M. (2006). Katanin's severing activity favors bundling of cortical microtubules in plants. Plant J. 46 1009–1017. [DOI] [PubMed] [Google Scholar]
- Sugimoto, K., Williamson, R.E., and Wasteneys, G.O. (2000). New techniques enable comparative analysis of microtubule orientation, wall texture, and growth rate in intact roots of Arabidopsis. Plant Physiol. 124 1493–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, D., Asenjo, A.B., Mennella, V., Sharp, D.J., and Sasa, H. (2006). Kinesin-13s form rings around microtubules. J. Cell Biol. 175 25–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme, D., Bouget, F.-Y., Van Poucke, K., Inze, D., and Geelen, D. (2004. a). Molecular dissection of plant cytokinesis and phragmoplast structure: A survey of GFP-tagged proteins. Plant J. 40 386–398. [DOI] [PubMed] [Google Scholar]
- Van Damme, D., Van Poucke, K., Boutant, E., Ritzenthaler, C., Inze, D., and Geelen, D. (2004. b). In vivo dynamics and differential microtubule-binding activities of MAP65 proteins. Plant Physiol. 136 3956–3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasteneys, G.O. (2002). Microtubule organization in the green kingdom: Chaos or self-order? J. Cell Sci. 115 1345–1354. [DOI] [PubMed] [Google Scholar]
- Wasteneys, G.O., and Fujita, M. (2006). Establishing and maintaining axial growth: Wall mechanical properties and the cytoskeleton. J. Plant Res. 119 5–10. [DOI] [PubMed] [Google Scholar]
- Waterman-Storer, C.M., and Salmon, E.D. (1997). Actomyosin-based retrograde flow of microtubules in the lamella of migrating epithelial cells influences microtubule dynamic instability and turnover and is associated with microtubule breakage and treadmilling. J. Cell Biol. 139 417–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann, S., Wang, H.-W., Avila-Sakar, A., Drubin, D.G., Nogales, E., and Barnes, G. (2006). The Dam1 kinetochore ring complex moves processively on depolymerising microtubule ends. Nature 440 565–569. [DOI] [PubMed] [Google Scholar]
- Wicker-Planquart, C., Stoppin-Mellet, V., Blanchoin, L., and Vantard, M. (2004). Interactions of tobacco microtubule-associated protein MAP65-1b with microtubules. Plant J. 39 126–134. [DOI] [PubMed] [Google Scholar]
- Wightman, R., and Turner, S.R. (2007). Severing at sites of microtubule crossover contributes to microtubule alignment in cortical arrays. Plant J. 52 742–751. [DOI] [PubMed] [Google Scholar]
- Williamson, R.E. (1991). Orientation of cortical microtubules in interphase plant cells. Int. Rev. Cytol. 129 135–206. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.