Abstract
Background
The neotropical butterfly Heliconius heurippa has a hybrid colour pattern, which also contributes to reproductive isolation, making it a likely example of hybrid speciation. Here we used phylogenetic and coalescent-based analyses of multilocus sequence data to investigate the origin of H. heurippa.
Results
We sequenced a mitochondrial region (CoI and CoII), a sex-linked locus (Tpi) and two autosomal loci (w and sd) from H. heurippa and the putative parental species, H. cydno and H. melpomene. These were analysed in combination with data from two previously sequenced autosomal loci, Dll and Inv. H. heurippa was monophyletic at mtDNA and Tpi, but showed a shared distribution of alleles derived from both parental lineages at all four autosomal loci. Estimates of genetic differentiation showed that H. heurippa is closer to H. cydno at mtDNA and three autosomal loci, intermediate at Tpi, and closer to H. melpomene at Dll. Using coalescent simulations with the Isolation-Migration model (IM), we attempted to establish the incidence of gene flow in the origin of H. heurippa. This analysis suggested that ongoing introgression is frequent between all three species and variable in extent between loci.
Conclusion
Introgression, which is a necessary precursor of hybrid speciation, seems to have also blurred the coalescent history of these species. The origin of Heliconius heurippa may have been restricted to introgression of few colour pattern genes from H. melpomene into the H. cydno genome, with little evidence of genomic mosaicism.
Background
Homoploid hybrid speciation, in which a hybrid lineage becomes established as a novel species without a change in chromosome number, is thought to be rare and has only been well documented in a handful of plant and animals species [1-7]. Indeed, most of what is known is based on detailed studies of just one group, the Helianthus sunflowers. Notably, the species Helianthus anomalus has a clearly hybrid genome in which large blocks of the genome are derived from one or other of the parental species, Helianthus petiolaris and Helianthus annus [8]. The most convincing evidence comes from the fact that synthetic hybrids between the two species show a similar genomic composition and some ecological characteristics of the natural hybrid lineage [9]. This work has led to a view of homoploid hybrid speciation in which the derived hybrid lineage has a genome comprising similar proportions of the two parental genomes [4].
Nonetheless, this is not the only way in which hybridization might contribute to speciation. In cichlid fishes and Darwin's finches for example, it has been suggested that adaptive radiation might have been facilitated by evolutionary novelty generated through hybridization [10,11]. There is also strong evidence for gene flow at adaptive loci in the Hawaiian silverswords and in smelt fishes [12,13]. Thus, novel traits that directly affect adaptive divergence and/or reproductive isolation might become established as a result of introgression [14,15]. If the traits also have pleiotropic effects on reproductive isolation (i.e. 'magic traits' sensu Gavrilets 2004 [16]), hybridization could make a direct contribution to reproductive isolation of a novel lineage and hence, to speciation [5,17]. Under this scenario, if establishment of the hybrid trait involved many generations of backcrossing, then the genome of the novel hybrid linage could be predominantly derived from one of the parental species [17]. Regions of the genome not under selection might also be subject to gene flow through occasional ongoing hybridization between hybrid and parental species [18,19]. In Heliconius butterflies, Müllerian mimicry is common, often between related but non-sister species [20,21]. This implies repeated parallel evolution of shared colour patterns from an ancestral phenotype [20]. However, an alternative is that similar colour elements might be homologous and transferred between species through occasional hybridization and backcrossing events [20]. Natural hybrids show that 27% of species have inter-specific hybridization and that backcrosses are fairly common [20]. Thus, Heliconius pattern diversity could be facilitated by the movement of pre-established colour pattern adaptations [20,22,23].
Heliconius heurippa appears to have speciated via the hybrid origin of its novel colour pattern, which shares elements derived from both H. melpomene and H. cydno [23-26]. When H. c. cordula and H. m. melpomene, subspecies that occur near the current range of H. heurippa on the eastern slope of the Colombian Andes are crossed, a virtually identical colour pattern to that of H. heurippa can be created in the laboratory after just three generations [26]. Crosses between H. heurippa and these artificial hybrids show that the pattern breeds true implying genetic homology between the different forms, although this remains to be proven at a molecular genetic level [26]. Heliconius colour patterns are aposematic and often mimetic, such that rare colour pattern hybrids are selected against by predators [27,28]. Thus, divergent colour pattern probably contributes to post-mating isolation between the species. In addition, behavioural experiments show that the combined hybrid colour pattern of H. heurippa is critical for mate recognition [26]. Removal of either the red element derived from H. melpomene, or the yellow element derived from H. cydno, results in the pattern being less attractive to H. heurippa males. These data therefore provide strong evidence that the Heliconius heurippa colour pattern is a hybrid trait that causes reproductive isolation.
Nonetheless, it remains unclear whether H. heurippa arose via a hybrid founding of the genome, similar to Helianthus anomalus, or through introgression of a few H. melpomene colour pattern alleles into the genome of H. cydno. In between these extremes, a variety of intermediate scenarios could be envisaged with varying levels of ongoing gene flow during speciation. Ecological and genetic studies indicate that H. heurippa is most closely related to H. cydno. Ecologically, H. heurippa is a geographic replacement of H. cydno, with similar habitat and altitudinal preferences. Crosses show female hybrid sterility between H. heurippa and H. melpomene, but complete compatibility between H. heurippa and H. cydno [24]. However, microsatellite data show that H. heurippa is a distinct species and not simply a geographic race of H. cydno; H. heurippa is considerably more differentiated than any other geographic populations of H. melpomene or H. cydno sampled in Panama, Colombia and Venezuela [26]. Additionally, H. heurippa had an intriguing pattern at two nuclear loci (invected and Distal-less). In both H. heurippa shares haplotypes with H. melpomene and H. cydno [26,29]. Here we examine the incidence of gene flow in the speciation history of H. heurippa, using sequences of these genes and fragments of five additional genes.
Results
Description of gene regions
We sequenced a mitochondrial region of 1572 bp representing 799 bp of CoI, 62 bp of the tRNALEU and 711 bp of CoII from 11 individuals. Including sequences from GenBank, the alignment had 1572 nucleotide sites examined from 69 individuals, of which 155 (10%) were variable. We obtained 584 bp of the Z-linked Triose phosphate isomerase exon 3 (31 bp), intron 3 (430 bp) and exon 4 (123 bp), for 11 alleles from 11 individuals. Including sequences from GenBank the total alignment had 25 alleles from 22 individuals. Identity was confirmed by comparison with reference sequences for H. c. chioneus (AF413788) and H. m. rosina (AF413790).
The four autosomal regions were portions of nuclear loci initially chosen for their potential role in the development of butterfly wing patterns [30]. Three of these, Distal-less, invected and scalloped, are transcription factors [31-33], while the fourth, white, is member of the ommochrome biosynthesis pathway that generates the yellow, orange and red pigments in Heliconius [34]. However, linkage mapping has shown that none of these genes are linked to the switch genes that control pattern divergence in Heliconius [35,36], so there is currently no evidence that they are involved in pattern evolution. We included 558 bp of Distal-less, corresponding to exon 4/5 (175 bp), intron 5 (333 bp) and exon 6 (50 bp) of Drosophila (NM166689- intron 4 is absent in Heliconius) for 32 alleles from 20 individuals; 439 bp of invected exon 2 (67 bp), intron 2 (265 bp) and exon 3 (107 bp) for 42 alleles from 23 individuals. We obtained 499 bp of white exon 4 (49 bp), intron 4 (397 bp) and exon 5 (53 bp) for 25 alleles from 17 individuals; 483 bp of the scalloped gene from exon 7 (13 bp), intron 7 (334 bp) and exon 8 (136 bp) for 22 alleles from 12 individuals.
Species relationships and population genetic parameters
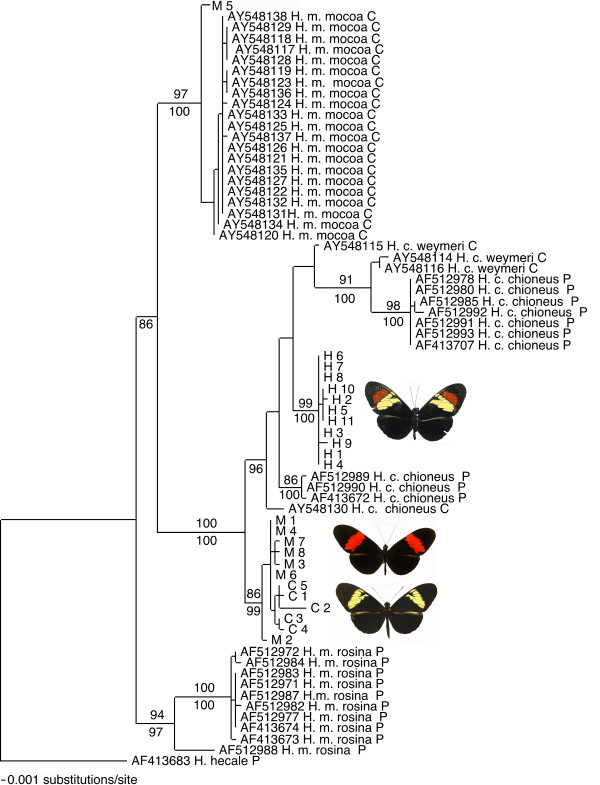
None of the three species formed a monophyletic group at all of the genes sampled. In the phylogeny derived from mitochondrial DNA, individuals of the three species fell into three well-supported monophyletic clades (Figure 1): 1) an eastern melpomene clade; 2) the cydno clade including all the H. cydno and H. heurippa sampled and seven H. m. melpomene from the eastern Andean foothills; 3) a western melpomene clade. Within the cydno clade, the H. heurippa haplotypes form a monophyletic group with five fixed differences from H. cydno (0.92% net divergence). Genetic diversity in H. heurippa was the lowest of the three species (Table 1). Tajima's D was not significantly different from zero (Table 1), suggesting that a small but constant population size rather than a recent bottleneck is more likely to explain the lack of variation in H. heurippa. Interestingly Tajima's D estimates were significantly negative for two of the three populations of H. melpomene (H. m. rosina and H. m. mocoa) included here for comparison (Table 1), possibly reflecting a recent bottleneck or mtDNA selective sweep.
Figure 1.
mtDNA Phylogenetic tree. Phylogenetic relationships of H. heurippa (H) with other populations of H. melpomene and H. cydno based on CoI and CoII sequences. C and M identify H. m. melpomene and H. c. cordula individuals of the putative parental species. Sequence ID's beginning with AF and AY indicate GenBank accession numbers. Branch lengths and probability values (under branches) were estimated using Bayesian analysis and bootstrap support (over branches) derived from a Maximum Parsimony analysis. Countries of origin are identified using the following abbreviations: P = Panama and C = Colombia; Abbreviations of species names are m. = melpomene, c. = cydno.
Table 1.
Summary of genetic polymorphism data for mtDNA sequences in each population
| Population | Na | Sb | θWc | θW 95% CId | DTe | DT |null |
|---|---|---|---|---|---|---|
| H. m. mocoa | 22 | 9 | 0.0016 | 0.0014–0.0018 | -1.8296 | < 0.05* |
| H. c. chioneus | 10 | 28 | 0.0064 | 0.0020–0.0108 | 0.3504 | >0.1 |
| H. heurippa | 11 | 4 | 0.0009 | 0.0001–0.0017 | -0.83418 | >0.1 |
| H. m. melpomene | 8 | 39 | 0.0099 | 0.0045–0.0152 | -1.5379 | >0.05 |
| H. c. cordula | 5 | 9 | 0.0028 | 0.0025–0.0031 | -0.1974 | >0.1 |
| H. m. rosina | 10 | 26 | 0.0060 | 0.0037–0.0083 | -1.8174 | < 0.05* |
H. c. weymeri and H. c. chioneus sequences from Colombia were excluded in the polymorphism analysis, because of the low number individuals available in each case (3 and 1, respectively). * Indicates statistical significance for departure of DT from neutral expectation at 95%. a Number of alleles. b Segregating sites. c Genetic diversity. d Genetic diversity 95% confidence interval. e Tajima's parameter.
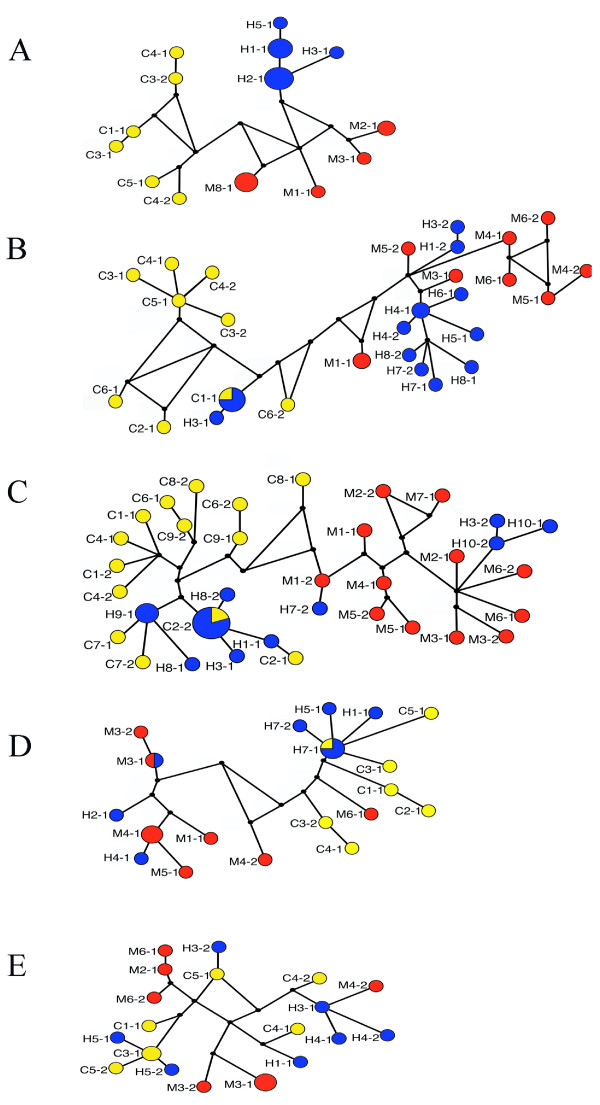
Among the five nuclear loci, the sex-linked locus Tpi was the only marker that clearly separated all three species. H. c. cordula and H. m. melopomene alleles formed distinct clades separated by five fixed differences and one shared polymorphism, with 1.3% net divergence (Figure 2A). H. heurippa alleles also formed a distinct cluster (Figure 2A) separated by five fixed differences from H. m. melpomene (1.3% net divergence) and by six from H. c. cordula (1.4% net divergence). In concordance with the network groups, FST values showed the species as three distinct populations, with H. heurippa showing greater differentiation from H. c. cordula (FST = 0.791) and H. m. melpomene (FST = 0.719) than that observed between H. c. cordula and H. m. melopomene (FST = 0.498 P < 0.05; Table 3; Figure 2A).
Figure 2.
Allele networks for nuclear genes. Yellow, Red and Blue are H. c. cordula, H. m. melpomene and H. heurippa alleles. Respective alleles are also identified with the letters C, M and H, followed by the individual number and allele number. Black dots are hypothetical ancestors. Sizes of the circles reflect allele frequencies in the population. (A) Tpi, (B) Dll, (C)inv, (D) w and (E) sd.
Table 3.
Genetic structure (FST) values for comparisons between the three populations
| mtDNA | |||
|---|---|---|---|
| Population | H. c. cordula | H. heurippa | H. m. melpomene |
| H. c. cordula (5) | - | ||
| H. heurippa (11) | 0.905 | ||
| 0.0001* | - | ||
| H. m. melpomene (8) | 0.058 | 0.739 | - |
| 0.44 | 0.0001* | ||
| Tpi | |||
| Population | H. c. cordula | H. heurippa | H. m. melpomene |
| H. c. cordula (6) | - | ||
| H. heurippa (11) | 0.791 | ||
| 0.0001* | - | ||
| H. m. melpomene (8) | 0.498 | 0.719 | - |
| 0.0001* | 0.0001* | ||
| Dll | |||
| Population | H. c. cordula | H. heurippa | H. m. melpomene |
| H. c. cordula (9) | - | ||
| H. heurippa (14) | 0.479 | ||
| 0.0001* | - | ||
| H. m. melpomene (9) | 0.621 | 0.085 | - |
| 0.0001* | 0.066 | ||
| Inv | |||
| Population | H. c. cordula | H. heurippa | H. m. melpomene |
| H. c. cordula (14) | - | ||
| H. heurippa (16) | 0.107 | ||
| 0.022* | - | ||
| H. m. melpomene (12) | 0.593 | 0.419 | - |
| 0.0001* | 0.0001* | ||
| W | |||
| Population | H. c. cordula | H. heurippa | H. m. melpomene |
| H. c. cordula (7) | - | ||
| H. heurippa (9) | 0.004 | ||
| 0.383 | - | ||
| H. m. melpomene (9) | 0.327 | 0.176 | - |
| 0.003* | 0.019* | ||
| Sd | |||
| Population | H. c. cordula | H. heurippa | H. m. melpomene |
| H. c. cordula (7) | - | ||
| H. heurippa (7) | 0.037 | - | |
| 0.196 | |||
| H. m. melpomene (8) | 0.148 | 0.119 | - |
| 0.006* | 0.020* | ||
* Significance at 95% was obtained by bootstrapping (10000 subsamples).
Three of the autosomal loci, Dll, inv and w, show a striking pattern in which H. c. cordula and H. m. melpomene are clearly differentiated, but H. heurippa shares variation with both species. At Dll, H. c. cordula and H. m. melpomene (Figure 2B) are separated by 10 fixed differences (3% net divergence) and share three polymorphisms. Similarly at inv, H. c. cordula and H. m. melpomene have ten fixed differences (4.6% net divergence) and two shared polymorphisms (Figure 2C). At w, one allele of H. m. melpomene was shared with H. c. cordula (M6-1; Figure 2D), such that there was only one fixed difference (2% net divergence) and eight shared polymorphisms. Nonetheless, estimates of FST between H. c. cordula and H. m. melpomene were high for all three loci (Dll, 0.621, inv, 0.593 and w, 0.327). In contrast, H. heurippa did not have any fixed differences with either H. c. cordula or H. m. melpomene at any of these three loci. At Dll, H. heurippa was more similar to H. m. melpomene (net divergence 0.41%) than H. c. cordula (net divergence 2.28%). At the other two loci, H. heurippa was more similar to H. cydno, with inv showing 0.56% and 2.76% divergence with H. c. cordula and H. m. melpomene respectively, while w showed 0.024% and 1% for the same two comparisons. Estimates of FST supported these observations, with Dll showing H. heurippa not significantly differentiated from H. m. melpomene but strongly differentiated from H. c. cordula (Table 3), while inv and w showed the opposite pattern with H. heurippa more similar to H. c. cordula (Table 3).
In contrast with the patterns observed in the other five loci, the sd locus showed no fixed differences among any of the three species (Figure 2E). H. c. cordula and H. m. melpomene had seven shared polymorphisms and net divergence of 0.62%. H. heurippa shared 14 and ten polymorphisms with H. c. cordula and H. m. melpomene respectively, representing net divergence of 0.10% and 0.5%. Genetic diversity was generally high in all three species (Table 2). Tajima's D was significantly negative in H. c. cordula (Table 2). FST values showed that H. heurippa was more similar to H. c. cordula than to H. m. melpomene (Table 3).
Table 2.
Summary of genetic polymorphism data for nuclear sequences, in H. c. cordula, H. heurippa and H. m. melpomene populations.
| H. c. cordula | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| locus | na | Sb | θWc | θW 95% CId | DTe | DT 95% CIf | P | DT rec 95% CIg | P |
| Tpi | 6 | 9 | 0.0083 | 0.0037–0.0129 | -0.1548 | (-1.4347 to1.5822) | 0.4850 | (-0.8862 to 0.9428) | 0.4150 |
| Dll | 9 | 23 | 0.0193 | 0.0101–0.0285 | -0.5311 | (-1.6696 to1.6159) | 0.3300 | (-0.7913 to 0.7376) | 0.0760 |
| inv | 14 | 51 | 0.0394 | 0.0177–0.0571 | -1.1859 | (-1.6520 to1.6498) | 0.1000 | (-0.6055 to 0.5822) | 0.000001* |
| W | 7 | 26 | 0.0281 | 0.0064–0.0498 | -0.3275 | (-1.5502 to1.5575) | 0.4070 | (-0.6332 to 0.6404) | 0.1710 |
| Sd | 7 | 20 | 0.0171 | 0.0128–0.0213 | -0.8328 | (-1.5533 to1.7872) | 0.2440 | (-0.7018 to 0.8047) | 0.0090* |
| H. heurippa | |||||||||
| locus | na | Sb | θWc | θW 95% CId | DTe | DT 95% CIf | P | DT rec 95% CIg | P |
| Tpi | 11 | 2 | 0.0014 | 0.0008–0.0036 | 0.199 | (-1.4296 to1.8276) | 0.5920 | (-1.4296 to 1.6648) | 0.5320 |
| Dll | 14 | 29 | 0.0229 | 0.0126–0.0331 | -0.642 | (-1.7616 to1.6671) | 0.2950 | (-0.7520 to 0.6874) | 0.0420 |
| inv | 16 | 42 | 0.0307 | 0.0198–0.0416 | 0.0026 | (-1.8007 to1.7247) | 0.5590 | (-0.6635 to 0.6064) | 0.5060 |
| W | 9 | 29 | 0.0340 | 0.0246–0.0434 | 0.1089 | (-1.6834 to1.5982) | 0.5850 | (-0.6231 to 0.6390) | 0.6450 |
| Sd | 7 | 25 | 0.0215 | 0.0127–0.0302 | -0.0604 | (-1.5947 to1.6325) | 0.5260 | (-0.6953 to 0.62731) | 0.4530 |
| H. m. melpomene | |||||||||
| locus | na | Sb | θWc | θW 95% CId | DTe | DT 95% CIf | P | DT rec 95% CIg | P |
| Tpi | 8 | 9 | 0.0078 | 0.0043–0.0113 | 1.2504 | (-1.6740 to1.7414) | 0.9120 | (-1.0680 to 0.9984) | 0.9890 |
| Dll | 9 | 21 | 0.0169 | 0.0048–0.0217 | -0.6685 | (-1.6864 to1.5708) | 0.3010 | (-0.7703 to 0.7226) | 0.0510 |
| inv | 12 | 29 | 0.0235 | 0.0170–0.0300 | -1.7198 | (-1.7129 to1.6628) | 0.024* | (-0.6769 to 0.6816) | 0.000001* |
| W | 9 | 31 | 0.0377 | 0.0341–0.0413 | -1.1847 | (-1.6591 to1.6624) | 0.1130 | (-0.6389 to 0.6184) | 0.000001* |
| Sd | 8 | 33 | 0.0266 | 0.0161–0.0371 | -0.3359 | (-1.5973 to1.6506) | 0.3790 | (-0.6258 to 0.5921) | 0.1320 |
* Indicates a P=Pr(D≤ observed value) statistically significant deviation of Tajima's D from Hudson 1990 panmixia model [72]. 95% CIs under the model were calculated by coalescent simulations (with fixed S). The two-tailed probability test of significance was derived from simulations (P = 0.025). a Number of alleles. b Segregating sites. c Genetic diversity. d Genetic diversity 95% confidence interval. e Tajima's D. f Tajima's D, 95% confidence interval calculated without recombination. g Tajima's D, 95% confidence interval calculated with recombination.
Recombination analysis and IM input files
In order to select the 'basic' dataset required by the IM software, indel-free alignments were investigated to search for evidence of recombination for each species pair comparison [37]. In the H. cydno – H. melpomene comparison, recombination was only detected at Dll with its maximum significant breakpoint found between sequences C6-2 and M5-2 (p = 0.007) between 16th and the 17th sites, as was indicated using the maximum chi-square method. A recombination free block of 206 bp was selected in the 5' to 3' direction from the site of probable recombination (Table 4).
Table 4.
IM blocks and mutation rate estimates for each species comparison.
| 95% credibility interval | |||||
|---|---|---|---|---|---|
| Comparison | L (pb) | Estimate | Lower Limit | Upper Limit | |
| H. cydno-H. melpomene | μCo | 1517 | 2.88 × 10-9 | 1.7 × 10-9 | 6.6 × 10-9 |
| μTpi | 416 | 0.642 × 10-9 | 0.377 × 10-9 | 1.3 × 10-9 | |
| μDll | 206 | 0.491 × 10-9 | 0.282 × 10-9 | 0.868 × 10-9 | |
| μinv | 391 | 1.6 × 10-9 | 0.792 × 10-9 | 2.3 × 10-9 | |
| μsd | 473 | 1.1 × 10-9 | 0.739 × 10-9 | 1.9 × 10-9 | |
| μW | 296 | 0.793 × 10-9 | 0.489 × 10-9 | 1.4 × 10-9 | |
| H. cydno-H. heurippa | μCo | 1517 | 2.88 × 10-9 | 1.7 × 10-9 | 7.4 × 10-9 |
| μTpi | 416 | 0.683 × 10-9 | 0.372 × 10-9 | 1.6 × 10-9 | |
| μDll | 412 | 1.5 × 10-9 | 0.712 × 10-9 | 4.1 × 10-9 | |
| μinv | 250 | 1.8 × 10-9 | 0.865 × 10-9 | 2.8 × 10-9 | |
| μsd | 308 | 0.61 × 10-9 | 0.29 × 10-9 | 1.7 × 10-9 | |
| μW | 296 | 1.3 × 10-9 | 0.702 × 10-9 | 4 × 10-9 | |
| H. melpomene-H. heurippa | μCo | 1517 | 2.88 × 10-9 | 1.7 × 10-9 | 8.1 × 10-9 |
| μTpi | 416 | 0.184 × 10-9 | 0.122 × 10-9 | 0.829 × 10-9 | |
| μDll | 412 | 5.4 × 10-9 | 1.5 × 10-9 | 7.1 × 10-9 | |
| μinv | 125 | 0.583 × 10-9 | 0.215 × 10-9 | 1.2 × 10-9 | |
| μsd | 308 | 5.5 × 10-9 | 2.7 × 10-9 | 1.09 × 10-8 | |
| μW | 296 | 1.1 × 10-9 | 0.399 × 10-9 | 3.5 × 10-9 | |
μx: mutation rate per base pair per generation for each locus.
In the H. cydno – H. heurippa comparison, inv had recombination (p = 0.015) with the most significant breakpoint found between sequences H3-1 and C8-2 between 52nd and 53rd sites. A non recombining block of 250 bp preceding the sites involved in the recombination was selected, in the 5' to 3' direction (Table 4). In this comparison, sd also showed significant recombination between sequences H1-1 and H4-1 with the most significant breakpoint at the 16th and 17th sites (p = 0.0001). For this locus, a 308 bp region (5' to 3') lacking recombination was included in further analysis (Table 4). In the H. melpomene – H. heurippa comparison, the only locus with recombination was inv in which, the exchange was between the sequences H7-2 and M7-1, with the most significant breakpoint between 12th and 13th sites (p = 0.005). A 125 bp 5' to 3' region after the sites involved in recombination was selected for the IM analysis (Table 4).
History of divergence between H. melpomene and H. cydno
Isolation-Migration model [38] was used to infer the population history of H. melpomene melpomene and H. cydno cordula. The IM program uses a coalescent model to estimate effective population size, time since divergence, and ongoing migration parameters from multilocus sequence data sampled from two sister species. Here, H. cydno cordula was estimated to have a two-fold greater population size compared to H. melpomene melpomene (Table 5). This result is consistent with other studies involving different H. cydno and H. melpomene geographical races [37,39]. The ancestral population size was 1.23 × 106 (0.76 × 106-1.98 × 106) individuals and the speciation event probably took place around 2 million years ago, similar to a previous estimate using a different dataset [37]. However, because of the weak scalar estimation for this parameter, this estimated time is a very approximate value (Table 5).
Table 5.
Genealogical parameters estimated under the IM model
| Species comparison | θ1 | θ2 | t | mtDNA | Tpi | inv | Dll | w | sd | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| a | b | a | b | a | B | a | b | a | b | a | b | ||||
| H. melpomene | 0.017 | 0.041 | 0.01 | 1.32 × 10-6 | ~0 | ~0 | ~0 | ~0 | ~0 | ~0 | ~0 | 0.61 × 10-6 | ~0 | ~0 | ~0 |
| 0.022 | 0.028 | 0.005 | 0.17 × 10-6 | ~0 | ~0 | ~0 | ~0 | ~0 | ~0 | ~0 | ~0 | ~0 | ~0 | ~0 | |
| H. cydno | 0.041 | 0.12 | 0.04 | 1.68 × 10-5 | 8.68 × 10-5 | 5.74 × 10-5 | 2.24 × 10-4‡ | 7.99 × 10-5 | 2.31 × 10-5 | 1.32 × 10-4‡ | 3.45 × 10-4‡ | 8.7 × 10-5 | 2.7 × 10-4‡ | 5.44 × 10-5 | 1.68 × 10-4‡ |
| 1,121,086 | 2,042,530 | 1,971,860‡ | |||||||||||||
| 0.041 | 0.0059 | ~0 | ~0 | ~0 | ~0 | 2.64 × 10-5 | 2.96 × 10-6 | 4.12 × 10-6 | ~0 | 1.05 × 10-5 | 0.016 × 10-6 | 2.25 × 10-6 | 2.21 × 10-5 | ||
| H. cydno | 0.028 | 0.0053 | ~0 | ~0 | ~0 | ~0 | 5.88 × 10-6 | 0.01 × 10-6 | 1.70 × 10-6 | ~0 | 1.89 × 10-6 | ~0 | 0.90 × 10-6 | 4.37 × 10-6 | |
| H. heurippa | 0.12 | 0.011 | ---- | 7.17 × 10-5 | 3.59 × 10-6 | 1.29 × 10-4‡ | 9.24 × 10-6 | 3.47 × 10-4‡ | 2.24 × 10-5 | 3.22 × 10-4‡ | 2.12 × 10-5 | 3.94 × 10-4‡ | 4.12 × 10-5 | 2.38 × 10-4‡ | 4.32 × 10-5 |
| 1,903,673 | 274,516 | ||||||||||||||
| 0.0060 | 0.014 | ~0 | ~0 | ~0 | ~0 | 4.48 × 10-6 | 0.07 × 10-6 | 0.027 × 10-6 | 2.48 × 10-5 | 1.01 × 10-5 | 0.029 × 10-6 | 1.36 × 10-6 | 5.67 × 10-5 | ||
| H. heurippa | 0.0056 | 0.007 | ---- | ~0 | ~0 | ~0 | ~0 | 0.03 × 10-6 | ~0 | ~0 | 5.45 × 10-6 | 0.018 × 10-6 | 0.015 × 10-6 | 0.011 × 10-6 | 4.7 × 10-6 |
| H. melpomene | 0.010 | 0.033 | 3.15 × 10-6 | 4.71 × 10-6 | 7.64 × 10-6 | 1.09 × 10-5 | 4.42 × 10-5 | 8.71 × 10-5 | 7.15 × 10-6 | 7.97 × 10-5 | 3.22 × 10-5 | 8.81 × 10-5 | 1.70 × 10-5 | 1.34 × 10-4‡ | |
| 295,349 | 701,033 | ||||||||||||||
For each cell the first value is the parameter estimate, second is the lower limit and third is upper limit at 95% level. ---- indicates that no reliable ML estimate was obtained for a parameter, ‡ unreliable estimate or limit due to flat or incomplete posterior probability distribution sampled and ~0 effectively zero, although the lowest 'bin' does not actually include zero (i.e. low gene flow is probable). a, refers to the forward migration rate (per generation per locus) of haplotypes from population 2 into population 1. b, refers to the forward migration rate (per generation per locus) of haplotypes from population 1 into population 2.
Two loci (mtDNA and w) showed evidence of gene flow. In both cases it was asymmetric (Table 5). All remaining genes had estimates asymptotically near to zero in both directions (Table 5). The estimated migration rate from H. c. cordula to H. m. melpomene (m1 = 1.32 × 10-6 per generation per locus) was high at mtDNA, as was expected from the pattern of shared variation seen in the mtDNA tree. Gene flow estimated in the other direction was nearly zero (Table 5). The w locus had a 0.61 × 10-6 per generation per locus migration rate in the same direction, a result that is consistent with the presence of the M6-1 H. melpomene haplotype in the H. cydno allele cluster.
Isolation-Migration model including H. heurippa
The IM model was then used to compare H. heurippa with each of the parental taxa. The estimated H. heurippa population size was similar (0.27 – 0.30 × 106) in each comparison and smaller than those estimated for the other species (Table 5). Unlike the comparison between H. m. melpomene and H. c. cordula, both comparisons involving H. heurippa failed to yield precise estimates of either the ancestral population size or the divergence time. The likelihood surfaces obtained for these parameters were either flat or rising over the parameter range investigated.
More importantly here, was the incidence of gene flow observed at several loci between H. heurippa and the other species, the only exceptions being mtDNA and Tpi, where the migration rate was not significantly different to zero in both directions (Table 5). At all other loci migration parameters were significantly greater than zero in at least one direction, with evidence for gene flow both from parental species into H. heurippa and vice-versa (Table 5).
Discussion
Genealogical pattern and introgression
Heliconius heurippa was initially identified as a putative hybrid species based on its intermediate colour pattern, which shows a striking similarity to phenotypes produced after just a few generations of hybridization between H. c. cordula and H. m. melpomene [26]. The sequence data presented here provides independent evidence that hybridization has played an important, and ongoing, role in the evolution of the H. heurippa genome. All four autosomal loci showed a pattern in which H. heurippa shares similar alleles with both H. m. melpomene and H. c. cordula. At three of these loci (inv, w and sd), H. heurippa was most closely related to H. c. cordula, while at the fourth (Dll) it was closer to H. m. melpomene. Nonetheless, our analysis suggests that the pattern of shared alleles between H. heurippa and its relatives is mainly due to introgression in the fairly recent past. The results are consistent with historical gene exchange playing an important role in the origin of H. heurippa, but do not provide evidence for a 'mosaic' hybrid genome as has been demonstrated in other examples of hybrid speciation.
Gene flow between H. c. cordula and H. m. melpomene
The comparison of H. c. cordula and H. m. melpomene under the IM model indicates ongoing introgressive hybridization at two loci (Table 5). In particular, there are very closely related mtDNA haplotypes shared between the species (gene flow from H. c. cordula to H. m. melpomene, m1 = 1.32 × 10-6 per generation per locus; Figure 1) and asymmetric moderate gene flow in w (m1 = 0.61 × 10-6 per generation per locus, gene flow from H. c. cordula to H. m. melpomene and 0 in the other direction). These species are known to hybridize in the wild and previously, shared alleles have been observed at another autosomal locus, Mpi for which a symmetrical migration rate of 1.54 ×10-6 per generation per locus was estimated in Panamanian populations [37,40]. Furthermore, substantial shared DNA sequence variation at 16 loci was observed in H. cydno and H. melpomene from Costa Rica [39].
Notably, shared mtDNA variation between H. cydno and H. melpomene in eastern Colombia (Figure 1) suggests recent introgression. The shared mtDNA haplotypes may have been retained as an ancient polymorphism since the speciation of H. melpomene and H. cydno, but this seems unlikely given the evolutionary distance between the species (1.5–2% divergence between the two mtDNA clades). Female hybrid sterility following Haldane's rule [41] would be expected to prevent introgression of mtDNA, although limited fertility of female hybrids has been documented and would provide a route for infrequent introgression [41].
The history of H. heurippa
Pairwise IM analyses of H. heurippa and either one of the other taxa, suggest a small species population size (Table 5). The absence of consistent negative Tajima's D across loci (Table 2) indicates a historically small constant size instead of a short bottleneck. No reliable estimates for divergence time and ancestral population size were obtained in either comparison. Two loci mtDNA and Tpi did not show gene flow for any pair evaluated that involves H. heurippa. The remaining loci showed recent gene interchange among H. heurippa and the two other species. There is no hybrid sterility between H. cydno and H. heurippa, and insectary mate choice experiments produce frequent matings in one direction (between H. heurippa males and H. cydno females). It seems likely that there is a hybrid zone between these species somewhere in the Andes between Villavicencio (Colombia) and San Cristobal (Venezuela; Linares Pers. Obs.), which would explain the observed levels of gene flow.
In the H. melpomene – H. heurippa species pair, inv (64) and w (348.2) in particular showed high rates of gene flow from H. melpomene to H. heurippa. These two species are broadly sympatric, which might facilitate hybridization. Nonetheless, although hybrid females are sterile in one direction of cross, female offspring of backcrossed males segregate for sterility which should allow some gene flow [24].
In a broad sense, our Isolation-Migration analysis suggests that different parts of the H. heurippa genome are subject to very different levels of gene flow with the two sister taxa. We also carried out linkage disequilibrium tests for introgression [42], which should be sensitive to very recent introgression events, but these were not significant for any of the loci, suggesting that gene flow is sufficiently ancient for linkage disequilibrium to have been lost. There are a number of possible explanations for such interlocus variation. One is that some of the loci sequenced for this study are either themselves subject to natural selection or else are linked to such regions. Selection does seem plausible as we know that there is a strong correlation between the Tpi locus and hybrid sterility in interracial H. melpomene, H. cydno x H. melpomene and H. melpomene x H. heurippa crosses [24,41,43]. If Tpi were associated with genes causing hybrid sterility, this might explain the clear lack of gene flow among the three species at this locus. This is also consistent with the fact that a disproportionate number of species differences, including morphology, physiology, oviposition preference, mate selection and pheromones, map to the Z chromosome in other Lepidoptera species [44].
At the other extreme, sd shows far more allelic mixing between species than the other loci studied. A similar pattern in the cydno-melpomene group races from Panama and Costa Rica has been observed at other loci [37,39]. This has led to suggestion that balancing selection could be maintaining diversity and perhaps promoting introgressive hybridization [37,39,40]. It is possible that this process is occurring at the sd haplotypes obtained here. There is currently no evidence for the role of sd, or any of the other loci studied here playing a role in the evolution of colour pattern, so it is unclear what the selection pressures on this locus might be.
In addition, analysis of protein coding sequence did not provide any direct evidence for positive or balancing selection on any of the loci, although the power of this analysis was limited by the fact that most of our sequence data is for non-coding regions (analysis using CODEML, data not shown, [45]). The genealogical pattern and IM analysis do suggest that there is both ancestral variation and recent gene flow at the sd locus (Table 5).
Overall, the species relationships are consistent with the H. heurippa genome containing a greater contribution from H. cydno than H. melpomene. The H. heurippa mtDNA haplotypes fall within an H. cydno clade, and at three of the five nuclear loci FST values show H. heurippa closer to H. cydno (Table 3). This is consistent with what is known about the three species. H. heurippa is a geographic replacement of H. cydno that flies in similar habitats. Where H. cydno flies sympatrically with H. melpomene, the former is associated more with closed canopy forest and tends to be found at higher altitudes [46], both characteristics also observed in H. heurippa populations in eastern Colombia (Linares and Jiggins Pers. Obs.). Furthermore, laboratory reconstruction of the H. heurippa colour pattern from H. c. cordula and H. m. melpomene involves backcrossing F1 male hybrids to H. cydno, with a correspondingly greater contribution from the H. cydno genome. Finally, patterns of hybrid sterility show that H. heurippa is more compatible with H. cydno [24,26]. Nonetheless, the Tpi genealogy contrasts with this general pattern, in that H. heurippa is similarly differentiated from both of the other species, suggesting a more contemporaneous divergence of all three species. This contrasts with the other genetic and ecological evidence placing H. heurippa as a closer relative of H. cydno, and might indicate an even more complex evolutionary history.
In summary, the data clearly imply that introgressive hybridization has occurred between the three species, but this does not distinguish between two alternative scenarios for the origin of H. heurippa. First, a branching speciation scenario with ongoing gene flow, and second a hybrid founding speciation scenario. Thus, these data alone cannot be taken as providing strong evidence in support of hybrid speciation. Indeed, the observed pattern of allelic variation contrasts markedly with the pattern seen in hybrid Helianthus sunflowers [7,47]. The latter species show genomes consisting of genetic "blocks" derived from one or the other of the parental species. These would be seen in our data as a clustering of the hybrid species entirely within one or the other parental species for each locus. However, this is not the general pattern seen in H. heurippa, where we observe allelic variation shared with both parental species at several loci. Intuitively, the latter pattern seems more consistent with high ongoing rates of introgression at many loci, and species differences maintained by selection at the remaining loci. This contrast is perhaps unsurprising given that the hybrid sunflowers are largely allopatric to their progenitors, while H. heurippa is still able to exchange genes with both of its parental species.
Superficially, the data seems to support the hybrid speciation hypothesis, with FST analyses showing H. heurippa more related to H. cydno at some loci and more related to H. melpomene at others. This is the kind of pattern that has been used previously to argue the case for hybrid speciation in other groups [2,48,49]. Furthermore, there are clearly alleles in H. heurippa that could be considered diagnostic for one or other of the parental species at different loci, similar to the 'private alleles' found in the 'Lonicera fly' and 'alpine Lycaeides' that were apparently derived from one or other putative parent [50,51]. Thus, our data are similar in several aspects to previous examples that have been proposed as good cases for hybrid speciation. Nevertheless, we would argue that such evidence needs to be combined with analysis of traits involved in causing reproductive isolation in order to argue convincingly that hybridization played a causative role in speciation.
Conclusion
Our results highlight the difficulty of clearly proving the case for hybrid speciation. If hybrid speciation is important, it must often occur in taxa with significant rates of introgressive hybridization, such that where shared variation is observed the alternative hypotheses of hybrid founding versus introgressive hybridization need to be rigorously tested. The evidence for the role of hybridization in the speciation of H. heurippa comes primarily from crossing and mate choice experiments that have addressed the origin of its colour pattern, and the role of that pattern in reproductive isolation [26]. That colour pattern is controlled by a small number of genes of major effect, so the hybrid speciation hypothesis does not make specific predictions regarding the rest of the genome. Here we have clearly shown that H. heurippa does not have a hybrid genome that is comparable to the sunflower Helianthus anomalus, with blocks of genes derived from one or other parent. Instead, the genome resembles that of other groups of closely related taxa in which hybridization is frequent, such as Anopheles gambie [52,53] and the Drosophila pseudoobscura group [38,42,54].
Discordant patterns are found at different markers such that particular alleles cannot be considered 'diagnostic' of a particular species. Thus, in this case hybrid speciation has resulted from the origin of a novel trait with a key role in speciation. Thus, in the case of H. heurippa, sequence analysis of the genes controlling the different pattern elements will be needed to uncover the genealogical history of the original speciation event.
Methods
Specimen collection, PCR and sequencing
Butterflies were collected in Colombia and Venezuela (see additional file 1: Specimen collection list), wings removed and stored in glassine envelopes. The bodies were preserved with 100% EtOH in the Universidad de Los Andes (M code) and in DMSO in the Jiggins collection (stri-b code) for H. heurippa (n = 11), H. m. melpomene (n = 8) and H. c. cordula (n = 9) from the eastern Colombian and Venezuelan Andes (see additional file 1: Specimen collection list). Cytochrome oxidase subunit 1 (CoI)/tRNALEU/cytochrome oxidase subunit 2 (CoII) region and the Z-linked locus, Triose-phosphate isomerase (Tpi), were amplified and sequenced from individual genomic DNA using PCR primers and conditions described previously [40]. All nuclear gene products were cloned before sequencing using pGEM-T Easy Vector System (Promega). For each individual 3–5 clones were sequenced to identify distinct alleles. Previously Distal-less (Dll) and invected (inv) sequences obtained by CS [26,29] and the newest white (w) and scalloped (sd) sequences were amplified from individual genomic DNA using PCR primers and conditions already described [30]. Sequences included in the analysis were generally represented by at least two identical clones. All loci sequences were deposited in GenBank under accessions [DQ674383-DQ674451, DQ445385-DQ445415 and DQ445416-DQ445457].
Phylogenetic analysis
Bayesian and parsimony phylogenetic trees were reconstructed as described previously for mtDNA data [40]. Recombination violates the assumption of a bifurcating genealogy [54], so for nuclear loci we constructed haplotype networks that take into account the presence of persistent ancestral nodes, multifurcations and reticulation. The presence of loops in these networks might reflect recombination events [55]. Networks for nuclear loci were constructed with statistical parsimony in TCS v 1.21 [56], considering gaps as missing data and adjusting the parsimony limit to the respective data set.
Population genetic analysis
For each species, the per site population mutation rate ΘW[57] with 95% confidence intervals was estimated with DnaSP v4.10.3 [58]. Deviation from a neutral model, and hence the effectiveness of ΘW in reflecting the effective population size (Ne) was tested by estimating Tajima's D [59]. Significant departure of Tajima's D from zero was evaluated both assuming recombination and without recombination. Both tests were carried out because presence of recombinant sites in the nuclear genes leads to a loss of power to reject neutrality [60]. For between species comparisons, the program SITES [61] was used to estimate net divergence between species [62] and the number of shared polymorphisms and fixed differences. Genetic differentiation between pairs of populations was measured using Wright's FST [63] adapted for DNA sequence data [64,65] and estimated using DnaSP v4.10.3 [58]. Statistical significance was obtained by bootstrapping i.e. randomly sampling with replacement the values of within population diversity, πS, and the values of the between population divergence, πB, 10000 times and recalculating FST for each replicate using SEQUENCER v6.1.0 [66].
In order to estimate the role of gene flow in shaping the pattern of shared alleles between species, the Isolation-Migration (IM) bayesian model was used [38]. This model considers a scenario where a population gives rise to two populations, after which there may be gene exchange between the two new populations [38]. The program relies on a metropolis-coupled Monte Carlo Markov chain (MCMC) genealogical sampling for Bayesian estimation of six major demographical parameters: recent population sizes, ancestral population size, time at which the ancestral population bifurcated and two or more migration rates according to whether gene flow is evaluated at the population or gene level [38]. The genealogical coalescence parameter estimation follows the classical Kingman theory in which recent alleles of two daughter populations will be traced to an ancestral pool in a Wright-Fisher fashion [38]. In this context, the genetic processes of mutation and drift occurs on a time scale of generations. Because of this, the parameters must be scaled based on a known mutation or drift rate.
We carried out analyses on three modified couplet datasets for each species pair: (i) H. melpomene-H. cydno (ii) H. melpomene-H. heurippa and (iii) H. cydno-H. heurippa. For the former comparison we expected that IM model fits well to the data. However, in the couplet data sets involving H. heurippa, IM could overestimate the divergence time and underestimate the migration rates between this species and either parent species. This is a consequence of the presence within H. heurippa of divergent alleles that relate it with the species not included in the IM comparison. Nonetheless, the coalescence process in the IM simulations is able to recover information about the ancestry of shared and divergent alleles between H. heurippa and any of the other two species and provided an approximation of the magnitude of gene flow.
Additionally, in concordance with the typical assumptions and limitations of the IM algorithm (i.e. no gap polymorphism, no recombination within loci), we looked for regions in our sequences that met those conditions. For each alignment, we obtained a dataset that was as complete as possible after deleting highly polymorphic indel regions. Over these data sets, evidence for recombination was evaluated by the Hudson four gamete test implemented in SITES [67]. However, this test assumes an infinite sites mutation model that is not realistic for our data due to observation of repeated changes at the same site under the HKY model [68]. Hence, recombination was also tested using a model-neutral test based on a bootstrapped correlation of linkage disequilibrium (R2) with physical distance and with a maximum chi-square test [69,70]. Both analyses were used to subsample the indel-free files in order to remove clearly recombinant regions by searching from the 5' region of each locus until a probable recombinant pattern emerge [37]. The same procedure was made from the 3' region. From 5' and 3' analysis, we selected a maximum block without recombination [37]. Effective population size scalars were assigned to each locus relative to autosomal inheritance. Specifically, the values were set at 0.25 for CO, 0.75 for Tpi and 1 for the other loci. Individual species population sizes and ancestral population size (θ = 4Nμ), divergence time, relative mutation rate μ and per locus directional migration rates (m) were estimated with all loci. Demographic values were obtained through a molecular clock scalar calibration to obtain parameters per base pair and per generation, taking as reference Brower's 2.3% divergence in mtDNA per million years estimated for insects [71] and with an assumption of four generations per year (Table 4). Migration rates (m1 y m2) were allowed to vary between loci and between species (i.e. asymmetrical gene flow, option -j6). After search for parameter range using preliminary runs, each pairwise comparison were run for at least 30 million steps from a 200,000 burn-in period under the HKY model [68] with 5 chains per set, with linear heating increment h of 0.033.
Authors' contributions
CS carried out laboratory work and genetic analyses. CDJ helped in data analysis and, with CS, drafted the manuscript. JET helped with statistical analysis. MRK carried out nuclear primer designing and provided reference sequences. ML participated in specimen collection and in the design of the study. All authors participated in the conception of the study and approval of the final manuscript.
Supplementary Material
Specimen collection list. Populations sampled with information about the genes sequenced for each individual.
Contributor Information
Camilo Salazar, Email: salazar.camilo@gmail.com.
Chris D Jiggins, Email: c.jiggins@zoo.cam.ac.uk.
Jesse E Taylor, Email: jtaylor@stats.ox.ac.uk.
Marcus R Kronforst, Email: mkronforst@cgr.harvard.edu.
Mauricio Linares, Email: mlinares@uniandes.edu.co.
Acknowledgements
The authors thank Instituto Colombiano para el Desarrollo de la Ciencia y la Tecnología, Francisco José de Caldas grant 1204-05-11414 and grant 7155-CO, the Smithsonian Tropical Research Institute, where part of the laboratory work was carried out, the Biotechnology and Biological Sciences Research Council and the Royal Society (fellowship to C. Jiggins) for financial support. C. Arias and M. Melo are gratefully acknowledged for their excellent collaboration in the laboratory.
References
- Arnold ML, Bennett BD. In: Hybrid Zones and the Evolutionary Process. Harrison RG, editor. New York, Oxford University Press ; 1993. Natural hybridization in Louisiana Irises: genetic variation and ecological determinants; pp. 115–139. [Google Scholar]
- Howarth DG, Baum DA. Genealogical evidence of homoploid hybrid speciation in an adaptive radiation of Scaevola (Goodeniaceae) in the Hawaiian islands. Evolution. 2005;59:948–961. [PubMed] [Google Scholar]
- Abbott R. Sex, sunflowers, and speciation. Science. 2003;301:1189–1190. doi: 10.1126/science.1089292. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH. Hybrid origins of plant species. Ann Rev Ecol Syst. 1997;28:359–389. doi: 10.1146/annurev.ecolsys.28.1.359. [DOI] [Google Scholar]
- Mallet J. Hybrid speciation. Nature. 2007;446:279–283. doi: 10.1038/nature05706. [DOI] [PubMed] [Google Scholar]
- Coyne JA, Orr HA. Speciation. Sunderland, Mass. , Sinauer Associates; 2004. [Google Scholar]
- Rieseberg LH, Raymond O, Rosenthal DM, Lai Z, Living-stone K, Nakazato T, Durphy JL, Schwatzbach AE, Donovan LA, Lexer C. Major ecological transitions in wild sunflowers facilitated by hybridization. Science. 2003;301:1211–1216. doi: 10.1126/science.1086949. [DOI] [PubMed] [Google Scholar]
- Ungerer MC, Baird SJE, Pan J, Rieseberg LH. Rapid hybrid speciation in wild sunflowers. Proc Natl Acad Sci USA. 1998;95:11757–11762. doi: 10.1073/pnas.95.20.11757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal DM, Rieseberg LH, Donovan LA. Re-creating ancient hybrid species complex phenotypes from early-generation synthetic hybrids: three examples using wild sunflowers. Am Nat. 2005;166:26–41. doi: 10.1086/430527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzburger W, Baric S, Sturmbauer C. Speciation via introgressive hybridization in East African cichlids? Molec Ecol. 2002;11:619–625. doi: 10.1046/j.0962-1083.2001.01438.x. [DOI] [PubMed] [Google Scholar]
- Grant PR, Grant BR. Hybridization of bird species. Science. 1992;256:193–197. doi: 10.1126/science.256.5054.193. [DOI] [PubMed] [Google Scholar]
- Lawton-Rauh A, Robichaux RH, Purugganan MD. Diversity and divergence patterns in regulatory genes suggest differential gene flow in recent-derived species of the Hawaiian silversword alliance adaptive radiation (Asteraceae) Molec Ecol. 2007;16:3995–4013. doi: 10.1111/j.1365-294X.2007.03445.x. [DOI] [PubMed] [Google Scholar]
- Saint-Laurent R; Legault M; Bernatchez L. Divergent selection maintains adaptive differentiation despite high gene flow between sympatric rainbow smelt ecotypes (Osmerus mordax Mitchill) Molec Ecol. 2003;12:315–330. doi: 10.1046/j.1365-294X.2003.01735.x. [DOI] [PubMed] [Google Scholar]
- Seehausen O. Hybridization and adaptive radiation. Trends Ecol Evol. 2004;19:198–207. doi: 10.1016/j.tree.2004.01.003. [DOI] [PubMed] [Google Scholar]
- Bell M, Travis MP. Hybridization, transgressive segregation, genetic covariation, and adaptive radiation. Trends Ecol Evol. 2005;20:358–361. doi: 10.1016/j.tree.2005.04.021. [DOI] [PubMed] [Google Scholar]
- Gavrilets S. In: Fitness Landscapes and the Origin of Species: Monographs in Population Biology vol. 41. Levin, S. A., Horn, H. S., editor. Vol. 41. Princeton, NJ., Princeton University Press ; 2004. [Google Scholar]
- Baack EJ, Rieseberg LH. A genomic view of introgression and hybrid speciation. Curr Opin Genet Dev. 2007. in press . [DOI] [PMC free article] [PubMed]
- Martinsen GD, Whitham TG, Turek RJ, Keim P. Hybrid populations selectively filter gene introgression between species. Evolution. 2001;55:1325–1335. doi: 10.1111/j.0014-3820.2001.tb00655.x. [DOI] [PubMed] [Google Scholar]
- Yatabe Y, Kane NC, Scotti-Saintagne C, Rieseberg LH. Rampant gene exchange across a strong reproductive barrier between the annual sunflowers Helianthus annus and H. petiolaris. Genetics. 2007;175:1883–1893. doi: 10.1534/genetics.106.064469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallet J, Beltrán M, Neukirchen W, Linares M. Natural hybridization in Heliconiine butterflies: the species boundary is a continuum. BMC Evol Biol. 2007;7:28. doi: 10.1186/1471-2148-7-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltrán M, Jiggins CD, Brower AVZ, Bermingham E, Mallet J. Do pollen feeding and pupal-mating have a single origin in Heliconius? Inferences from multilocus sequence data . Biol J Linn Soc. 2007;92:221–239. doi: 10.1111/j.1095-8312.2007.00830.x. [DOI] [Google Scholar]
- Linares M. The ghost of mimicry past: laboratory reconstruction of an extinct butterfly "race". Heredity. 1997;78:628–635. [Google Scholar]
- Gilbert LE. In: Ecology and Evolution Taking Flight: Butterflies as Model Systems. Boggs CL, Watt WB, Ehrlich PR, editor. Chicago, University of Chicago Press ; 2003. Adaptive novelty through introgression in Heliconius wing patterns: evidence for shared genetic "tool box" from synthetic hybrid zones and a theory of diversification; pp. 281–318. [Google Scholar]
- Salazar C, Jiggins CD, Arias CF, Tobler A, Bermingham E, Linares M. Hybrid incompatibility is consistent with a hybrid origin of Heliconius heurippa Hewitson from its close relatives, Heliconius cydno Doubleday and Heliconius melpomene Linnaeus. J Evol Biol. 2005;18:247–256. doi: 10.1111/j.1420-9101.2004.00839.x. [DOI] [PubMed] [Google Scholar]
- Mallet J. Causes and consequences of a lack of coevolution in Müllerian mimicry. Evol Ecol. 1999;13(7-8):777–806. doi: 10.1023/A:1011060330515. [DOI] [Google Scholar]
- Mavárez J, Salazar C, Bermingham E, Salcedo C, Jiggins CD, Linares M. Speciation by hybridization in Heliconius butterflies. Nature. 2006;441:868–871. doi: 10.1038/nature04738. [DOI] [PubMed] [Google Scholar]
- Mallet J, Barton N, Lamas MG, Santisteban CJ, Muedas MM, Eeley H. Estimates of selection and gene flow from measures of cline width and linkage disequilibrium in Heliconius hybrid zones. Genetics. 1990;124:921–936. doi: 10.1093/genetics/124.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapan D. Three-butterfly system provides a field test of Müllerian mimicry. Nature. 2001;409:338–340. doi: 10.1038/35053066. [DOI] [PubMed] [Google Scholar]
- Kronforst M, Salazar C, Linares M, Gilbert L. No genomic mosaicism in a putative hybrid butterfly species. Proc R Soc Lond B. 2007;274:1255–1264. doi: 10.1098/rspb.2006.0207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronforst M. Primers for the amplification of nuclear introns in Heliconius butterflies. Molec Ecol Notes. 2005;5:158–162. doi: 10.1111/j.1471-8286.2004.00873.x. [DOI] [Google Scholar]
- Carroll SB, Gates J, Keys DN, Paddock SW, Panganiban GEF, Selegue JE, Williams JA. Pattern formation and eyespot determination in butterfly wings. Science. 1994;265:109–114. doi: 10.1126/science.7912449. [DOI] [PubMed] [Google Scholar]
- Keys DN, Lewis DL, Selegue JE, Pearson BJ, Goodrich LV, Johnson RL, Gates J, Scott MP, Carroll SB. Recruitment of a hedgehog regulatory circuit in butterfly eyespot evolution. Science. 1999;283:532–534. doi: 10.1126/science.283.5401.532. [DOI] [PubMed] [Google Scholar]
- Brunetti CR, Selegue JE, Monteiro A, French V, Brakefield PM, Carroll SB. The generation and diversification of butterfly eyespot color patterns. Curr Biol. 2001;11:1578–1585. doi: 10.1016/S0960-9822(01)00502-4. [DOI] [PubMed] [Google Scholar]
- Nijhout HF. The Development and Evolution of Butterfly Wing Patterns. Washington, DC , Smithsonian Institution Press; 1991. [Google Scholar]
- Joron M, Jiggins CD, Papanicolaou A, McMillan WO. Heliconius wing patterns: an evo-devo model for understanding phenotypic diversity. Heredity. 2006;97:157–167. doi: 10.1038/sj.hdy.6800873. [DOI] [PubMed] [Google Scholar]
- Beldade P, McMillan WO, Papanicolaou A. Butterfly genomics eclosing. Heredity. 2008;100:150–157. doi: 10.1038/sj.hdy.6800934. [DOI] [PubMed] [Google Scholar]
- Bull V, Beltrán M, Jiggins CD, McMillan WO, Bermingham E, Mallet J. Polyphyly and gene flow between non-sibling Heliconius species. BMC Biol. 2006;4:11. doi: 10.1186/1741-7007-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey J, Nielsen R. Multilocus methods for estimating population sizes, migration rates and divergence time, with applications to the divergence of Drosophila pseudoscura and D. persimilis. Genetics. 2004;167:747–760. doi: 10.1534/genetics.103.024182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronforst M, Young LG, Blume LM, Gilbert LE. Multilocus analyses of admixture and introgression among hybridizing Heliconius butterflies. Evolution. 2006;60:1254–1268. [PubMed] [Google Scholar]
- Beltrán M, Jiggins CD, Bull V, McMillan WO, Bermingham E, Mallet J. Phylogenetic discordance at the species boundary: gene genealogies in Heliconius butterflies. Mol Biol Evol. 2002;19:2176–2190. doi: 10.1093/oxfordjournals.molbev.a004042. [DOI] [PubMed] [Google Scholar]
- Naisbit RE, Jiggins CD, Linares M, Salazar C, Mallet J. Hybrid sterility, Haldane’s rule and speciation in Heliconius cydno and H. melpomene. Genetics. 2002;161:1517–1526. doi: 10.1093/genetics/161.4.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado CA, Kliman RM, Markert JA, Hey J. Inferring the history of speciation from multilocus DNA sequence data: the case of Drosophila pseudoobscura and close relatives. Molec Biol Evol. 2002;19:472–488. doi: 10.1093/oxfordjournals.molbev.a004103. [DOI] [PubMed] [Google Scholar]
- Jiggins CD, Linares M, Naisbit RE, Salazar C, Yang Z, Mallet J. Sex-linked hybrid sterility in a butterfly. Evolution. 2001;55(8):1631–1638. doi: 10.1111/j.0014-3820.2001.tb00682.x. [DOI] [PubMed] [Google Scholar]
- Prowell DP. In: Endless Forms Species and Speciation. Howard DJ, Berlocher SH, editor. New York , Oxford University Press; 1998. Sex Linkage and Speciation in Lepidoptera; pp. 309–329. [Google Scholar]
- Yang Z, Nielsen R, Goldman N, Pedersen AMK. Codon-substitution models for heterogeneus selection pressure at amino acid sites. Genetics. 2000;155:431–449. doi: 10.1093/genetics/155.1.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada C, Jiggins CD. Patterns of pollen feeding and habitat preference among Heliconius species. Ecol Entomol. 2002;27:448–456. doi: 10.1046/j.1365-2311.2002.00434.x. [DOI] [Google Scholar]
- Rieseberg LH, Sinervo B, Linder CR, Ungerer MC, Arias DM. Role of gene interactions in hybrid speciation: evidence from ancient and experimental hybrids. Science. 1996;272:741–745. doi: 10.1126/science.272.5262.741. [DOI] [PubMed] [Google Scholar]
- Meyer A, Salzburger W, Schartl M. Hybrid origin of swordtail species (Teleostei: Xiphophorus clemenciae) driven by sexual selection. Molec Ecol. 2006;15:721–731. doi: 10.1111/j.1365-294X.2006.02810.x. [DOI] [PubMed] [Google Scholar]
- Maddison WP. Gene trees in species trees. Syst Biol. 1997;46(3):523–536. doi: 10.2307/2413694. [DOI] [Google Scholar]
- Schwarz D, Matta BM, Shakir-Botteri NL, McPheron BA. Host shift to an invasive plant triggers rapid animal hybrid speciation. Nature. 2005;436:546–549. doi: 10.1038/nature03800. [DOI] [PubMed] [Google Scholar]
- Gompert Z, Fordyce JA, Forister ML, Shapiro AM, Nice CC. Homoploid hybrid speciation in an extreme habitat. Science. 2006;314:1923–1925. doi: 10.1126/science.1135875. [DOI] [PubMed] [Google Scholar]
- Turner TL, Hahn MW, Nuzhdin SV. Genomic islands of speciation in Anopheles gambiae. PLoS Biol. 2005;3:1572–1578. doi: 10.1371/journal.pbio.0030285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besansky NJ, Krzywinski J, Lehmann T, Simard F, Kern M, Mukabayie O, Fontenille D, Touré Y, Sagnon NF. Semipermeable species boundaries between Anopheles gambie and Anopheles arabiensis: evidence from multilocus DNA sequence variation. Proc Natl Acad Sci USA. 2003;100:10818–10823. doi: 10.1073/pnas.1434337100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado CA, Hey J. The causes of phylogenetic conflict in a classic Drosophila species group. Proc Roy Soc Lond B. 2003;270:1193–1202. doi: 10.1098/rspb.2003.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D, Crandall KA. Intraspecific gene genealogies: trees grafting into networks. Trends Ecol Evol. 2001;16:37–45. doi: 10.1016/S0169-5347(00)02026-7. [DOI] [PubMed] [Google Scholar]
- Clement M, Posada D, Crandall KA. TCS: a computer program to estimate gene genealogies. Mol Ecol. 2000;9:1657–1660. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- Watterson GA. On the number of segregating sites in genetical models without recombination. Theor Popul Biol. 1975;7:256–275. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
- Rozas J, Sánchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorhism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall JD. A comparison of estimators or the population recombination rate. Molec Biol Evol. 2000;17:156–163. doi: 10.1093/oxfordjournals.molbev.a026228. [DOI] [PubMed] [Google Scholar]
- Hey J, Wakeley J. A coalescent estimator of the population recombination rate. Genetics. 1997;145:833–846. doi: 10.1093/genetics/145.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. Molecular Evolutionary Genetics. New York , Columbia University Press; 1987. [Google Scholar]
- Wright S. The genetical structure of populations. Ann Eugen. 1951;15:323–354. doi: 10.1111/j.1469-1809.1949.tb02451.x. [DOI] [PubMed] [Google Scholar]
- Lynch M, Crease TJ. The analysis of population survey data on DNA sequence variation. Molec Biol Evol. 1990;7:377–394. doi: 10.1093/oxfordjournals.molbev.a040607. [DOI] [PubMed] [Google Scholar]
- Charlesworth B. Measures of divergence between populations and the effect of forces that reduce variability. Molec Biol Evol. 1998;15:538–543. doi: 10.1093/oxfordjournals.molbev.a025953. [DOI] [PubMed] [Google Scholar]
- Kessing B. In: SEQUENCER, Version 6.1.0. Distributed by the author. Naos Marine Laboratories STRI, editor. Panama ; 2000. [Google Scholar]
- Hudson RR, Kaplan NL. Statistical proprieties of the number of recombination events in the history of a sample of DNA sequences. Genetics. 1985;111:147–164. doi: 10.1093/genetics/111.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa M, Kishino K, Yano T. Dating the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol. 1985;22:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- Piganeau G, Eyre-Walker A. A reanalysis of the indirect evidence for recombination in human mitochondrial DNA. Heredity. 2004;92:282–288. doi: 10.1038/sj.hdy.6800413. [DOI] [PubMed] [Google Scholar]
- Piganeau G, Gardner M, Eyre-Walker A. A broad survey of recombination in animal mitochondria. Molec Biol Evol. 2004;21:2319–2325. doi: 10.1093/molbev/msh244. [DOI] [PubMed] [Google Scholar]
- Brower AVZ. Rapid morphological radiation and convergence among races of the butterfly Heliconius erato inferred from patterns of mitochondrial DNA evolution. Proc Natl Acad Sci USA. 1994;91:6491–6495. doi: 10.1073/pnas.91.14.6491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR. In: Oxford Surveys in Evolutionary Biology. Futuyma D, Antonovics J, editor. Vol. 7. Oxford, Oxford University. Press ; 1990. Gene genealogies and the coalescent process; pp. 1–44. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Specimen collection list. Populations sampled with information about the genes sequenced for each individual.