Abstract
3D cell cultures are rapidly becoming the method of choice for the physiologically relevant modeling of many aspects of non‐malignant and malignant cell behavior ex vivo. Nevertheless, only a limited number of distinct cell types have been evaluated in this assay to date. Here we report the first large scale comparison of the transcriptional profiles and 3D cell culture phenotypes of a substantial panel of human breast cancer cell lines. Each cell line adopts a colony morphology of one of four main classes in 3D culture. These morphologies reflect, at least in part, the underlying gene expression profile and protein expression patterns of the cell lines, and distinct morphologies were also associated with tumor cell invasiveness and with cell lines originating from metastases. We further demonstrate that consistent differences in genes encoding signal transduction proteins emerge when even tumor cells are cultured in 3D microenvironments.
Keywords: Breast cancer, 3D culture, Extracellular matrix, Gene expression profiling, Signal transduction
1. Introduction
Mechanistic studies of human cancer are largely reliant on ex vivo culture of established cell lines. The overwhelming majority of these studies are performed using immortalized cell lines cultured on two‐dimensional plastic substrata. Non‐malignant mammary epithelial cells, as well as other differentiated cell types, rapidly lose many aspects of the differentiated state upon dissociation and culture on plastic substrata (Bissell et al., 1973; Bissell and Tilles, 1971; Bissell, 1981; Emerman and Pitelka, 1977). Over many decades, we and others have proposed (Bissell et al., 1982) and demonstrated (Barcellos‐Hoff et al., 1989, 1994, 2003, 1992, 1991, 1990) that signals from the extracellular matrix play crucial roles in the establishment and maintenance of tissue specificity of non‐malignant mammary cells. We have shown that functional and morphological differentiation can be largely restored by growing cells in a reconstituted basement membrane which provides in culture the crucial cues from extracellular matrix proteins to which these cells respond in vivo (Barcellos‐Hoff et al., 1989; Li et al., 1987; Petersen et al., 1992) and these culture techniques are now being used to study differentiated function in several tissues (reviewed in Kleinman and Martin, 2005; Schmeichel and Bissell, 2003).
We extended these studies to malignant human breast cells and reported in 1992 that non‐malignant and malignant cells can be distinguished rapidly and reliably when grown in three‐dimensional (3D) laminin‐rich extracellular matrix (lrECM) cultures (Petersen et al., 1992). Non‐malignant cells (e.g. HMT‐3522 S1) undergo a small number of rounds of cell division, after which they organize into polarized, growth‐arrested colonies with many of the morphological features of mammary acini (Petersen et al., 1992). This ability to correctly sense the cues from the basement membrane and organize into acini is shared by the other non‐malignant breast epithelial cells which we have studied: MCF‐10A (Muthuswamy et al., 2001; Petersen et al., 1992) and 184 (Fournier et al., 2006). In contrast, malignant cells – both established cell lines and cells from primary tumors – adopt a variety of colony morphologies but share some common aspects – loss of tissue polarity, a disorganized architecture and a failure to arrest growth (Park et al., 2006; Petersen et al., 1992).
Crucially, our studies have shown that signal transduction pathways in non‐malignant cells are integrated in 3D lrECM cultures in ways not observed when cells are cultured as monolayers. Initially, we reported that the expression and activity of β1‐integrin and EGFR are reciprocally downregulated in breast cancer cells treated with various signaling inhibitors, but only when cultured on 3D substrata (Wang et al., 1998). In another example, T4‐2 cells treated with PI3‐Kinase inhibitors undergo a reversion of the malignant phenotype in 3D culture, with downregulation of EGFR, β1‐integrin and upregulation of PTEN – changes which are only seen in cells grown on lrECM – while proximal markers of drug efficacy (e.g. pAkt and pGSK3β) responded similarly in cells grown on both substrata (Liu et al., 2004). We have also shown crucial differences in apoptotic sensitivity in response to chemotherapeutic agents for non‐malignant and malignant breast cell lines in 2D and 3D culture (Weaver et al., 2002), further underscoring the relative value of 3D models over more conventional approaches. More recently we have defined a gene expression signature from acini formed from non‐malignant breast epithelial cells in 3D lrECM and showed that human breast tumors sharing this pattern had a significantly better prognosis (Fournier et al., 2006). These 3D culture models also have played a key role in our validation of two new molecular targets in breast cancer, β1‐integrin (Park et al., 2006; Weaver et al., 1997) and TACE/ADAM17 (Kenny and Bissell, 2007). These data have raised the question of the extent to which monolayer cultures may be failing to recapitulate signaling in vivo (Bissell et al., 2003, 1999).
Whereas there are dramatic morphological (and hence biochemical) differences between normal and malignant cells in 2D and 3D (Bissell et al., 2005; Petersen et al., 1992), the cancer cells are much less sensitive to environmental perturbations. Nevertheless, it is becoming clear that even tumor cells respond to chemotherapy and other factors differently in different microenvironments. Much is known about the gene expression patterns of different breast cancer cell lines on tissue culture plastic (Neve et al., 2006; Perou et al., 1999), yet a comprehensive analysis of the phenotype of a large panel of breast cancer cell lines in 3D culture has been lacking to date. We asked whether we could detect differences in morphology of the breast cancer cell lines in 3D and whether these differences could be correlated with aspects of gene expression. We focused on a large panel of breast cancer cell lines which largely recapitulate the diversity of gene expression patterns and mutational and genomic aberrations found in breast tumors in vivo (Chin et al., 2006; Neve et al., 2006). Here, we report the morphological phenotype of 25 of these breast cell lines grown in 2D and 3D cultures, and their gene expression profiles under these same conditions. These data reveal that breast cancer cell lines generally form colonies with one of four distinctive morphologies and that culture in a 3D microenvironment results in significant and reproducible gene expression changes, even in malignant cells.
2. Results
Twenty‐five established epithelial cell lines originally derived from reduction mammoplasty or breast tumors were cultured in a 3D culture assay in which cells are seeded singly on top of a thin gel comprised of laminin‐rich extracellular matrix (Lee et al., 2007; Park et al., 2006), overlaid with medium containing a small amount of additional lrECM and analyzed after 4 days. In this study we compare 25 cell lines using the following endpoints: 3D morphology, proliferation index at this time point, and expression level and activity of several key signaling proteins. Additionally, we performed Affymetrix gene expression analysis of cells cultured on both substrata.
2.1. Cell lines exhibit distinct morphologies when cultured in the 3D lrECM on the ‘on‐top’ assay
Whereas the panel of cell lines adopted largely non‐distinct morphologies when cultured as monolayers, dramatic differences emerged when grown on a 3D lrECM substratum. The 3D morphologies of these cell lines were characterized by phase contrast microscopy and localization of F‐actin in colonies at the culture endpoint. We classified the cell lines into four distinct morphological groups referred to as: Round, Mass, Grape‐like and Stellate. Representative examples of each class are shown in Figure 1, and the morphology of each of the cell lines in the panel is shown in Figure 2. The Round cell class include HCC1500, MCF‐12A, MDA‐MB‐415, MPE‐600, and S1; these form round colonies on top of gels as viewed by phase contrast microscopy and have nuclei that are organized in a regular manner around the center of the colony as assessed by confocal microscopy. The Mass class, BT‐474, BT‐483, HCC70, HCC1569, MCF‐7, T4‐2 and T‐47D, form colonies which may also have round colony outlines by phase contrast microscopy, but are distinguished from the Round morphological grouping by their disorganized nuclei and filled colony centers (colonies of the Round morphology occasionally form lumens, although the frequency varies greatly between the cell lines). The Grape‐like class, AU565, CAMA‐1, MDA‐MB‐361, MDA‐MB‐453, MDA‐MB‐468, SK‐BR‐3, UACC‐812, ZR‐75‐1 and ZR‐75‐B, form colonies with poor cell–cell contacts and are distinguished by their grape‐like appearance. This phenotype is readily apparent by phase contrast microscopy in most of the cell lines grouped in this morphology but is particularly obvious when F‐actin localization is visualized using phalloidin staining. Although by phase contrast microscopy, UACC‐812 and ZR‐75‐B appear mass‐like, F‐actin staining of these colonies clearly shows a lack of robust cell–cell adhesion and they were therefore classed in the Grape‐like morphology. Lastly, the Stellate class, BT‐549, Hs578T, MDA‐MB‐231 and MDA‐MB‐436, are distinguished by their invasive phenotype in 3D culture, with stellate projections that often bridge multiple cell colonies.
Figure 1.
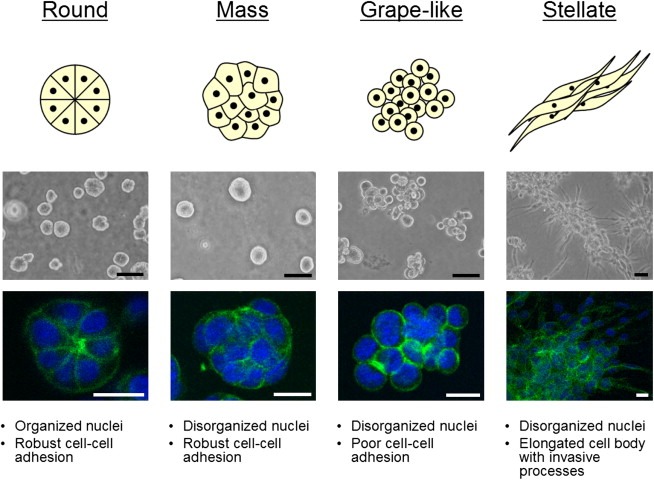
Breast cell line colony morphologies in 3D culture fall into four distinct groups. A panel of 25 breast cell lines were cultured in three‐dimensions and grouped into four distinct morphologies. A schematic and key descriptors of each morphology is shown in addition to phase contrast and F‐actin and nuclear fluorescence images of representative cell lines of each morphology: for Round, S1 is shown; Mass, BT‐474; Grape‐like, SK‐BR‐3; and Stellate, MDA‐MB‐231. Scale bars: phase contrast, 50μm; fluorescence, 20μm.
Figure 2.
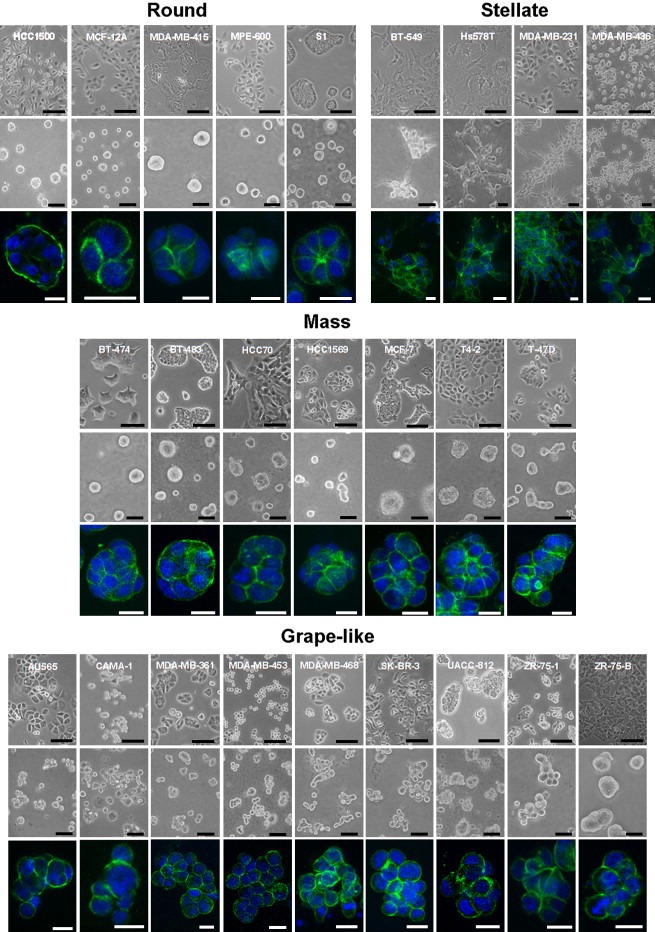
Morphologies of breast cell lines cultured in two‐ and three‐dimensions. Phase contrast images of the complete panel of 25 breast cell lines cultured on tissue culture plastic (top panel) and in the 3D lrECM assay (middle panel) are shown grouped by 3D morphological classification: Round, Mass, Grape‐like and Stellate. 3D cultures were stained for F‐actin and nuclei were counterstained with DAPI. Optical sections of representative colonies are shown for all cell lines (bottom panel) with the exception of MDA‐MB‐468 for which a Z projection with maximum intensity projection (Image J) is shown (because of the high degree of dimensionality of the colonies generated by this cell line, optical sections appear to show single cells not evidently part of the same colony). Scale bars: top panel, 100μm; middle panel, 50μm; bottom panel, 20μm.
2.2. Proliferation analysis
We determined the proliferation index of these 25 cell lines at the endpoint of the 3D lrECM assay by indirect immunofluorescence against Ki67 antigen (Table 1). The proportion of nuclei staining positive for Ki67 at this time point (Table 1) did not show any significant correlation to colony morphology (ANOVA, P=0.92), Sorlie/Perou tumor class (ANOVA, P=0.30), ER status (t‐test, P=0.23) or whether the cell line was derived from a primary tumor or a metastasis (t‐test, P=0.35).
Table 1.
Breast cell line characteristics
| 3D morphology | Cell line | ER | PgR | ERBB2amp. | Mutational status | Tumor type | Tissue source | Tumorigenicity | Tumor classification | Proliferation index (3D) (%) |
|---|---|---|---|---|---|---|---|---|---|---|
| Round | HCC1500 | + | + | N/A | DC | P | Unknown | Basal B | 85.9 | |
| MCF‐12A | − | − | N/A | F | P | No | Basal B | 9.42 | ||
| MDA‐MB‐415 | + | − | TP53 | AC | M (PE) | No | Luminal | 6.02 | ||
| MPE‐600a | + | − | N/A | IDC | Unknowna | Unknown | Luminal | 35.3 | ||
| S1b | − | − | N/A | F | P | No | Basal B | 31 | ||
| Mass | BT‐474 | + | + | + | PIK3CA | IDC | P | Yes | Luminal | 19.2 |
| BT‐483 | + | + | N/A | PIDC | P | Yes | Luminal | 25 | ||
| HCC70 | − | − | N/A | DC | P | Unknown | Basal A | 6.82 | ||
| HCC1569 | − | − | PTEN, TP53 | MC | P | Unknown | Basal A | 43.1 | ||
| MCF‐7 | + | + | CDKN2A, PIK3CA | IDC | M (PE) | Yes | Luminal | 56.7 | ||
| T4‐2b | − | − | TP53e | F | P | Yes | Basal B | 61.6 | ||
| T‐47D | + | + | PIK3CA, TP53 | IDC | M (PE) | Yes | Luminal | 53.4 | ||
| Grape‐like | AU565c | − | − | + | None | AC | M (PE) | Unknown | Luminal | 53.36 |
| CAMA‐1 | + | + | PTEN, VHL | AC | M (PE) | Yes | Luminal | 53.4 | ||
| MDA‐MB‐361 | + | + | + | None | AC | M (Br) | Yes | Luminal | 4.43 | |
| MDA‐MB‐453 | − | − | + | CDH1, PIK3CA | AC | M (PE) | No | Luminal | 72.5 | |
| MDA‐MB‐468 | − | − | MADH4, PTEN, RB1, TP53 | AC | M (PE) | Yes | Basal A | 82.4 | ||
| SK‐BR‐3c | − | − | + | N/A | AC | M (PE) | Yes | Luminal | 35.2 | |
| UACC‐812 | + | − | + | None | IDC | P | Unknown | Luminal | 3.95 | |
| ZR‐75‐1d | + | + | N/A | IDC | M (As) | Yes | Luminal | 5.88 | ||
| ZR‐75‐Bd | + | + | N/A | IDC | M (As) | Unknown | Luminal | 22.1 | ||
| Stellate | BT‐549 | − | − | RB1, TP53 | PIDC | P | Yes | Basal B | 23 | |
| Hs578T | − | − | CDKN2A, HRAS, TP53 | CS | P | Yes | Basal B | 44.2 | ||
| MDA‐MB‐231 | − | − | BRAF, CDKN2A, KRAS, TP53 | AC | M (PE) | Yes | Basal B | 49.5 | ||
| MDA‐MB‐436 | − | − | N/A | AC | M (PE) | Yes | Basal B | 66.5 | ||
Relevant characteristics of non‐malignant and malignant breast cell lines used are organized by 3D morphology and summarized here. Estrogen receptor (ER), progesterone receptor (PgR), ERBB2 amplification status (ERBB2 amp.), primary tumor type, tissue source, experimental tumorigenicity, defined as the ability of a cell line to form tumors in an immunecompromised mouse model, and tumor classification, based on previously published data (Lacroix and Leclercq, 2004; Neve et al., 2006), are indicated. Data on mutations were extracted from the Cancer Genome Project at the Sanger Institute (http://www.sanger.ac.uk/genetics/CGP/CellLines/) for all available cell lines unless otherwise indicated. Cell lines for which mutational status was not available are indicated as N/A. Genes tested in all cell lines: APC, BRAF, CDH1, CDKN2A, CTNNB1, EGFR, HRAS, KRAS, MADH4, NRAS, PIK3CA, PTEN, RB1, STK11, TP53, VHL; additional genes tested in BT‐549, Hs578T, MCF‐7, MDA‐MB‐231 and T‐47D: BRCA1, BRCA2, ERBB2, FLT3, PDGFRA. The proliferation index of cell lines cultured for 4 days in 3D defined as the percentage of nuclei staining positive for Ki67 antigen is also shown.
Abbreviations: AC, adenocarcinoma; As, ascites; Br, brain; CS, carcinosarcoma; DC, ductal carcinoma; F, fibrocystic disease; IDC, invasive ductal carcinoma; M, metastasis; MC, metaplastic carcinoma; P, primary; PE, pleural effusion; PIDC, papillary invasive ductal carcinoma.
MPE‐600 was developed by Vysis International, Inc.
T4‐2 is a tumorigenic cell line derived from S1 in vitro.
AU565 and SK‐BR‐3 were derived from the same patient.
ZR‐75‐B was cloned from ZR‐75‐1.
T4‐2 is not included in the Sanger Institute Cancer Genome Project, but a TP53 mutation in the cell line was reported in (Moyret et al., 1994).
2.3. Expression of signaling proteins
Protein lysates were isolated from cell colonies for western blotting against proteins involved in key signaling processes and in cell–cell interactions (Figure 3). Many of the known major aspects of protein expression in the panel of cell lines are reflected in this analysis. MDA‐MB‐468 had the highest levels of EGFR, consistent with their reported amplification of this gene (Filmus et al., 1985). This cell line also had the highest levels of EGFR activity, the second highest being found in T4‐2 cells, in which this gene is also amplified (Briand et al., 1996).
Figure 3.
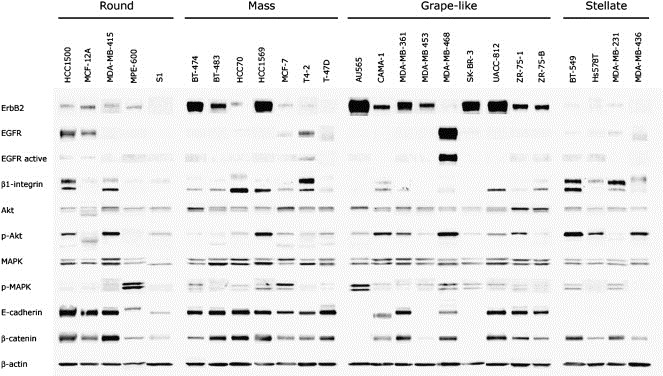
Western blot analysis of breast cell lines cultured in 3D. Western blot analysis of the indicated proteins is shown. Equal amounts of protein were loaded per lane, and lysates of S1 and T4‐2 were run on all separate blots as to control for equal loading and exposure time. Signal from control cell lines were semi‐quantitatively normalized across blots using S1 or T4‐2 signal for each protein in Adobe Photoshop. Normalized protein profiles for each cell line were then aligned and grouped by morphology in this composite figure (see Section 4 for additional detail).
Two immunoreactive bands were seen for β1‐integrin; the lower and upper bands have been reported to correspond to the partially and fully glycosylated forms, respectively (Bellis, 2004). Interestingly, the protein recognized in the upper band was more highly expressed in six cell lines, while the variant of lower apparent molecular weight was the predominant isoform in 14 of the cell lines. The upper band was most common in the Stellate cells.
Several cell lines had significant levels of phosphorylated Akt, including HCC1569, CAMA‐1, BT‐549, ZR‐75‐1 and MDA‐MB‐468, all of which have no functional PTEN. Other lines, which have activating mutations in the gene encoding PI3‐Kinase have somewhat lower, but still substantial levels of pAkt (BT‐474, MCF‐7 and T‐47D) suggesting that PTEN loss may have a more potent effect on activation of Akt than PIK3CA activation. MDA‐MB‐415 also expressed high levels of pAkt, although to our knowledge defects in this pathway in this cell line have not yet been reported.
Most cell lines had similar levels of β‐catenin, with the exception of some of the E‐Cadherin negative cells such as SK‐BR‐3 and AU565. This is consistent with the role of E‐cadherin in stabilizing a pool of β‐catenin in adherens junctions, with the loss of E‐Cadherin leading to the rapid turnover of cytosolic β‐catenin in these cell lines which express the CTNNB1 gene but have a high activity of the β‐catenin turnover pathway (Orford et al., 1997).
Each of the Stellate cell lines lacked E‐cadherin, and the Grape‐like cell lines generally had lower levels than the other two groups, most probably reflecting the more limited cell–cell interactions observed in colonies with this morphology. In many cases, E‐cadherin negativity was consistent with the reported deletions in this panel of cell lines (Hiraguri et al., 1998). Two cell lines, CAMA‐1 and MPE‐600, were positive for E‐cadherin, but the immunoreactive bands were of a slightly different size to the 120KDa band found in the other cell lines. MPE‐600 has been reported to have a deletion of exon 9, while CAMA‐1 has a mutation in the splice acceptor site of exon 12 (van de Wetering et al., 2001).
The highest levels of ErbB2 were found in cell lines of the Grape‐like and Mass categories. Cells of the Round category expressed moderate levels of ErbB2, while this protein was not detected in Stellate cells. AU565, BT‐474, HCC1569, SK‐BR‐3, MDA‐MB‐361, MDA‐MB‐453, and UACC‐812 have been reported to have amplified this gene (Lacroix and Leclercq, 2004 and references therein; Neve et al., 2006). Of the remaining cell lines with high levels of ErbB2, BT‐483 and ZR‐75‐1 are not amplified at the ErbB2 locus (Kallioniemi et al., 1992; Kraus et al., 1987), however, both are known to overexpress ErbB2 (Gazdar et al., 1998; Kraus et al., 1987).
2.4. Gene expression profiling of breast epithelial cells grown in two‐ and three‐dimensions
We determined the gene expression profile of 24 of the 25 cell lines in 2D and 3D cultures. During the period in which these studies were performed, the array platform being utilized was upgraded from the Affymetrix high‐density oligonucleotide array cartridge system to the newer, Affymetrix high throughput array (HTA) GeneChip system. To ensure the validity of our inter‐platform comparison (for a detailed description, please see Section 4), we analyzed a number of samples on both platforms. These samples clustered together following the cross‐platform normalization, indicating that the approach is valid and robust. In total, 89 samples from 24 different cell lines were analyzed, 47 in 2D and 42 in 3D. Replicates were not averaged and are presented as individual samples to provide additional confidence in the robustness of our inter‐platform comparison and clustering approach. These data have been deposited in the Array Express database (http://www.ebi.ac.uk/arrayexpress/; accession number E‐TABM‐244). Figure 4 represents unsupervised hierarchical clustering of the 89 samples.
Figure 4.
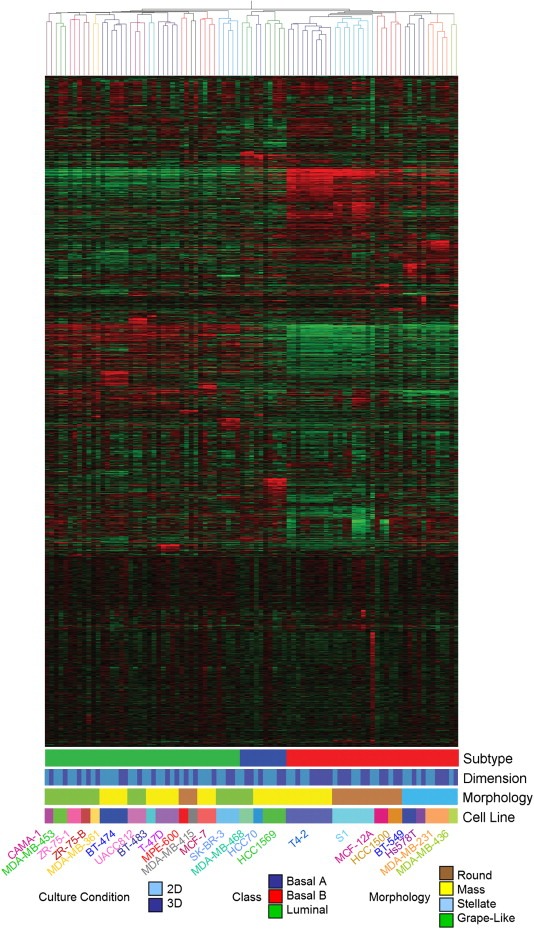
Gene expression profiling of breast cell lines grown in two‐ and three‐dimensions. Unsupervised hierarchical clustering of 89 samples representing 24 non‐malignant and malignant breast cell lines cultured on either tissue culture plastic (2D) or lrECM (3D). Tree branches are colored to indicate cell line identity (see key on bottom of figure). Also colored in the key are the morphological group and the tumor classification to which each cell line belongs.
In agreement with the earlier report from one of our laboratories (Neve et al., 2006), the cell lines grouped into two broad clusters which have similarities to the Luminal and Basal subclasses described by Perou et al. (2000) in human breast cancer cases. As described by Neve et al. (2006) the Basal cell lines could be further subdivided into two groups, Basal A and Basal B. The cell lines of the HMT‐3522 series (non‐malignant S1 and malignant T4‐2) cluster with the non‐malignant MCF‐12A cell line in the Basal B subclass. Given that the malignant T4‐2 cells were derived from S1 cells by selection in culture (rather than over many years of mutation and evolution under the complex selective pressures in a cancer patient), it is perhaps unsurprising, but also intriguing that it clusters with those of a more normal phenotype. The HCC1500 cell line is an outlier of this group, distinct from the other cells with a basal phenotype. In 3D culture, this cell line (although tumor‐derived) still retains some aspects of the normal mammary gland morphology and has been occasionally observed to form colonies with nuclear organization around a central lumen with apical F‐actin localization (data not shown).
While the Basal/Luminal delineation seems to be the strongest driver of the clustering pattern, cell lines of similar morphologies within these sub‐types frequently clustered together suggesting that the gene expression pattern of the cells is a strong determinant of colony morphology (Figure 4). All four of the Stellate cell lines are more similar to each other than to cell lines of any other morphology, while all of the Round cell lines of the Basal subtype clustered together. Among the Luminal subtype, five of the seven Grape‐Like cell lines cluster together and there is a sub‐cluster of two Round cell lines in a larger cluster of mostly Mass cell lines.
2.5. 2D versus 3D expression comparison
From the hierarchical clustering, it is clear that the 2D and 3D expression profiles of the individual cell lines cluster together, independently of the substratum on which they were cultured. These data indicate that 3D culture did not effect a consistent and widespread change in gene expression causing each cell line to adopt a substantially different and common program of gene expression. Nevertheless, it is also clear that the 3D microenvironment did effect significant changes in the gene expression profiles of these cancer cell lines.
To identify those genes which respond consistently to the 3D culture microenvironment in all of the cell lines, we averaged all of the replicates for each cell line under each culture condition and tested whether there was a set of genes which distinguished cell lines grown in 2D versus those grown in 3D (ANOVA, cutoff P<0.00025). We identified 96 Affymetrix probes which were strongly up/downregulated consistently across the majority of the cell lines (Figure 5). These data indicate that, although cell line identity and the Luminal/Basal phenotype make a strong contribution to the gene expression profiles, the culture microenvironment and substratum also exert significant effects.
Figure 5.
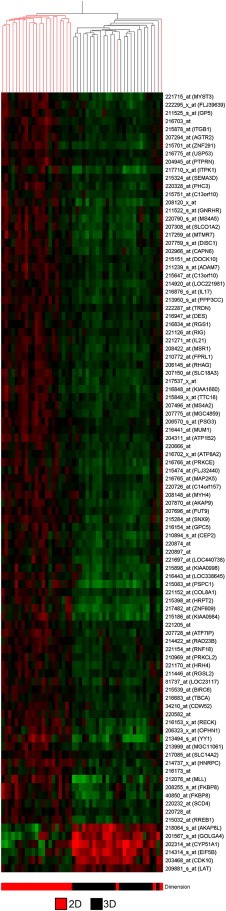
Genes which distinguish 2D and 3D culture conditions. All replicates were averaged so that each condition represents one cell line in either 2D or 3D culture. The 22215 Affymetrix probes were tested for association with the parameter, Dimension, using a cutoff of P < 0.00025, corrected for multiple comparisons using the Benjamini and Hochberg test. Ninety‐six probes significantly distinguished the expression profiles of cells grown on plastic from those grown on lrECM at this level of significance.
Of these 96 Affymetrix probes, 41 corresponded to genes with annotated functions. We used Gene Ontology annotations (Ashburner et al., 2000) to determine whether particular molecular functions were statistically overrepresented in this set. These genes and their Gene Ontology classifications are shown in Figure 6. Of the eight classifications found, one – “signal transducer activity”– was statistically significantly overrepresented in the set of genes which differ between 2D and 3D culture (P=0.0201). The “enzyme regulator activity” class almost reached statistical significance (P=0.0509). Given that many signal transduction molecules are regulated at the level of phosphorylation and other protein modifications rather than at transcriptional levels, we expect that the actual changes at the level of cellular signaling would be even more striking than the gene expression arrays indicate. These data provide additional support for our contention that regulation of signal transduction is substantially different in cells cultured on basement membrane gels (Liu et al., 2004, 2002, 1998, 2002).
Figure 6.
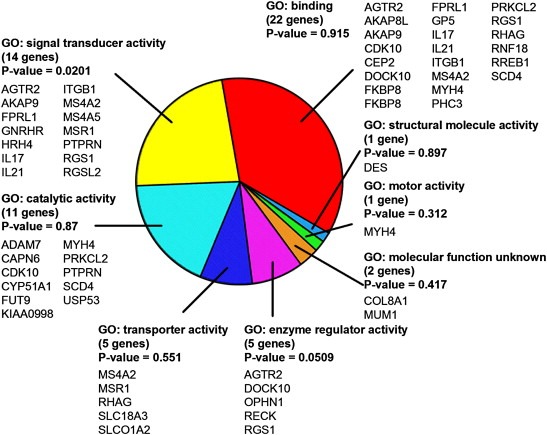
Gene Ontology analysis of genes distinguishing cells grown in 2D and 3D culture conditions. Gene Ontology analysis of the 41 of the 96 genes shown in Figure 5 for which Gene Ontology annotations were available. Genes encoding proteins involved in signal transduction are significantly overrepresented in this set (P=0.0201), while genes encoding proteins involved in the regulation of enzyme activity almost achieved statistical significance (P=0.0509).
3. Discussion
The utility of the 3D culture models to explore aspects of the normal and malignant phenotypes in culture has now been widely recognized and many workers have adopted these approaches to study cancers of the breast and of other tissues (reviewed in Schmeichel and Bissell, 2003). One recent fruitful avenue has been to take non‐malignant cells, e.g. S1 or MCF‐10A which form acini in 3D cultures, and specifically overexpress or ablate expression of potentially cancer‐relevant genes and analyze the effects of the manipulation on the ability of these cells to execute acinar morphogenesis (Debnath et al., 2002, 2003, 2005, 2005, 2005, 2001, 2006, 2006, 1997, 2007). Many of these reports have shown that the effects of activation of oncogenes or inactivation of tumor‐suppressor genes are profoundly different in cells cultured in different microenvironments. Setting the “oncogenic lesion” a priori has allowed the dissection of the potential contribution of individual components to various aspects of the malignant phenotype and has provided a wealth of useful information. In these systems it is possible to examine one, two or perhaps three factors acting in concert, but the technical difficulties with these experiments, and the intrinsic gene selection bias in experimental design resulting from our incomplete knowledge of cancer cell biology and of the endogenous aberrations in the host cells, impose a significant constraint on the utility of this widely used approach.
Unlike such models developed in tissue culture, the cell lines in our panel were selected for the ability to proliferate to form clinically relevant and often invasive and metastatic tumors in women. The exposure to these strong evolutionary pressures in vivo, promoting such phenotypes as angiogenic potential, resistance to growth inhibitory cues and ability to survive in circulation and at sites distant from the primary tumor – may be more likely to represent relevant cancer models than those constructed in the laboratory. This panel recapitulates, in large part, the full spectrum of interacting mutations and aberrations found in human breast cancer cases (Neve et al., 2006). These include those alterations of which we already have a good understanding and many alterations – “unknown unknowns” – of which we are still ignorant. In this way, our experimental approaches are not biased strongly by prior assumptions, and the sample set is sufficiently diverse to allow us and other workers to choose individual cell lines based on the data provided here to test specific hypotheses. More importantly, perhaps, we should consider these assays and paradigms as we explore ways of testing and selecting current and future chemotherapeutics.
We had previously reported, in a much smaller sample set, that breast cancer cell lines adopt a variety of morphologies in 3D culture and share a number of common properties which distinguish them from non‐malignant breast epithelial cells – specifically, a failure to arrest growth and form organized, polarized colonies in response to cues from the extracellular matrix. Here we not only expand on the previous work, but show the plasticity and the diversity of the malignant cells themselves when grown on top of 3D gels. We describe four distinct morphological classes which show correlations with the underlying gene and protein expression patterns. Interestingly, eight of the nine Grape‐Like cells were isolated from tumor metastases. In general, these cells formed less closely associated colonies with reduced cell–cell adhesion compared to cell lines of the other morphologies. This may reflect, in part, the acquisition of the ability to break away from their neighbors in the primary tumor over the course of their evolution as they acquired the ability to metastasize.
We previously reported that breast cancer cell lines belonging to the Basal B category were much more invasive by Boyden chamber assays than those belonging to the Basal A and Luminal categories (Neve et al., 2006). The two outliers in this correlation were MCF‐10A and MCF‐12A, both Basal B but highly un‐invasive. Interestingly, both of these cell lines fall under the Round class by 3D morphology (Muthuswamy et al., 2001 and Figure 2). Of the other eight Basal B cell lines which are highly invasive, six cell lines are of the Stellate 3D morphology: three are described in full here (BT‐549, Hs578T, MDA‐MB‐231) and we have also characterized the other three (HBL‐100, MDA‐MB‐157, SUM 159PT) as Stellate; data not shown). MDA‐MB‐436 has been shown to be invasive by similar in vitro assays (Albini et al., 1987). Thus far, all of the cell lines we have characterized as Stellate have been shown to be invasive in commonly used in vitro assays, suggesting the utility of the 3D colony morphology as a functional assay for invasive potential.
In the model we have studied most intensively, malignant T4‐2 of the HMT‐3522 series, we have used the 3D culture assay to identify a number of signaling nodes which regulate proliferation and architecture of the cells in response to a number of signaling inhibitors (for review see Bissell et al., 2003). In this assay, the disorganized, apolar phenotype of the T4‐2 cells can be ‘reverted’ to a phenotype much more closely approximating that of non‐malignant breast acini (and the reverted structures show a profound reduction in tumorigenicity in vivo). We determined that the level of signaling downstream of both EGFR and β1‐integrin are critical for the maintenance or reversion of the malignant phenotype in this cell line (Wang et al., 1998; Weaver et al., 1997) and have made substantial inroads into understanding how the key signals may be elicited (Kenny and Bissell, 2007) and transduced (Liu et al., 2004). We expanded this analysis to a small number of other breast cancer cell lines in 3D cultures (MDA‐MB‐231, MCF‐7 and Hs578T) and demonstrated the feasibility of using combinatorial approaches to identify the key deregulated pathways in these cell lines (Wang et al., 2002). Large scale studies of tumor cells in this culture context offer the possibility of rapidly identifying the key molecular vulnerabilities in ways not requiring intensive gene expression or proteomic analyses. The demonstration by Nevins and co‐workers that gene expression profiles can be used to predict sensitivity and resistance to different chemotherapeutic agents (Bild et al., 2006; Potti et al., 2006) is exciting and could be used to shortlist compounds for testing in this assay. Since we have shown that resistance to a number of chemotherapeutic agents are dramatically increased by tissue (acinar) polarity in 3D cultures, it would be interesting to explore if signatures from 3D models are better at predicting drug sensitivity in vivo than profiles derived from cells cultured on 2D plastic substrata.
In addition to providing important physical and biochemical cues to adhesion receptors, the gelatinous extracellular matrices provide significantly more pliable microenvironments than tissue culture plastic and are thus, in these very important respects, considerably closer to the microenvironments within which cells exist in vivo. We and others have repeatedly demonstrated that this environment allows cells to self‐organize transcriptionally (Schmidhauser et al., 1992; Xu et al., in press), architecturally (Petersen et al., 1992; Roskelley et al., 1994) and at the level of nuclear organization (Lelievre et al., 1998; Maniotis et al., 2005) to express differentiated functions lost in cells grown on tissue culture plastic. We have provided much data suggesting that signal transduction is integrated in cells grown on laminin‐containing gels in ways which are fundamentally different from cells grown on plastic. That the differentiated state of so many non‐malignant cell types is restored ex vivo under the 3D conditions we have defined argues that the signaling events observed are significantly closer to the cognate events in vivo. These concepts are now supported further by the systematic differences observed in 2D versus 3D in the expression of genes encoding signaling proteins in malignant cells. The data support our contention that even malignant cells are plastic and dependent on their microenvironmental signals for survival, growth and metastasis (Bissell and Radisky, 2001). This study, reporting the initial characterization of the growth kinetics, morphology and gene expression patterns of this panel of cell lines in physiologically relevant culture substrata, represents an important initial step to realizing the full potential of thinking in three‐dimensions.
4. Experimental procedures
4.1. Cell culture
HMT‐3522 S1 (S1) and HMT‐3522 T4‐2 (T4‐2) mammary epithelial cells were maintained on tissue culture plastic as previously described (Briand et al., 1996; Briand et al., 1987; Petersen et al., 1992; Weaver et al., 1997). The following human breast cell lines were maintained on tissue culture plastic in the following manners: CAMA‐1, Hs578T, MCF‐7, MDA‐MB‐231, MDA‐MB‐361, MDA‐MB‐415, MDA‐MB‐436, MDA‐MB‐453, MDA‐MB‐468, MPE‐600, SK‐BR‐3, UACC‐812 were propagated in DMEM/H‐21 (Invitrogen) with 10% fetal bovine serum (Gemini); AU565, BT‐474, BT‐483, BT‐549, HCC70, HCC1500, HCC1569, T‐47D, ZR‐75‐1, ZR‐75‐B were propagated in RPMI 1640 (Invitrogen) with 10% fetal bovine serum; and MCF‐12A was propagated in DMEM/F‐12 (Invitrogen) with 10ng/ml insulin, 100ng/ml cholera toxin, 500ng/ml hydrocortisone, 20ng/ml EGF (Sigma), and 5% fetal bovine serum. Three‐dimensional laminin‐rich extracellular matrix (3D lrECM) on‐top cultures (Lee et al., 2007) were prepared by trypsinization of cells from tissue culture plastic, seeding of single cells on top of a thin gel of Engelbreth‐Holm‐Swarm (EHS) tumor extract (Matrigel: BD Biosciences; Cultrex BME: Trevigen), and addition of medium containing 5% EHS. Cell lines with Round 3D morphology were seeded at a density of 3.1×104 cells per cm2; cell lines with Stellate 3D morphology were seeded at 1.6×104 cells per cm2; all other cell lines were seeded at 2.1×104 cells per cm2. All 3D lrECM cell cultures were maintained in H14 medium with 1% fetal bovine serum, with the exception of S1 and T4‐2 which were maintained in their propagation medium, for 4 days with media change every 2 days.
4.2. Expression analysis
Colonies were isolated from 3D cultures by dissolution in PBS/EDTA as previously described (Lee et al., 2007). Purified total cellular RNA was extracted using RNeasy Mini Kit with on‐column DNase digestion (Qiagen). RNA was quantified by measuring optical density at A260 and quality was verified by agarose gel electrophoresis. Affymetrix microarray analysis was performed using either the Affymetrix high‐density oligonucleotide array human HG‐U133A chip cartridge system or the Affymetrix High Throughput Array (HTA) GeneChip system, in which HG‐U133A chips are mounted on pegs arranged in a 96 well format.
4.2.1. Data processing
A multi‐step process was used in order to be able to perform a robust comparison of expression data generated at different times on two separate array platforms. Robust multi‐array analysis (RMA) (Irizarry et al., 2003) was performed separately on data generated on both platforms. The data sets from each platform were then row‐centered separately by subtracting a constant from all probesets across samples, such that the mean of each probeset equaled zero. The row‐centered data sets were then combined and row‐centered together. These data have been deposited in the Array Express database (http://www.ebi.ac.uk/arrayexpress/; accession number E‐TABM‐244).
4.3. Immunofluorescence and image acquisition
Immunofluorescence assays were performed as previously described (Lee et al., 2007). In brief, cells were either isolated from 3D cultures and fixed onto glass slides with 4% paraformaldehyde or 1:1 methanol:acetone or fixed directly in culture with 4% paraformaldehyde. Cells were blocked and stained with either fluorescein‐labeled phalloidin (Molecular Probes) diluted 1:500 or anti‐Ki67 antigen (Vector Laboratories) diluted 1:500 followed by Texas Red‐labeled secondary antibody. Nuclei were counterstained with diaminophenylindole (DAPI; Sigma). Slides were mounted with VECTASHIELD Hardset Mounting Medium (Vector Laboratories).
Proliferation index was assessed by quantification of proportion of cells positive for Ki67 antigen. The number of nuclei positive for Ki67 antigen was scored by eye and divided by the total number of nuclei. The mean number of total nuclei counted per cell line was 441, with a minimum of 119.
Confocal analysis was performed using either a Zeiss LSM 410 laser scanning confocal system or a Solamere Technology Group spinning disk confocal system comprised of a Zeiss Axiovert 200M inverted microscope, Yokagawa CSU10 confocal scan head and Stanford Photonics XR/Mega‐10 ICCD camera, run by QED InVivo software (Media Cybernetics). Images were analysed using ImageJ (National Institutes of Health) and Adobe Photoshop.
4.4. Western blotting
Cells were isolated from 3D cultures as described above, lysed in 2% SDS in PBS containing 1mM sodium orthovanadate, 1.5mM sodium fluoride (Sigma), and 1× protease inhibitor cocktail set I (Calbiochem), and homogenized by sonication. Equal amounts of protein were fractionated by SDS‐PAGE, transferred to nitrocellulose membranes and probed with antibodies against the following proteins: Akt, E‐cadherin, β‐catenin, EGFR, activated EGFR, β1‐integrin (BD Biosciences), phosphorylated S473 Akt, ErbB2, MAPK, phosphorylated T202 and T204 MAPK (Cell Signaling Technology). β‐actin (Sigma) was used as a loading control. Blots were developed with SuperSignal West Femto Maximum Sensitivity Substrate (Pierce) and visualized with a FluorChem 8900 imager (Alpha Innotech). Lysates from S1 and T4‐2 cells were run on all separate blots in order for signals from cell lines run at different times to be semi‐quantitatively compared. Normalization across separate blots was performed in Adobe Photoshop. First, the sizes of all protein bands were normalized across blots by measuring the width of all T4‐2 protein bands with the Ruler tool and resizing blots to equalize the widths while maintaining a fixed aspect ratio. Next, normalization of signal of each protein blotted was performed by using the Eyedropper tool to measure and equalize maximum band intensities of either S1 or T4‐2 across all blots. Signal from T4‐2 (being generally stronger than S1) was used for normalization except for phosphorylated MAPK and phosphorylated Akt. For normalization of saturated β1‐integrin and β‐actin signal, the size of the saturated bands was used for normalization. After normalization was complete, protein bands were aligned and all layers were flattened.
Acknowledgements
These investigations were initially made possible by an Innovator award from the Department of Defense Breast Cancer Research Program (DAMD17‐02‐1‐0438) to M.J.B. and additionally by a Distinguished Scientist Award and a grant from the US Department of Energy, Office of Biological and Environmental Research (DE‐AC03 SF0098), and in part by the National Cancer Institute (R01 CA064786 to M.J.B and O.W.P. and U54 CA112970 to J.W.G). P.A. Kenny was supported by postdoctoral training fellowships from the Susan G. Komen Breast Cancer Foundation (#2000‐223) and the Department of Defense Breast Cancer Research Program (DAMD17‐00‐1‐0224).
Kenny Paraic A., Lee Genee Y., Myers Connie A., Neve Richard M., Semeiks Jeremy R., Spellman Paul T., Lorenz Katrin, Lee Eva H., Barcellos-Hoff Mary Helen, Petersen Ole W., Gray Joe W., Bissell Mina J., (2007), The morphologies of breast cancer cell lines in three-dimensional assays correlate with their profiles of gene expression, Molecular Oncology, 1, doi:10.1016/j.molonc.2007.02.004.
References
- Albini, A. , Iwamoto, Y. , Kleinman, H.K. , Martin, G.R. , Aaronson, S.A. , Kozlowski, J.M. , McEwan, R.N. , 1987. A rapid in vitro assay for quantitating the invasive potential of tumor cells. Cancer Research. 47, 3239–3245. [PubMed] [Google Scholar]
- Ashburner, M. , Ball, C.A. , Blake, J.A. , Botstein, D. , Butler, H. , Cherry, J.M. , Davis, A.P. , Dolinski, K. , Dwight, S.S. , Eppig, J.T. , 2000. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature Genetics. 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcellos-Hoff, M.H. , Aggeler, J. , Ram, T.G. , Bissell, M.J. , 1989. Functional differentiation and alveolar morphogenesis of primary mammary cultures on reconstituted basement membrane. Development. 105, 223–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellis, S.L. , 2004. Variant glycosylation: an underappreciated regulatory mechanism for beta1 integrins. Biochimica et Biophysica Acta. 1663, 52–60. [DOI] [PubMed] [Google Scholar]
- Bild, A.H. , Yao, G. , Chang, J.T. , Wang, Q. , Potti, A. , Chasse, D. , Joshi, M.B. , Harpole, D. , Lancaster, J.M. , Berchuck, A. , 2006. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 439, 353–357. [DOI] [PubMed] [Google Scholar]
- Bissell, D.M. , Hammaker, L.E. , Meyer, U.A. , 1973. Parenchymal cells from adult rat liver in nonproliferating monolayer culture. I. Functional studies. Journal of Cell Biology. 59, 722–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissell, D.M. , Tilles, J.G. , 1971. Morphology and function of cells of human embryonic liver in monolayer culture. Journal of Cell Biology. 50, 222–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissell, M.J. , 1981. The differentiated state of normal and malignant cells or how to define a “normal” cell in culture. International Review of Cytology. 70, 27–100. [DOI] [PubMed] [Google Scholar]
- Bissell, M.J. , Hall, H.G. , Parry, G. , 1982. How does the extracellular matrix direct gene expression?. Journal of Theoretical Biology. 99, 31–68. [DOI] [PubMed] [Google Scholar]
- Bissell, M.J. , Kenny, P.A. , Radisky, D.C. , 2005. Microenvironmental regulators of tissue structure and function also regulate tumor induction and progression: the role of extracellular matrix and its degrading enzymes. Cold Spring Harbor Symposia on Quantitative Biology. 70, 343–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissell, M.J. , Radisky, D. , 2001. Putting tumours in context. Nature Reviews Cancer. 1, 46–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissell, M.J. , Rizki, A. , Mian, I.S. , 2003. Tissue architecture: the ultimate regulator of breast epithelial function. Current Opinion in Cell Biology. 15, 753–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissell, M.J. , Weaver, V.M. , Lelievre, S.A. , Wang, F. , Petersen, O.W. , Schmeichel, K.L. , 1999. Tissue structure, nuclear organization, and gene expression in normal and malignant breast. Cancer Research. 59, 1757–1763s. (Discussion 1763s-1764s) [PubMed] [Google Scholar]
- Briand, P. , Nielsen, K.V. , Madsen, M.W. , Petersen, O.W. , 1996. Trisomy 7p and malignant transformation of human breast epithelial cells following epidermal growth factor withdrawal. Cancer Research. 56, 2039–2044. [PubMed] [Google Scholar]
- Briand, P. , Petersen, O.W. , Van Deurs, B. , 1987. A new diploid nontumorigenic human breast epithelial cell line isolated and propagated in chemically defined medium. In Vitro Cellular and Developmental Biology. 23, 181–188. [DOI] [PubMed] [Google Scholar]
- Chin, K. , Devries, S. , Fridlyand, J. , Spellman, P.T. , Roydasgupta, R. , Kuo, W.L. , Lapuk, A. , Neve, R.M. , Qian, Z. , Ryder, T. , 2006. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 10, 529–541. [DOI] [PubMed] [Google Scholar]
- Debnath, J. , Mills, K.R. , Collins, N.L. , Reginato, M.J. , Muthuswamy, S.K. , Brugge, J.S. , 2002. The role of apoptosis in creating and maintaining luminal space within normal and oncogene-expressing mammary acini. Cell. 111, 29–40. [DOI] [PubMed] [Google Scholar]
- Debnath, J. , Walker, S.J. , Brugge, J.S. , 2003. Akt activation disrupts mammary acinar architecture and enhances proliferation in an mTOR-dependent manner. Journal of Cell Biology. 163, 315–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerman, J.T. , Pitelka, D.R. , 1977. Maintenance and induction of morphological differentiation in dissociated mammary epithelium on floating collagen membranes. In Vitro. 13, 316–328. [DOI] [PubMed] [Google Scholar]
- Filmus, J. , Pollak, M.N. , Cailleau, R. , Buick, R.N. , 1985. MDA-468, a human breast cancer cell line with a high number of epidermal growth factor (EGF) receptors, has an amplified EGF receptor gene and is growth inhibited by EGF. Biochemical and Biophysical Research Communications. 128, 898–905. [DOI] [PubMed] [Google Scholar]
- Fournier, M.V. , Martin, K.J. , Kenny, P.A. , Xhaja, K. , Bosch, I. , Yaswen, P. , Bissell, M.J. , 2006. Gene expression signature in organized and growth-arrested mammary acini predicts good outcome in breast cancer. Cancer Research. 66, 7095–7102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazdar, A.F. , Kurvari, V. , Virmani, A. , Gollahon, L. , Sakaguchi, M. , Westerfield, M. , Kodagoda, D. , Stasny, V. , Cunningham, H.T. , Wistuba, I.I. , 1998. Characterization of paired tumor and non-tumor cell lines established from patients with breast cancer. International Journal of Cancer. 78, 766–774. [DOI] [PubMed] [Google Scholar]
- Gunawardane, R.N. , Sgroi, D.C. , Wrobel, C.N. , Koh, E. , Daley, G.Q. , Brugge, J.S. , 2005. Novel role for PDEF in epithelial cell migration and invasion. Cancer Research. 65, 11572–11580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraguri, S. , Godfrey, T. , Nakamura, H. , Graff, J. , Collins, C. , Shayesteh, L. , Doggett, N. , Johnson, K. , Wheelock, M. , Herman, J. , 1998. Mechanisms of inactivation of E-cadherin in breast cancer cell lines. Cancer Research. 58, 1972–1977. [PubMed] [Google Scholar]
- Irie, H.Y. , Pearline, R.V. , Grueneberg, D. , Hsia, M. , Ravichandran, P. , Kothari, N. , Natesan, S. , Brugge, J.S. , 2005. Distinct roles of Akt1 and Akt2 in regulating cell migration and epithelial-mesenchymal transition. Journal of Cell Biology. 171, 1023–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry, R.A. , Bolstad, B.M. , Collin, F. , Cope, L.M. , Hobbs, B. , Speed, T.P. , 2003. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Research. 31, e15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isakoff, S.J. , Engelman, J.A. , Irie, H.Y. , Luo, J. , Brachmann, S.M. , Pearline, R.V. , Cantley, L.C. , Brugge, J.S. , 2005. Breast cancer-associated PIK3CA mutations are oncogenic in mammary epithelial cells. Cancer Research. 65, 10992–11000. [DOI] [PubMed] [Google Scholar]
- Kallioniemi, O.P. , Kallioniemi, A. , Kurisu, W. , Thor, A. , Chen, L.C. , Smith, H.S. , Waldman, F.M. , Pinkel, D. , Gray, J.W. , 1992. ERBB2 amplification in breast cancer analyzed by fluorescence in situ hybridization. Proceedings of the National Academy of Sciences of the United States of America. 89, 5321–5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny, P.A. , Bissell, M.J. , 2007. Targeting TACE-dependent EGFR ligand shedding in breast cancer. Journal of Clinical Investigation. 117, 337–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinman, H.K. , Martin, G.R. , 2005. Matrigel: basement membrane matrix with biological activity. Seminars in Cancer Biology. 15, 378–386. [DOI] [PubMed] [Google Scholar]
- Kraus, M.H. , Popescu, N.C. , Amsbaugh, S.C. , King, C.R. , 1987. Overexpression of the EGF receptor-related proto-oncogene erbB-2 in human mammary tumor cell lines by different molecular mechanisms. EMBO Journal. 6, 605–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroix, M. , Leclercq, G. , 2004. Relevance of breast cancer cell lines as models for breast tumours: an update. Breast Cancer Research and Treatment. 83, 249–289. [DOI] [PubMed] [Google Scholar]
- Lee, G.Y. , Kenny, P.A. , Lee, E.H. , Bissell, M.J. , 2007. 3D culture models of normal and malignant breast epithelial cells. Nature Methods. 4, 359–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelievre, S.A. , Weaver, V.M. , Nickerson, J.A. , Larabell, C.A. , Bhaumik, A. , Petersen, O.W. , Bissell, M.J. , 1998. Tissue phenotype depends on reciprocal interactions between the extracellular matrix and the structural organization of the nucleus. Proceedings of the National Academy of Sciences of the United States of America. 95, 14711–14716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, M.L. , Aggeler, J. , Farson, D.A. , Hatier, C. , Hassell, J. , Bissell, M.J. , 1987. Influence of a reconstituted basement membrane and its components on casein gene expression and secretion in mouse mammary epithelial cells. Proceedings of the National Academy of Sciences of the United States of America. 84, 136–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H. , Radisky, D.C. , Wang, F. , Bissell, M.J. , 2004. Polarity and proliferation are controlled by distinct signaling pathways downstream of PI3-kinase in breast epithelial tumor cells. Journal of Cell Biology. 164, 603–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniotis, A.J. , Valyi-Nagy, K. , Karavitis, J. , Moses, J. , Boddipali, V. , Wang, Y. , Nunez, R. , Setty, S. , Arbieva, Z. , Bissell, M.J. , 2005. Chromatin organization measured by AluI restriction enzyme changes with malignancy and is regulated by the extracellular matrix and the cytoskeleton. American Journal of Pathology. 166, 1187–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyret, C. , Madsen, M.W. , Cooke, J. , Briand, P. , Theillet, C. , 1994. Gradual selection of a cellular clone presenting a mutation at codon 179 of the p53 gene during establishment of the immortalized human breast epithelial cell line HMT-3522. Experimental Cell Research. 215, 380–385. [DOI] [PubMed] [Google Scholar]
- Muthuswamy, S.K. , Li, D. , Lelievre, S. , Bissell, M.J. , Brugge, J.S. , 2001. ErbB2, but not ErbB1, reinitiates proliferation and induces luminal repopulation in epithelial acini. Nature Cell Biology. 3, 785–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neve, R.M. , Chin, K. , Fridlyand, J. , Yeh, J. , Baehner, F.L. , Fevr, T. , Clark, L. , Bayani, N. , Coppe, J.P. , Tong, F. , 2006. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 10, 515–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orford, K. , Crockett, C. , Jensen, J.P. , Weissman, A.M. , Byers, S.W. , 1997. Serine phosphorylation-regulated ubiquitination and degradation of beta-catenin. Journal of Biological Chemistry. 272, 24735–24738. [DOI] [PubMed] [Google Scholar]
- Overholtzer, M. , Zhang, J. , Smolen, G.A. , Muir, B. , Li, W. , Sgroi, D.C. , Deng, C.X. , Brugge, J.S. , Haber, D.A. , 2006. Transforming properties of YAP, a candidate oncogene on the chromosome 11q22 amplicon. Proceedings of the National Academy of Sciences of the United States of America. 103, 12405–12410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, C.C. , Zhang, H. , Pallavicini, M. , Gray, J.W. , Baehner, F. , Park, C.J. , Bissell, M.J. , 2006. Beta1 integrin inhibitory antibody induces apoptosis of breast cancer cells, inhibits growth, and distinguishes malignant from normal phenotype in three dimensional cultures and in vivo. Cancer Research. 66, 1526–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perou, C.M. , Jeffrey, S.S. , van de Rijn, M. , Rees, C.A. , Eisen, M.B. , Ross, D.T. , Pergamenschikov, A. , Williams, C.F. , Zhu, S.X. , Lee, J.C. , 1999. Distinctive gene expression patterns in human mammary epithelial cells and breast cancers. Proceedings of the National Academy of Sciences of the United States of America. 96, 9212–9217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perou, C.M. , Sorlie, T. , Eisen, M.B. , van de Rijn, M. , Jeffrey, S.S. , Rees, C.A. , Pollack, J.R. , Ross, D.T. , Johnsen, H. , Akslen, L.A. , Fluge, O. , Pergamenschikov, A. , Williams, C. , Zhu, S.X. , Lonning, P.E. , Borresen-Dale, A.L. , Brown, P.O. , Botstein, D. , 2000. Molecular portraits of human breast tumors. Nature. 406, 747–752. [DOI] [PubMed] [Google Scholar]
- Petersen, O.W. , Ronnov-Jessen, L. , Howlett, A.R. , Bissell, M.J. , 1992. Interaction with basement membrane serves to rapidly distinguish growth and differentiation pattern of normal and malignant human breast epithelial cells. Proceedings of the National Academy of Sciences of the United States of America. 89, 9064–9068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potti, A. , Dressman, H.K. , Bild, A. , Riedel, R.F. , Chan, G. , Sayer, R. , Cragun, J. , Cottrill, H. , Kelley, M.J. , Petersen, R. , 2006. Genomic signatures to guide the use of chemotherapeutics. Nature Medicine. 12, 1294–1300. [DOI] [PubMed] [Google Scholar]
- Reginato, M.J. , Mills, K.R. , Paulus, J.K. , Lynch, D.K. , Sgroi, D.C. , Debnath, J. , Muthuswamy, S.K. , Brugge, J.S. , 2003. Integrins and EGFR coordinately regulate the pro-apoptotic protein Bim to prevent anoikis. Nature Cell Biology. 5, 733–740. [DOI] [PubMed] [Google Scholar]
- Roskelley, C.D. , Desprez, P.Y. , Bissell, M.J. , 1994. Extracellular matrix-dependent tissue-specific gene expression in mammary epithelial cells requires both physical and biochemical signal transduction. Proceedings of the National Academy of Sciences of the United States of America. 91, 12378–12382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roskelley, C.D. , Srebrow, A. , Bissell, M.J. , 1995. A hierarchy of ECM-mediated signalling regulates tissue-specific gene expression. Current Opinion in Cell Biology. 7, 736–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeichel, K.L. , Bissell, M.J. , 2003. Modeling tissue-specific signaling and organ function in three dimensions. Journal of Cell Science. 116, 2377–2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidhauser, C. , Casperson, G.F. , Myers, C.A. , Sanzo, K.T. , Bolten, S. , Bissell, M.J. , 1992. A novel transcriptional enhancer is involved in the prolactin- and extracellular matrix-dependent regulation of beta-casein gene expression. Molecular Biology of the Cell. 3, 699–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streuli, C.H. , Bailey, N. , Bissell, M.J. , 1991. Control of mammary epithelial differentiation: basement membrane induces tissue-specific gene expression in the absence of cell–cell interaction and morphological polarity. Journal of Cell Biology. 115, 1383–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streuli, C.H. , Bissell, M.J. , 1990. Expression of extracellular matrix components is regulated by substratum. Journal of Cell Biology. 110, 1405–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streuli, C.H. , Schmidhauser, C. , Bailey, N. , Yurchenco, P. , Skubitz, A.P. , Roskelley, C. , Bissell, M.J. , 1995. Laminin mediates tissue-specific gene expression in mammary epithelia. Journal of Cell Biology. 129, 591–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Wetering, M. , Barker, N. , Harkes, I.C. , van der Heyden, M. , Dijk, N.J. , Hollestelle, A. , Klijn, J.G. , Clevers, H. , Schutte, M. , 2001. Mutant E-cadherin breast cancer cells do not display constitutive Wnt signaling. Cancer Research. 61, 278–284. [PubMed] [Google Scholar]
- Wang, F. , Hansen, R.K. , Radisky, D. , Yoneda, T. , Barcellos-Hoff, M.H. , Petersen, O.W. , Turley, E.A. , Bissell, M.J. , 2002. Phenotypic reversion or death of cancer cells by altering signaling pathways in three-dimensional contexts. Journal of the National Cancer Institute. 94, 1494–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, F. , Weaver, V.M. , Petersen, O.W. , Larabell, C.A. , Dedhar, S. , Briand, P. , Lupu, R. , Bissell, M.J. , 1998. Reciprocal interactions between beta1-integrin and epidermal growth factor receptor in three-dimensional basement membrane breast cultures: a different perspective in epithelial biology. Proceedings of the National Academy of Sciences of the United States of America. 95, 14821–14826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver, V.M. , Lelievre, S. , Lakins, J.N. , Chrenek, M.A. , Jones, J.C. , Giancotti, F. , Werb, Z. , Bissell, M.J. , 2002. Beta4 integrin-dependent formation of polarized three-dimensional architecture confers resistance to apoptosis in normal and malignant mammary epithelium. Cancer Cell. 2, 205–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver, V.M. , Petersen, O.W. , Wang, F. , Larabell, C.A. , Briand, P. , Damsky, C. , Bissell, M.J. , 1997. Reversion of the malignant phenotype of human breast cells in three-dimensional culture and in vivo by integrin blocking antibodies. Journal of Cell Biology. 137, 231–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrobel, C.N. , Debnath, J. , Lin, E. , Beausoleil, S. , Roussel, M.F. , Brugge, J.S. , 2004. Autocrine CSF-1R activation promotes Src-dependent disruption of mammary epithelial architecture. Journal of Cell Biology. 165, 263–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, R. , Spencer, V.A. , Bissell, M.J. , 2007. Extracellular matrix-regulated gene expression requires cooperation of SWI/SNF and transcription factors. Journal of Biological Chemistry, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan, L. , Xiang, B. , Muthuswamy, S.K. , 2006. Controlled activation of ErbB1/ErbB2 heterodimers promote invasion of three-dimensional organized epithelia in an ErbB1-dependent manner: implications for progression of ErbB2-overexpressing tumors. Cancer Research. 66, 5201–5208. [DOI] [PubMed] [Google Scholar]


