Abstract
Transforming growth factor β (TGF-β) regulates a broad range of biological processes, including cell growth, development, differentiation, and immunity. TGF-β signals through its cell surface receptor serine kinases that phosphorylate Smad2 or Smad3 proteins. Because Smad3 and its partner Smad4 bind to only 4-bp Smad binding elements (SBEs) in DNA, a central question is how specificity of TGF-β-induced transcription is achieved. We show that Smad3 selectively binds to two of the three SBEs in PE2.1, a TGF-β-inducible fragment of the plasminogen activator inhibitor-1 promoter, to mediate TGF-β-induced transcription; moreover, a precise 3-bp spacer between one SBE and the E-box, a binding site for transcription factor μE3 (TFE3), is essential for TGF-β-induced transcription. Whereas an isolated Smad3 MH1 domain binds to TFE3, TGF-β receptor-mediated phosphorylation of full-length Smad3 enhances its binding to TFE3. Together, these studies elucidate an important mechanism for specificity in TGF-β-induced transcription of the plasminogen activator inhibitor-1 gene.
Transforming growth factor β (TGF-β) regulates a broad range of biological processes, including cell growth, differentiation, and production of extracellular matrix proteins; mutations that disrupt the TGF-β signaling pathway contribute to tumor progression (1–3). TGF-β signals through sequential activation of two cell surface receptor serine-threonine kinases, which phosphorylate Smad2 and/or Smad3 (4–7). Phosphorylated Smad2 or Smad3, together with Smad4, translocates into the nucleus and associates with other transcription factors, leading to the activation of transcription of specific genes (4, 5, 8–12). Transcription factors that cooperate with Smad proteins to regulate transcription of certain genes include FAST-1, which mediates activin induction of the Mix.2 gene during embryonic frog development (13, 14), and c-Jun, c-Fos, ATF2, and vitamin D receptor, which interact with phosphorylated Smad3 to mediate TGF-β-induced transcription of various genes (11, 15–17).
Zawel et al. (18) identified a palindromic Smad binding element, GTCTAGAC, by selecting for Smad3 and Smad4 binding sequences from a pool of random oligonucleotides. The three-dimensional structure of the Smad3 MH1 domain indicates that an MH1 monomer binds precisely to a 4-bp sequence, AGAC (19). Mutation of the AGAC sequence in the TGF-β-inducible plasminogen activator inhibitor-1 (PAI-1) and c-Jun promoters abrogates TGF-β-inducible transcription (20–23). Together, these reports indicate that a AGAC sequence is sufficient for high-affinity binding of Smad3 and Smad4. Theoretically, this AGAC Smad binding element (SBE) should appear on average once every 256 bp in the genome (1:44); thus, most, if not all, genes that contain binding sites for Smad partner transcription factors, such as FAST-1, AP-1, and transcription factor μE3 (TFE3) (9, 11, 12, 21, 22), will have SBEs in their promoters, but not all genes with binding sites for such transcription factors are transcriptionally responsive to TGF-β. Therefore, it is unclear what controls the specificity of TGF-β signaling at the transcription level.
TFE3 is a member of the basic helix–loop–helix leucine zipper (bHLH-Zip) family of transcription factors (24). It contains an N-terminal and a C-terminal transcription activation domain and a central basic helix–loop–helix leucine zipper DNA binding domain (25), although additional N-terminal sequences may also exist (26). Data from its three-dimensional structure together with biochemical studies indicate that the basic helix–loop–helix leucine zipper domain of this family of transcription factors binds to a CACGTG sequence, the so-called E-box (21, 24, 27). We previously reported that Smad3 and TFE3 cooperatively activate TGF-β-induced transcription by binding to their cognate sites in a 36-bp TGF-β-inducible PE2.1 element, which was identified through a systematic truncation of the 800-bp TGF-β-inducible promoter of the PAI-1 gene (21). Here we used Smad3, TFE3, and the PE2.1 promoter as a model system to elucidate how the specificity of TGF-β-induced gene transcription of the PAI-1 gene is achieved.
Methods
Plasmid Construction.
Standard molecular biology techniques were used (28). To construct luciferase genes driven by various mutant PE2.1 promoters, we designed oligonucleotides that contained various mutations as indicated in Figs. 2 and 3 and then inserted the corresponding annealed mutant oligonucleotides into the KpnI/PstI site of PE2.1-Luc to generate the resulting reporter genes as indicated in Figs. 2 and 3. To make constructs for expressing glutathione S-transferase (GST) fusion proteins of human Smad3, Smad3 cDNA was amplified by PCR, and the resulting fragments were inserted into the BamHI/XhoI site of pGEX6–3 (Pharmacia). The junctions in all of the plasmids were sequenced, and at least two independent clones for luciferase assays were used and their promoters were sequenced to verify the mutations.
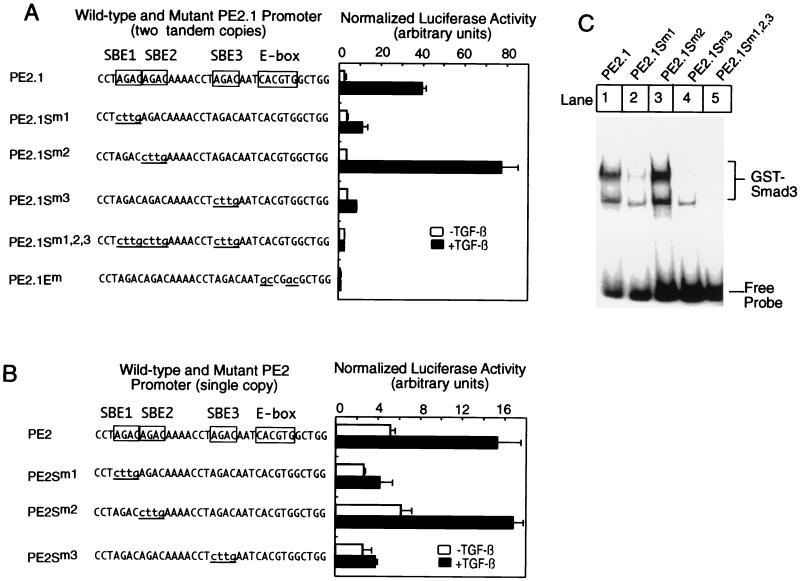
Figure 2.
Only two out of three potential Smad binding sites in the PE2.1 promoter are essential for TGF-β-induced transcription. (A) Luciferase reporter genes were transfected into HepG2 cells for luciferase assays as described in Fig. 1. The top sequence at the left part of A denotes the sequence from one of the two tandem copies of the PE2.1 promoter that drive expression of the luciferase gene. E-box, TFE3 binding site. (B) A luciferase reporter gene driven by a single copy of the wild-type or mutant PE2.1 promoter was transfected into HepG2 cells, and the luciferase assays were performed as described in Fig. 1. (C) Either a wild-type 32P-labeled DNA probe or probes with mutations as indicated in A were incubated with 1.0 μg of GST-Smad3, and the gel shift assay was performed as described (21). The gel was scanned in a phosphorimager.
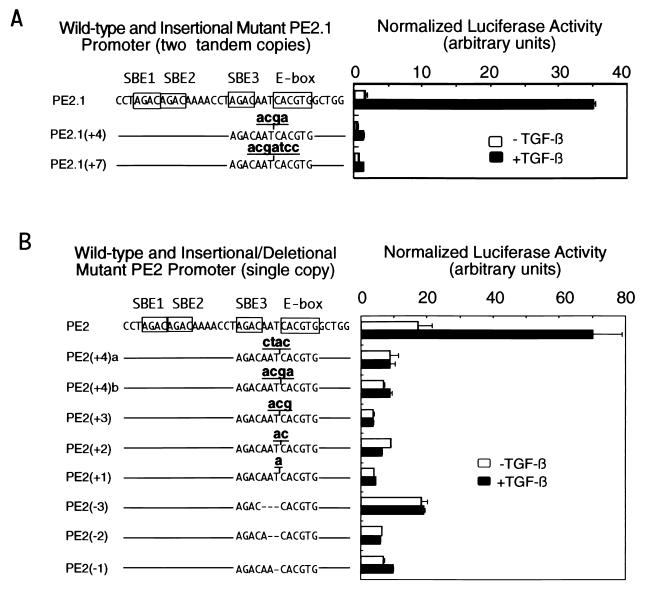
Figure 3.
The 3-bp spacer between the third SBE and the TFE3 binding site of the PE2.1 promoter is essential for the TGF-β-induced activation of the promoter. (A) Luciferase reporter genes driven by two tandem copies of the wild-type PE2.1 promoter or mutant PE2.1 promoters with an insertion between the SBE3 and the E-box are shown on the left. The line represents the wild-type DNA sequence depicted above the line. The luciferase reporter genes were transfected into HepG2 cells for luciferase assays as described in Fig. 1. A volume of 100 μl of cell lysate, instead of the usual 20 μl, was used for the β-galactosidase assays. (B) Luciferase reporter genes driven by a single copy of the wild-type, or mutant PE2.1 promoters were transfected into HepG2 cells for the luciferase assay as described in Fig. 1.
Tissue Culture, Transfection, and Luciferase Assays.
HepG2 cells and BOSC23 cells were cultured as described (21). Cells were transfected by the calcium phosphate precipitation method (28). For luciferase assays, cells were also cotransfected with 0.3 μg per well of pCMV-β encoding the LacZ gene (CLONTECH) as an internal control to normalize the luciferase activity. To transfect HepG2 cells, cells were seeded at a density of 100,000 cells per well in 12-well plates and transfected as described (21). All luciferase activities were normalized by the β-galactosidase activities and presented as an average from duplicate samples.
GST and Flag-Epitope Pulldown Assays.
Glutathione Sepharose beads (Pharmacia) coupled with GST-Smad3 or GST-Smad3 subfragments were incubated with in vitro translated 35S-labeled TFE3 at 4°C in 200 μl of binding buffer [20 mM Tris, pH 8.0/150 mM NaCl/1 mM EDTA/1% Nonidet P-40/12% glycerol/1× protease inhibitor mixture Complete (Boehringer Mannheim)] for 1 h. The beads were centrifuged and washed three times with 500 μl of binding buffer before addition of the SDS loading buffer and separation by SDS/PAGE. The gel was exposed to a phosphorimager plate and scanned in a phosphorimager. For Flag-epitope pulldown assays, cell lysates from the transfected BOSC23 cells were lysed in Nonidet P-40 lysis buffer (50 mM Tris, pH 8.0/100 mM NaCl/25 mM β-glycerolphosphate/1% Nonidet P-40/1× protease mixture Complete), and then the cell lysates were mixed with Sepharose beads conjugated with anti-Flag antibody M2 (Eastman Kodak) and rotated at 4°C for 1 h before removal of the supernatant. The remaining beads were mixed with 200 μl of binding buffer, and 1 μl of 35S-labeled TFE3 in reticulocyte lysate was added to the samples. The resulting samples were rotated at 4°C for 1 h being washed three times with 500 μl of binding buffer and separation on an 8–16% gradient SDS/PAGE gel. The gel was exposed to a phosphorimager plate and scanned in a phosphorimager (Fuji BAS 2000).
Gel-Shift Assays and Immunoblotting.
Gel-shift reactions were performed as described (21). Radiolabeled probes were synthesized by PCR amplification in the presence of [α-32P]dCTP. Signals for the gel shift assays were detected on a phosphorimager. The expression of Flag-tagged Smad3 or mutant Smad3A proteins in transfected BOSC23 cells were detected with anti-Flag (M2) antibody (Eastman Kodak) as described (21).
Results
Smad3 Selectively Binds to Multiple SBEs in the PE2.1 Fragment of the PAI-1 Promoter to Mediate TGF-β-Induced Transcription.
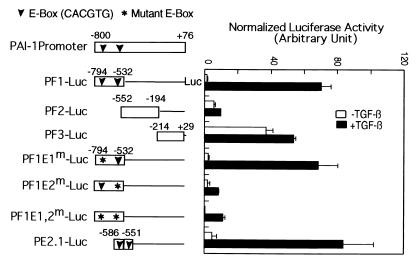
Of the 876 bases in the PAI-1 promoter, the segment from −532 to −794 (PF1, Fig. 1) is sufficient to confer inducibility by TGF-β in human HT1080 cells (21). Fig. 1 shows that the same holds for expression in HepG2 cells of luciferase reporter genes containing only a single copy of these segments of the PAI-1 promoter. Segment PF1, which contains two E-box sequences, mediates an approximately 40-fold induction by TGF-β. In contrast, PF2 and PF3 failed to mediate any TGF-β-induced transcription, although the PF3 element supported significant basal (i.e., TGF-β-independent) transcription. Mutation of the first E-box in PF1 did not diminish the promoter activity. In contrast, mutation of the second E-box sequence in PF1, or both E-box sequences together, markedly reduced all PF1 promoter activity, especially in the presence of TGF-β; thus, the second E-box in the PF1 promoter, and by implication in the full-length PAI-1 promoter, is essential for TGF-β inducibility. Fig. 1 also shows that PE2.1, containing two tandem copies of a 36-bp sequence surrounding the second E-box (PE2), mediated a 20-fold reporter gene induction by TGF-β. Together, these results indicate that the second E-box sequence is essential for the activity of the PAI-1 promoter and the position of the E-box in the natural promoter is critical for conferring TGF-β-induced transcription; moreover, the minimal PE2.1 promoter, containing the second E-box and surrounding sequences, mimics the full promoter activity and is well regulated by TGF-β.
Figure 1.
The E-box sequence is essential for TGF-β-induced transcription from the natural PAI-1 promoter. HepG2 cells were transfected with 2.5 μg of luciferase reporter genes as indicated and 0.3 μg of pCMV-gal containing the LacZ gene as an internal control. The transfected cells were treated with or without TGF-β (200 pM) for 20 h as described (21). The downward-pointing arrowheads indicate the position of E-boxes and the asterisks denote mutations of specific E-boxes. PE2.1-Luc contains two tandem copies of the indicated PE2 element (−586 to −551); all other constructs contain a single copy of the indicated promoter segment. The luciferase activity is normalized to the β-galactosidase activity, and the error bar indicates the standard deviation of the duplicate samples.
We noticed that a single PE2 element (−586 to −551) contained three SBEs and tested whether all three are essential for TGF-β-induced transcription. To this end, we generated luciferase reporter genes driven by two tandem copies of the PE2 promoter with mutations in various SBEs and transfected them into HepG2 cells for luciferase assays. Confirming data in Fig. 1, the PE2.1-Luc reporter gene with the wild-type promoter conferred a 20-fold induction of luciferase activities by TGF-β (Fig. 2A). Mutation of either the first or the third SBE (AGAC → cttg) in the PE2.1 element dramatically reduced TGF-β-induced transcription. In contrast, mutation of the second SBE in the promoter slightly enhanced TGF-β-induced transcription. Mutations in either all three SBEs or the TFE3 binding E-box completely abrogated TGF-β-induced transcription (Fig. 2A). These results suggest that two SBEs are essential for TGF-β-induced transcription from the PE2.1 promoter and the sequence adjacent to the first and the third SBE may be important for TGF-β-induced transcription. Indeed, mutation of the CT sequence 5′ of the first SBE to AC diminished the PE2.1 promoter activity by 50% (data not shown).
To rule out the possibility that the tandem mutant copies of the PE2 element caused an artificial response in TGF-β-induced transcription, we constructed luciferase reporter genes driven by a single copy of the wild-type or mutant PE2 element, and the resultant constructs were transfected into HepG2 cells for luciferase assays. Fig. 2B shows that a single copy of the PE2 element supported less TGF-β- induced transcription than did two copies in tandem. Importantly, mutation of either the first or the third SBE, but not the second, abolished TGF-β-induced gene transcription, fully consistent with the results obtained with two tandem copies of the PE2 promoter. As expected, mutation of any two of the three SBEs in the PE2 element also abolished TGF-β-induced transcription (data not shown).
Fig. 2C shows a gel shift assay with purified GST-Smad3 and 32P-labeled tandem copies of either the wild-type PE2.1 promoter or promoters with the indicated SBE mutations. Consistent with the luciferase assays shown in Fig. 2A, mutation of either the first or the third SBE in the PE2.1 element dramatically diminished its binding to GST-Smad3 (Fig. 2C, lanes 2 and 4). As expected, mutation of all three SBEs also completely abolished binding to GST-Smad3 (lane 5), whereas mutation of the second SBE did not reduce its binding to GST-Smad3 at all (lane 3). Similar results were obtained with Smad3 and Smad4 proteins from BOSC23 cells cotransfected with a constitutively active type I TGF-β receptor TβRI-T204D (data not shown).
Together, these results indicate (i) not all the SBEs in the natural PE2.1 promoter are essential for TGF-β-induced transcription as well as for binding to Smad3; (ii) more than one SBE in the promoter is essential for TGF-β-induced transcription of a natural promoter; and (iii) both the third SBE and the E-box in the 36-bp PE2 promoter, which are separated only by a 3-bp spacer, are essential for TGF-β-induced transcription.
The 3-bp Spacer Sequence Between the Third SBE and the TFE3 Binding Site Is Essential for TGF-β-Induced Transcription.
We further examined the role of the 3-bp spacer between the SBE and the E-box of the PE2.1 promoter in TGF-β-induced transcription (Fig. 3). We added 4 bp (acga) or 7 bp (acgatcc) to the 3-bp spacer of the two tandem copies of the PE2 promoter and transfected the resulting constructs into HepG2 cells for luciferase assays (Fig. 3A). Whereas expression of luciferase from the wild-type construct was induced 20-fold by TGF-β, insertion of either 4- or 7-bp random nucleotides into the spacer severely impaired TGF-β-induced gene expression.
To pinpoint the minimal perturbation in the spacer that still impairs TGF-β-induced transcription, we inserted 1, 2, 3, or 4 bp into the spacer of a single copy of the PE2 promoter, or deleted 3, 2, or 1 bp of the spacer. Fig. 3B shows that any insertion or deletion of nucleotides in the spacer region between the third SBE and the E-box completely abrogated TGF-β-induced transcription of the reporter gene; thus, there is an absolute requirement for a 3-bp spacer sequence between the third SBE and the E-box. Insertion of 4 bp, either CTAC or ACGA, in the spacer also abolished TGF-β-induced gene transcription (Fig. 3B), suggesting that it is not the particular inserted sequences that had this effect. Insertion or deletion of nucleotides in this spacer region may interfere with formation of a ternary complex of TFE3, Smad3, and the PE2 promoter or with recruitment of other transcriptional coactivators.
TFE3 Binds to the MH1 Domain of Smad3.
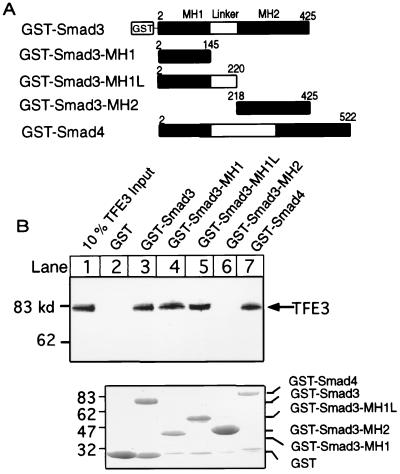
We then examined potential physical interactions between TFE3 and Smad3. First, GST-Smad3, GST-Smad4, or GST-Smad3 fragments (GST-Smad3-MH1; GST-Smad3-MH1L; and GST-Smad3-MH2), as shown in Fig. 4A, were incubated with in vitro synthesized 35S-labeled TFE3, and then TFE3 bound to GST-Smad3 or -4 or GST-Smad3 fragments were separated by SDS/PAGE and detected in a phosphorimager. Fig. 4B shows that about 10% of the added 35S-labeled TFE3 bound to GST-Smad3, GST-Smad3-MH1, GST-Smad3-MH1L, and GST-Smad4 (lanes 3–5 and 7), but not to the control GST protein or GST-Smad3-MH2 (lanes 2 and 6). Similarly, TFE3 also bound to GST-Smad4-MH1 but not Smad4-MH2 (data not shown). This result indicates that TFE3 binds to the MH1 domain of both Smad3 and Smad4, the same domain that also binds to DNA. As a control, an equivalent amount of GST-fusion proteins used for the GST-pulldown assays were separated by SDS/PAGE and stained by Coomassie blue (Fig. 4B).
Figure 4.
N-terminus of Smad3 binds to TFE3. (A) A diagram of various GST-Smad3 fusion proteins that were used in the GST-pulldown assays. (B, Upper) GST or GST-fusion proteins (6 μg) were bound to glutathione beads and incubated with 1.5 μl of 35S-labeled TFE3 synthesized in reticulocyte lysate with a TNT kit (Promega); the bound TFE3 was separated by SDS/PAGE, exposed to a phosphorimager plate for 16 h, and scanned in a phosphorimager. (Lower) Various GST-Smad fusion proteins (6 μg) as indicated were separated by SDS/PAGE and stained with Coomassie Brilliant Blue R-250.
Phosphorylation of Smad3 Enhances Its Association with TFE3.
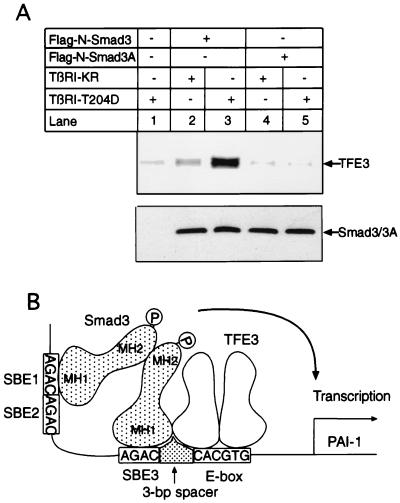
To determine whether phosphorylation of Smad3 induces its association with TFE3, we transfected BOSC23 cells with Flag-N-Smad3 or Flag-N-Smad3A, which contains mutations in the TGF-β-inducible phosphorylation sites, together with the constitutively active Type I TGF-β receptor TβRI-T204D. Flag-N-Smad3 or Flag-N-Smad3A were purified from the lysates of the transfected cells by binding to Sepharose beads conjugated with anti-Flag antibodies. Then 35S-labeled TFE3 was incubated with the beads and the bound TFE3 was detected in a phosphorimager. Fig. 5A shows that only a small amount of TFE3 bound to the control beads (lane 1), and the kinase-dead (null) mutant type I TGF-β receptor, TβRI (KR), did not support Smad3 binding to TFE3 (lane 2). In contrast, coexpression of the constitutively active receptor TβRI-T204D increased the ability of the wild-type Flag-N-Smad3 to bind 35S-labeled TFE3 10-fold (Fig. 5, lane 3); however, little TFE3 bound to the mutant Flag-N-Smad3A (lane 4), and coexpression of TβRI-T204D did not increase binding of Flag-N-Smad3A to TFE3 (lane 5), although the amounts of Flag-N-Smad3 and Smad3A used in the experiment were comparable (Fig. 5A). Taken together, these results demonstrate that phosphorylation of Smad3 by the type I receptor induces its association with TFE3; however, we do not know the exact stoichiometry of the complex formed between TFE3 and Smad3 and Smad4.
Figure 5.
Phosphorylation of Smad3 induces its association with TFE3. (A, Upper) BOSC23 cells were transfected with the indicated plasmids. Either Flag-N-Smad3 or mutant Flag-N-Smad3A in the cell lysate (200 μl) was bound to Sepharose beads conjugated with anti-Flag antibody M2 as described in Methods, and 1 μl of 35S-labeled TFE3 synthesized in reticulocyte lysate was added to the beads. Bound TFE3 was processed and detected in a phosphorimager as described in Methods. (Lower) An immunoblot with 3 μl (about 150 μg protein) of cell lysate for each lane was probed with an anti-Flag M2 antibody to detect the Flag epitope-tagged Smad3 or Smad3A. (B) A model for TFE3 and Smad3-mediated specificity in TGF-β-induced transcription. E-box, TFE3 binding site.
Discussion
An Essential 3-bp Spacer Between the E-box and a Smad-Binding Element Increases the Specificity in TGF-β-Induced Transcription.
Expression of the PAI-1 gene is controlled by multiple growth factors and cytokines in an array of cells and tissues, and its transcriptional control is likely to be complex. Here we focused on induction of the PAI-1 gene by TGF-β and demonstrated how phosphorylated Smad proteins cooperate with a widely expressed transcription factor, TFE3, to mediate specific gene induction.
Smad2 and/or Smad3 are thought to be required for induction of all genes induced by TGF-β. When phosphorylated by the activated Type I TGF-β receptor, these Smads bind to the common co-Smad, Smad4, translocate into the nucleus, and mediate induction or repression of specific genes (4, 5, 8–10, 12, 18). A 4-bp SBE DNA sequence, AGAC, is sufficient to bind Smad3 and Smad4 (5, 9, 10, 12, 18, 20). Similarly, a six-base palindromic E-box sequence, CACGTG, is sufficient for binding TFE3 (29). On average, there will be one CACGTG and 16 AGAC sequences (SBEs) in every 4,096 bp in the genome (1 × 46/4,096 and 16 × 44/4,096, respectively); therefore, simply having a combination of one or two SBEs and one TFE3 or AP-1 binding site in a promoter is not sufficient to explain the specificity of TGF-β-induced transcription.
Here we demonstrated that the second E-box in the full PAI-1 promoter and only two of the three 4-bp SBEs in the PE2 element are essential for TGF-β-induced promoter activity. Importantly, a 3-bp spacer between one of the SBEs and the E-box is essential. By simple calculation, the probability of having one AGAC and one CACGTG sequence separated by a spacer of fixed length is 1:410, or about one in a million base pairs. The combination of the E-box, multiple SBEs, and a requirement for a fixed spacer between the two binding sites thus markedly increases the specificity of TGF-β-induced transcription of the PAI-1 gene. To our knowledge, this is the first study that shows that the transcription apparatus selectively uses certain 4-bp SBEs in a TGF-β-inducible promoter and the 3-bp spacer between the SBE and the binding site for another transcription factor is essential for TGF-β-induced transcription. Similarly, the spacer between protein-binding sites affects the activities of receptors for thyroid hormone, vitamin D3, and retinoic acid (30).
Phosphorylation-Induced Interaction of TFE3 and Smad3 Enhances Specificity in TGF-β-Induced Transcription.
Whereas both GST-Smad3 and GST-Smad4 bind TFE3 (Fig. 4), phosphorylation of full-length Smad3 enhances its association with TFE3 about 10-fold (Fig. 5). Presumably, the phosphorylated Smad3 also becomes bound to endogenous Smad4. Because Smad4 is chemically unmodified during TGF-β signaling, we cannot readily determine whether it is Smad4 or phosphorylated Smad3 that physically interacts with TFE3; nonetheless, this phosphorylation-dependent association between Smad3 and TFE3 can account for the synergistic activation of TGF-β-induced transcription by TFE3 and Smad3. In its native state, Smad3 is unable to bind to DNA or may only bind DNA weekly, presumably because the C-terminal MH2 domain masks its N-terminal DNA-binding MH1 domain. Phosphorylation of the C-terminal serines (SSVS) on Smad3 by the type I receptor allows Smad3 to bind Smad4; phosphorylation apparently “opens” the protein and exposes its MH1 domain. In vitro, GST-Smad3 binds TFE3 when its C-terminal serines were presumably not phosphorylated (Fig. 4). The GST moiety in fusion proteins often forms a dimer (19), which may mimic the effect of phosphorylation of the C-terminal serine residues and lead to increased affinity between the nonphosphorylated Smad3 and TFE3. On the other hand, the relatively high concentration of the purified GST-Smad3 may also increase its binding to TFE3; nevertheless, our work showed a specific interaction between the N-terminal MH1 domain of Smad3 and TFE3 (Fig. 4).
Fig. 5B shows a model for TGF-β-induced transcription, with the TGF-β-inducible PE2 segment representing the entire PAI-1 promoter. First, TGF-β induces phosphorylation of Smad3, and phosphorylated Smad3 binds to TFE3. It is likely that TFE3 also binds to Smad4 in response to TGF-β, because TFE3 directly binds to the GST-Smad4 fusion protein (Fig. 4B). We do not know whether TFE3 binds Smad2 nor do we know the stoichiometry of the TFE3/Smad3/Smad4 complex. Second, a complex of Smad3, Smad4, and TFE3 cooperatively binds in a precise geometry to the PE2 promoter, such that 3 bp separate the E-box from one of the two essential Smad-binding sites. After binding to DNA, the Smad3/Smad4/TFE3 complex likely recruits transcription coactivators such as p300 and CREB binding protein (31–35), leading to enhanced transcription of the PAI-1 gene. This model suggests a critical role for the selective use of SBEs and a requirement for a fixed spacer between the SBE and the binding site for other transcription factors in achieving high specificity of TGF-β-induced gene transcription. As noted, other transcription factors, binding to the PF3 and perhaps other segments of the PAI-1 promoter, may enhance or otherwise modify the extent of TGF-β-induced transcription.
Thus, selective use of the multiple SBEs in the PE2 promoter segment and the spacer between the SBE and the E-box play a critical role in determining the specificity of TGF-β-induced PAI-1 induction. We have not found a second TGF-β-induced promoter with an essential TFE3 binding site adjacent to an SBE, and the conclusions we have drawn concerning the interactions of Smad proteins, TFE3, and promoter sequences may apply in detail only to the PAI-1 promoter. Several recent reports point to important crosstalk between Smad proteins and transcription factors such as FAST-1, AP-1, SP1, CREB, and Tinman in the expression of various TGF-β-inducible genes (9, 11, 12, 22, 36–39). We speculate that the number of SBEs as well as spacer segments of precise length will also be critical for the specificity of TGF-β-induced transcription of other promoters that employ these other Smad-partner transcription factors. It will be interesting to determine whether the spacing and number of SBEs in these promoters indeed are important for the specificity and magnitude of TGF-β- induced gene transcription.
Acknowledgments
TGF-β1 was a kind gift from R & D Systems. We thank members of the Lodish group for stimulating discussions. This work was supported by National Institutes of Health Grant CA63260 to H.F.L., a Burroughs Wellcome Career Award (1676) and a Howard Temin Award (CA78592-02) to X.H., and start-up funds from Princeton University to Y.S.
Abbreviations
- PAI-1
plasminogen activator inhibitor-1
- TFE3
transcription factor μE3
- SBE
Smad binding element
- TGF-β
transforming growth factor β
- GST
glutathione S-transferase
References
- 1.Attisano L, Wrana J L. Curr Opin Cell Biol. 1998;10:188–194. doi: 10.1016/s0955-0674(98)80141-5. [DOI] [PubMed] [Google Scholar]
- 2.Heldin C-H, Miyazono K, ten Dijke P. Nature (London) 1997;390:465–471. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- 3.Massague J. Annu Rev Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- 4.Liu X, Sun Y, Constantinescu S N, Karam E, Weinberg R A, Lodish H F. Proc Natl Acad Sci USA. 1997;94:10669–10674. doi: 10.1073/pnas.94.20.10669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Macias-Silva M, Abdollah S, Hoodless P A, Pirone R, Attisano L, Wrana J L. Cell. 1996;87:1215–1224. doi: 10.1016/s0092-8674(00)81817-6. [DOI] [PubMed] [Google Scholar]
- 6.Souchenlnytskyi S, Tamaki K, Engstrom U, Wernstedt C, ten Dijke P, Heldin C-K. J Biol Chem. 1997;272:28107–28115. doi: 10.1074/jbc.272.44.28107. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y, Feng X-H, Wu R-Y, Derynck R. Nature (London) 1996;383:168–172. doi: 10.1038/383168a0. [DOI] [PubMed] [Google Scholar]
- 8.Derynck R, Zhang Y, Feng X. Cell. 1998;95:737–740. doi: 10.1016/s0092-8674(00)81696-7. [DOI] [PubMed] [Google Scholar]
- 9.Labbe E, Silvestri C, Hoodless P A, Wrana J L, Attisano L. Mol Cell. 1998;2:109–120. doi: 10.1016/s1097-2765(00)80119-7. [DOI] [PubMed] [Google Scholar]
- 10.Lagna G, Hata A, Kemmati-Brivanlou A, Massague J. Nature (London) 1996;383:832–836. doi: 10.1038/383832a0. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y, Feng X-H, Derynck R. Nature (London) 1998;394:909–913. doi: 10.1038/29814. [DOI] [PubMed] [Google Scholar]
- 12.Zhou S, Zawel L, Lengauer C, Kinzler K W, Vogelstein B. Mol Cell. 1998;2:121–127. doi: 10.1016/s1097-2765(00)80120-3. [DOI] [PubMed] [Google Scholar]
- 13.Chen X, Rubock M J, Whitman M. Nature (London) 1996;383:691–696. doi: 10.1038/383691a0. [DOI] [PubMed] [Google Scholar]
- 14.Chen X, Weisberg E, Fridmacher V, Watanabe M, Naco G, Whitman M. Nature (London) 1997;389:85–89. doi: 10.1038/38008. [DOI] [PubMed] [Google Scholar]
- 15.Liberati N, Datto M, Frederick J, Shen X, Wong C, Rougier-Chapman E, Wang X. Proc Natl Acad Sci USA. 1999;96:4844–4849. doi: 10.1073/pnas.96.9.4844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sano Y, Harada J, Tashiro S, Gotoh-Mandeville R, Maekawa T, Ishii S. J Biol Chem. 1999;274:8949–8957. doi: 10.1074/jbc.274.13.8949. [DOI] [PubMed] [Google Scholar]
- 17.Yanagisawa J, Yanagi Y, Masuhiro Y, Suzawa M, Watanabe M, Kashiwagi K, Toriyabe T, Kawabata M, Miyazono H, Kato S. Science. 1999;283:1317–1321. doi: 10.1126/science.283.5406.1317. [DOI] [PubMed] [Google Scholar]
- 18.Zawel L, Dai J, Buckhaults P, Zhou S, Kinzler K, Vogelstein B, Kern S. Mol Cell. 1998;1:611–617. doi: 10.1016/s1097-2765(00)80061-1. [DOI] [PubMed] [Google Scholar]
- 19.Shi Y, Wang Y-F, Jayaraman L, Yang H, Massague J, Pavletich N P. Cell. 1998;94:585–594. doi: 10.1016/s0092-8674(00)81600-1. [DOI] [PubMed] [Google Scholar]
- 20.Dennler S, Itoh S, Vivien D, ten Dijke P, Huet S, Gauthier J. EMBO J. 1998;17:3091–3100. doi: 10.1093/emboj/17.11.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hua X, Liu X, Ansari D O, Lodish H F. Genes Dev. 1998;12:3084–3095. doi: 10.1101/gad.12.19.3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wong C, Rougier-Chapman E M, Frederick J P, Datto M B, Liberati N T, Li J-M, Wang X-F. Mol Cell Biol. 1998;19:1821–1830. doi: 10.1128/mcb.19.3.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wrana J L, Attisano L, Carcamo J, Zentella A, Doody J, Laiho M, Wang X-F, Massagué J. Cell. 1992;71:1003–1014. doi: 10.1016/0092-8674(92)90395-s. [DOI] [PubMed] [Google Scholar]
- 24.Beckmann H, Kadesch T. Genes Dev. 1991;5:1057–1066. doi: 10.1101/gad.5.6.1057. [DOI] [PubMed] [Google Scholar]
- 25.Artandi S E, Merrell K, Avitahl N, Wong K-K, Calame K. Nucleic Acids Res. 1995;23:3865–3871. doi: 10.1093/nar/23.19.3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weterman M A, Wilbrink M, Geurts van Kessel A. Proc Natl Acad Sci USA. 1996;93:15294–15298. doi: 10.1073/pnas.93.26.15294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferre-D’Amare A R, Prendergast G C, Ziff E B, Burley S K. Nature (London) 1993;363:38–45. doi: 10.1038/363038a0. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 29.Beckmann H, Su L-K, Kadesch T. Genes Dev. 1990;4:167–179. doi: 10.1101/gad.4.2.167. [DOI] [PubMed] [Google Scholar]
- 30.Umesono K, Murakami K, Thompson C, Evans R. Cell. 1991;5:1255–1266. doi: 10.1016/0092-8674(91)90020-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feng X, Zhang Y, Wu R, Derynck R. Genes Dev. 1998;15:2153–2163. doi: 10.1101/gad.12.14.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Janknecht R, Wells N J, Hunter T. Genes Dev. 1998;12:2114–2119. doi: 10.1101/gad.12.14.2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pouponnot C, Jayaraman L, Massague J. J Biol Chem. 1998;273:22865–22868. doi: 10.1074/jbc.273.36.22865. [DOI] [PubMed] [Google Scholar]
- 34.Shen X, Hu P P, Liberati N T, Datto M B, Frederick J P, Wang X-F. Mol Biol Cell. 1998;9:3309–3319. doi: 10.1091/mbc.9.12.3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Topper J N, DiChiara M R, Brown J D, Williams A J, Falb D, Collins T, Gimbrone M A., Jr Proc Natl Acad Sci USA. 1998;95:9506–9511. doi: 10.1073/pnas.95.16.9506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li J-M, Nichols M A, Chandrasekharan S, Xiong Y, Wang X-F. J Biol Chem. 1995;270:26750–26753. doi: 10.1074/jbc.270.45.26750. [DOI] [PubMed] [Google Scholar]
- 37.Moustakas A, Kardassis D. Proc Natl Acad Sci USA. 1998;95:6733–6738. doi: 10.1073/pnas.95.12.6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Szutes D, Eresh S, Bienz M. Genes Dev. 1998;12:2022–2035. doi: 10.1101/gad.12.13.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu X, Yin Z, Hudson J B, Ferguson E L, Frasch M. Genes Dev. 1998;12:2354–2370. doi: 10.1101/gad.12.15.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]