Abstract
NtrC (nitrogen regulatory protein C) is a bacterial enhancer-binding protein of 469 residues that activates transcription by σ54-holoenzyme. A region of its transcriptional activation (central) domain that is highly conserved among homologous activators of σ54-holoenzyme—residues 206–220—is essential for interaction with this RNA polymerase: it is required for contact with the polymerase and/or for coupling the energy from ATP hydrolysis to a change in the conformation of the polymerase that allows it to form transcriptionally productive open complexes. Several mutant NtrC proteins with amino acid substitutions in this region, including NtrCA216V and NtrCG219K, have normal ATPase activity but fail in transcriptional activation. We now report that other mutant forms carrying amino acid substitutions at these same positions, NtrCA216C and NtrCG219C, are capable of activating transcription when they are not bound to a DNA template (non-DNA-binding derivatives with an altered helix–turn–helix DNA-binding motif at the C terminus of the protein) but are unable to do so when they are bound to a DNA template, whether or not it carries a specific enhancer. Enhancer DNA remains a positive allosteric effector of ATP hydrolysis, as it is for wild-type NtrC but, surprisingly, appears to have become a negative allosteric effector for some aspect of interaction with σ54-holoenzyme. The conserved region in which these amino acid substitutions occur (206–220) is equivalent to the Switch I region of a large group of purine nucleotide-binding proteins. Interesting analogies can be drawn between the Switch I region of NtrC and that of p21ras.
The bacterial enhancer-binding protein NtrC (nitrogen regulatory protein C) activates transcription by the σ54-holoenzyme form of RNA polymerase (1–3). To do so it catalyzes the isomerization of closed complexes between this polymerase and a promoter to transcriptionally productive open complexes in a reaction that depends on hydrolysis of a purine nucleoside triphosphate. Because formation of open complexes is thermodynamically as well as kinetically unfavorable, NtrC and other activators of σ54-holoenzyme must function as molecular machines. To form the large oligomers that are required for ATP hydrolysis and, hence, transcriptional activation, NtrC must be phosphorylated at aspartate 54. Binding of the phosphorylated protein to an enhancer stimulates oligomer formation and, hence, stimulates ATPase activity and transcriptional activation (4, 5). Phosphorylated NtrC oligomers bound to an enhancer contact σ54-holoenzyme by means of DNA loops (1) and apparently catalyze an energy-dependent change in the conformation of the polymerase that allows it to form open complexes (6–10).
NtrC is composed of three functional domains (11–13). The N-terminal domain is the regulatory domain, which contains the site of phosphorylation. The C-terminal domain contains a helix–turn–helix DNA-binding motif and the major dimerization determinants of the protein. The central domain is the transcriptional activation domain—the domain responsible for biological output. It is composed of about 240 aa residues and shows a high degree of sequence conservation with all known transcriptional activators of σ54-holoenzyme. Secondary structure predictions coupled with the use of recognition algorithms for protein folds indicated that the central domain of NtrC and the corresponding domains of other activators adopt a mononucleotide-binding fold similar to those of p21ras and the G domain of the bacterial polypeptide elongation factor EF-Tu (14).
Mutations causing specific loss of transcriptional activation by the NtrC protein of Salmonella typhimurium affect residues in the seven conserved motifs within its central domain (14–17). Many result in decreased binding or hydrolysis of ATP. Perhaps the most interesting mutations causing loss of transcriptional activation affect what appears to be the Switch I region of NtrC (residues 206–220), which lies between its putative Walker A and B motifs. These lesions have little if any effect on the ATPase activity of NtrC but essentially eliminate biological output. By inference, they must affect some aspect of the interaction with σ54 holoenzyme—contact with the polymerase and/or energy coupling. Two amino acid substitutions in the corresponding region of the homologous activator DctD decrease its cross-linking to σ54 and the β-subunit of RNA polymerase (18).
To study the Switch I region of NtrC further we used in vitro mutagenesis to generate proteins with single cysteine substitutions in this region. Two of these, NtrCA216C and NtrCG219C, have more than sufficient ATPase activity for transcriptional activation and bind the enhancer normally. However, unlike the previously studied forms with amino acid substitutions at these positions—NtrCA216V and NtrCG219K—which fail to activate transcription, the new forms retain the ability to activate transcription poorly only at very high concentrations. Further studies of NtrCA216C indicated that DNA has become a negative allosteric effector: binding to an enhancer or “nonspecific” DNA inhibits transcriptional activation by this protein, although the enhancer remains a positive allosteric effector for ATP hydrolysis, as it is for wild-type NtrC (NtrCWT). By inference, inhibitory effects are exerted on some aspect of interaction with the polymerase.
Materials and Methods
Mutagenesis and Construction of Plasmids for NtrC Overproduction.
Plasmid pJES990 (19), a pTZ18U derivative that contains the cysteine-free (C30V and C364A) version of the ntrC coding region, was mutated with the following oligonucleotides: CATGAGAAAGGCTGCTTTACCGGGGCG (introduces the A216C substitution and a BbvI site) and GGCGCTTTTACCTGCGCAAATACCATCCGGC (introduces the G219C substitution and an FspI site). Mutagenesis was performed with the Muta-Gene phagemid in vitro mutagenesis kit (Bio-Rad). The sequences of 369-bp AgeI-BbsI fragments of the resulting plasmids, which cover the mutations, were confirmed. The AgeI-BbsI fragments were ligated into pJES990, digested similarly, to ensure that other regions of ntrC were error-free. The resulting plasmids were pJES1042 (A216C) and pJES1045 (G219C), respectively. The 1.9-kb KpnI-EcoRI fragments from pJES1042 and pJES1045 then were ligated into similarly digested pJES559 (20), which is the overexpression plasmid for maltose-binding protein (MBP) fusions to NtrC. The resulting plasmids were pJES1049 (MBP-NtrCA216C) and pJES1052 (MBP-NtrCG219C), respectively. To construct non-DNA-binding derivatives of MBP-NtrC, the 0.6-kb CspI-EcoRI fragments of pJES994 (cysteine-free version of MBP-NtrC) (19), pJES1049 and pJES1052 were replaced with the CspI-EcoRI fragment from pJES641 (overexpression plasmid for NtrC3ala) (21), resulting in pJES1135 (MBP-NtrC3ala), pJES1137 (MBP-NtrCA216C, 3ala), and pJES1138 (MBP-NtrCG219C, 3ala), respectively.
Protein Purification.
MBP-NtrC and its derivatives were purified essentially as described (20). Briefly, isopropyl β-d-thiogalactoside-induced cells were harvested and disrupted by passage through a French Pressure Cell. MBP-NtrC was precipitated with ammonium sulfate (35–70%) and then was chromatographed on Amylose (New England Biolabs), heparin agarose (Bethesda Research Laboratories), and Resource-Q (Pharmacia). Core RNA polymerase and σ54 were purified as described (22).
Transcription Assays.
The ability of NtrC proteins to catalyze open complex formation by σ54-holoenzyme was assessed in a single-cycle transcription assay as described (8, 23, 24). Templates were supercoiled plasmid pJES534 (carries a strong enhancer composed of two strong NtrC-binding sites) or pJES535 (lacks specific NtrC-binding site) (21) at a concentration of l nM. Final concentrations of reagents during open complex formation were: 30 nM core RNA polymerase, 50 nM σ54, 10 mM carbamoyl phosphate, 4 mM ATP, and 2–1,000 nM NtrC (always reported as dimer concentration). Open complex formation was terminated by addition of heparin to a final concentration of 0.1 mg/ml, and synthesis of transcripts was achieved by addition of GTP and CTP to final concentrations of 400 μM and 100 μM, respectively, together with 5 μCi [α-32P]CTP (1 Ci = 37 GBq) in each reaction.
ATPase Assays.
The release of Pi from [γ-32P]ATP was monitored as described (15, 24) by using essentially the same assay buffer as the one used in transcription assays (23). Phosphorylation of NtrC was achieved by incubating NtrC with 10 mM carbamoyl phosphate at 37°C for 10 min before ATP was added. A synthetic DNA oligonucleotide (69 bp) carrying the strong enhancer (24) was used to assess enhancer-stimulated ATPase activity. A 69-bp oligonucleotide lacking specific NtrC-binding sites was used as a control to demonstrate the absence of contaminating DNA-dependent ATPase activity.
DNA-Binding Assays.
Gel mobility-shift assays were performed as described (21), using an amplified PCR fragment (106 bp) from pJES464, which carries the natural glnA enhancer (25).
Gel Filtration.
Protein samples (100 μl) were sieved on a Bio-Silect Sec 400-5 column (300 × 7.8 mm; Bio-Rad) with a Bio-Silect 400 guard column (50 × 7.8 mm; Bio-Rad). The column equilibration and protein dilution buffer contained 10 mM Tris⋅HCl (pH 7.4), 50 mM KCl, 0.1 mM EDTA, 8 mM MgCl2 and 5% glycerol, with or without 10 mM carbamoyl phosphate. Phosphorylation of NtrC was achieved by incubating NtrC with 10 mM carbamoyl phosphate at room temperature for 10 min before sample injection. The column was run at 0.8 ml/min on a Gold Nouveau HPLC system (Beckman). Elution of proteins was detected at 214 nm, which gives a better signal/noise ratio than detection at 280 nm, with a bandwidth of 4 nm.
Results
Some Switch I Mutant Forms of NtrC Retain Residual Ability to Activate Transcription.
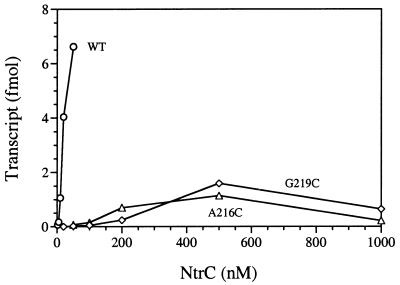
On a template carrying a strong enhancer upstream of the glnA promoter, phosphorylated mutant proteins (MBP-)NtrCA216C and NtrCG219C could activate transcription only at very high concentrations (Fig. 1). Whereas phosphorylated NtrCWT activates well at 20 nM, NtrCA216C and NtrCG219C yielded only trace amounts of transcripts at a concentration 10-fold higher (200 nM) and showed very poor template utilization even at best (at 500 nM). As discussed below, we considered two obvious explanations for why NtrCA216C and NtrCG219C could activate transcription only at very high concentrations: first, they had a defect in DNA binding, which is known to cause such a requirement (25), and second, they had a defect in oligomerization or enhancer-stimulated oligomerization and, hence, in ATPase activity.
Figure 1.
Transcriptional activation at the glnA promoter by Switch I mutant forms of NtrC. Single-cycle transcription assays were performed as described in Materials and Methods. The template was supercoiled plasmid pJES534 (1 nM), which carries a strong enhancer ≈460 bp upstream of the glnA promoter (21). Relevant NtrC proteins are indicated next to the curves: WT, MBP-NtrCWT; A216C, MBP-NtrCA216C; G219C, MBP-NtrCG219C.
NtrCA216C and NtrCG219C Bind the Enhancer Normally and Have Sufficient ATPase Activity to Activate Transcription at Low Concentrations.
Studies of the NtrCG219K protein, which carries an extra positive charge in the Switch I region, provide evidence that this region, although far removed from the helix–turn–helix DNA-binding motif of NtrC in the linear sequence, is nonetheless near the DNA. NtrCG219K bound better to the glnA enhancer than NtrCWT in a gel mobility-shift assay (16). Whereas NtrCWT gives only a single shifted species, which requires cooperative binding of two NtrC dimers to the two sites that constitute the enhancer, NtrCG219K yielded two shifted species. Unlike NtrCWT, NtrCG219K also could bind separately to the stronger of the two sites when they were separated. Subsequent studies of DNA-binding by using affinity coelectrophoresis indicated that the affinity of NtrCG219K for nonspecific DNA was ≈10-fold higher than that of NtrCWT (25) (A. K. North and S.K., unpublished data), which probably accounts for the improved binding of NtrCG219K to the glnA enhancer. By contrast to the case for NtrCG219K, binding of NtrCA216C and NtrCG219C to the glnA enhancer appeared to be normal. Like NtrCWT, both proteins gave a single shifted species in a gel mobility-shift assay and they did so at concentrations similar to those for NtrCWT (data not shown).
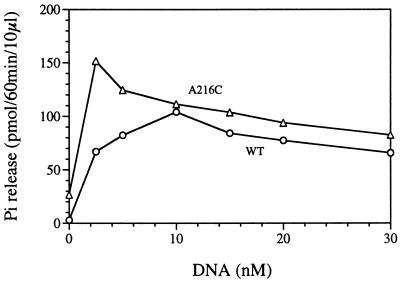
Because the oligomerization determinants of NtrC have not been localized within the central domain, it was possible that the NtrCA216C and NtrCG219C proteins failed to oligomerize normally or to show normal enhancer stimulation of oligomerization. To test this functionally, we measured the ATPase activities of these proteins in the absence and presence of a strong enhancer (Fig. 2 and Table 1). The activity of both phosphorylated NtrCA216 and NtrCWT (20 nM) was low in the absence of the enhancer (y intercepts), but the activity of NtrCA216C was higher than that of NtrCWT. For both proteins, the enhancer functioned as a positive allosteric effector. The ATPase activity of NtrCA216C was greater than that of NtrCWT at low enhancer concentrations (<10 nM DNA) but still as good as that of NtrCWT at high enhancer concentrations. A similar result was obtained for NtrCG219C (100 nM; Table 1). Thus, phosphorylation and oligomerization of these proteins appear to be intact and their ATPase activities are sufficient for transcriptional activation at low protein concentrations.
Figure 2.
Stimulation of the ATPase activity of NtrCA216C by an enhancer. ATPase activity of phosphorylated NtrCA216C (20 nM) was measured in the absence or presence of a synthetic DNA oligonucleotide that carries a strong enhancer (16, 24). It was compared with the ATPase activity of phosphorylated NtrCWT (20 nM). Pi release was measured at 60 min. Relevant NtrC proteins are indicated next to the curves: WT, MBP-NtrCWT; A216C, MBP-NtrCA216C.
Table 1.
ATPase activities of mutant NtrC proteins*
| Protein | Without enhancer DNA | With enhancer DNA† | With nonspecific DNA† |
|---|---|---|---|
| NtrCWT | 17 | 615 | 18 |
| NtrCA216C | 320 | 505 | 290 |
| NtrCG219C | 280 | 450 | 260 |
ATPase activity (pmol/60 min per 10 μl) was measured at 100 nM NtrC protein at 37°C. Proteins were phosphorylated as described in Materials and Methods.
†See Materials and Methods.
Increased ATPase Activities of NtrCA216C and NtrCG219C Appear to Be Due to Increased Oligomerization.
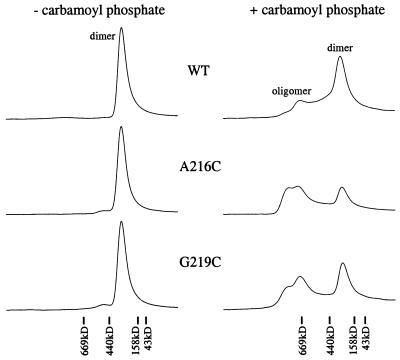
To determine whether the higher ATPase activities of NtrCA216C and NtrCG219C might be accounted for by an increased tendency to oligomerize, we examined the behavior of the phosphorylated proteins (1 μM) on an analytical gel-filtration column (Fig. 3). Whereas all proteins eluted as dimers (188 kDa for MBP-NtrC) when unphosphorylated, both mutant proteins yielded more oligomer than NtrCWT when phosphorylated with carbamoyl phosphate (10 mM) (>50% oligomer for mutant proteins and <20% for NtrCWT). Although oligomeric species eluted with standards of 600–800 kDa, it is not clear whether they are hexamers, octamers, or a mixture of the two. Functional studies are most easily rationalized if the active oligomer for NtrCWT is an octamer (1).
Figure 3.
Gel-filtration chromatography of unphosphorylated (− carbamoyl phosphate, Left) or phosphorylated (+ carbamoyl phosphate, Right) MBP-NtrCWT (WT), MBP-NtrCA216C (A216C), and MBP-NtrCG219C (G219C). The column was calibrated with a set of molecular mass standards: thyroglobulin, 669 kDa; ferritin, 440 kDa; aldolase, 158 kDa; ovalbumin, 43 kDa; whose positions of elution are shown as vertical bars. Proteins (100 μl at a concentration of 1 μM) were injected into the sieving column, and eluted proteins were detected by relative absorbance at 214 nm. Traces show elution profiles from 6 to 16 min after injection. Peak positions of dimer (≈200 kDa) and oligomer (≈600–800 kDa) of MBP-NtrC are marked.
The Defect in Transcriptional Activation by NtrCA216C Is Caused Solely by Its DNA Binding.
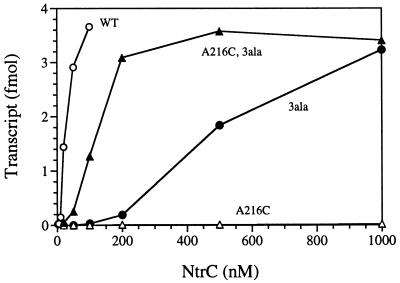
Because the enhancer-stimulated ATPase activity of NtrCA216C was intact, we suspected that the enhancer might prevent its interaction with σ54-holoenzyme and that its residual ability to activate transcription only at high protein concentrations might be due to the activity of a small number of molecules in solution rather than those tethered to the template. To assess inhibitory effects of binding to the enhancer, we tested a derivative of NtrCA216C that was incapable of DNA binding for its ability to activate transcription. Stable, soluble derivatives with this property carry three alanine substitutions in their helix–turn–helix DNA-binding motif and must activate transcription from solution (25). Without the stimulatory effects of the enhancer on nucleating oligomer formation and raising the local concentration of NtrC, the NtrC3ala protein activates transcription only at much higher concentrations (200 nM) than the NtrCWT protein from which it was derived (detectable activation at 5 nM) (Fig. 4). But at a high enough concentration (1000 nM), NtrC3ala yields as high a level of template utilization as NtrCWT. Surprisingly, NtrCA216C, 3ala activated transcription at lower concentrations (50 nM) than NtrC3ala itself and reached the same maximum template utilization as NtrC3ala at a concentration of only 200 nM. Better transcriptional activation by NtrCA216C, 3ala than NtrC3ala at low concentrations is commensurate with the increased ability of NtrCA216C to oligomerize and with its increased ATPase activity in solution (Figs. 2 and 3). The control protein NtrCA216C yielded barely detectable amounts of transcripts in this experiment even at high protein concentrations (Fig. 4). Lack of reproducibility of transcriptional activation by this DNA-binding form of the protein may be accounted for by the fact that it, too, must activate transcription from solution and can do so only when binding to the template is saturated.
Figure 4.
Transcriptional activation at the glnA promoter by DNA-binding or non-DNA-binding forms (3ala) of NtrC. Single-cycle transcription assays were performed as described in Materials and Methods. The template was supercoiled plasmid pJES534 (1 nM). Non-DNA-binding forms of NtrC carry three alanine substitutions in the helix–turn–helix motif and decrease binding affinity for the strong enhancer by >5,000-fold (25). Relevant NtrC proteins are indicated next to the curves: WT, MBP-NtrCWT; A216C, MBP-NtrCA216C; 3ala, MBP-NtrC3ala; A216C, 3ala, MBP-NtrCA216C, 3ala.
Like NtrCA216C, 3ala, NtrCG219C, 3ala activated transcription better than its DNA-binding form, NtrCG219C, but, unlike the case for NtrCA216C, 3ala, NtrCG219C, 3ala activated far less well than NtrC3ala itself (data not shown). Thus, the G219C substitution alone appears to cause a defect in the interaction with σ54-holoenzyme and the enhancer further inhibits this interaction.
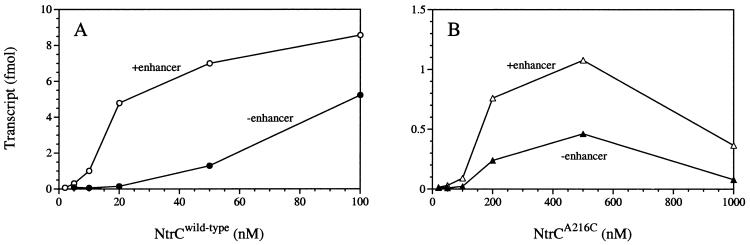
Inhibitory Effects of DNA Binding on Transcriptional Activation by NtrCA216C Are Not Enhancer-Specific.
To determine whether inhibitory effects of DNA binding were enhancer-specific, we compared transcriptional activation by NtrCA216C on a supercoiled template that lacked an enhancer with that on one which carried an enhancer. For the NtrCWT control, a higher protein concentration (50 nM) was required to reach the threshold for transcriptional activation on a template without the enhancer than on one with the enhancer (Fig. 5A). It should be noted that the concentration to reach this threshold (50 nM) was nonetheless lower than that for NtrC3ala (200 nM) (Fig. 4), presumably because of residual tethering effects of DNA binding in the former case (25). By contrast, there was no difference between the concentration of NtrCA216C required to reach the threshold for transcriptional activation on templates with or without the enhancer (Fig. 5B; note differences in scale from Fig. 5A). Moreover, NtrCA216C gave many fewer transcripts on either template than did its non-DNA-binding derivative, NtrCA216C, 3ala, which must activate from solution (Fig. 4). This indicates that both “nonspecific” DNA binding and enhancer binding inhibit transcriptional activation by NtrCA216C.
Figure 5.
Transcriptional activation by MBP-NtrCWT (A) and MBP-NtrCA216C (B) on supercoiled templates carrying or lacking an enhancer. Single-cycle transcription assays were performed as described in Materials and Methods on supercoiled plasmid pJES534 (1 nM), which carries two strong NtrC-binding sites (+enhancer), or pJES535 (1 nM), which lacks specific NtrC-binding sites (−enhancer).
Discussion
Roles of the Switch I Region for Activators of σ54-Holoenzyme.
The C-terminal portion of the Switch I region (215GAFTGA220 for NtrC) is one of the most highly conserved regions among activators of σ54-holoenzyme, and mutations affecting this small region arise very frequently upon screening for loss of transcriptional activation in vivo (16, 17, 26, 27). Phosphorylated forms of NtrC mutant proteins NtrCA216V, NtrCG219K, and NtrCA220T fail in transcriptional activation despite the fact that they show little if any loss of ATPase activity, and, hence, it was inferred that they fail in productive interaction with σ54-holoenzyme (16). Because the alanine and glycine residues that are altered in the mutant proteins are unlikely to be involved in specific contacts with the polymerase, it is probable that this portion of Switch I is important for energy coupling—an event triggering the conformational change in the polymerase that is essential for open complex formation. Similarly, mutant NtrC proteins with single cysteine substitutions for A216 and G219 have only a small residual capacity to activate transcription despite the fact that they have normal ATPase and enhancer-stimulated ATPase activities. In the case of NtrCA216C, defects in transcriptional activation are completely relieved in a derivative, NtrCA216C, 3ala, that is incapable of DNA binding. Hence, DNA binding appears to inhibit profoundly the interaction of NtrCA216C with σ54-holoenzyme, an interaction that is otherwise intact. The non-DNA-binding derivative of NtrCG219C (NtrCG219C, 3ala) had less activity than NtrCA216C, 3ala, and both NtrCA216V, 3ala (1, 5) and NtrCG219K, 3ala (A. K. North and S.K., unpublished data) lacked detectable activity. Unfortunately, direct assays for conformational changes in NtrC upon ATP hydrolysis and contact with polymerase have yet to be developed. Although the homologous activator DctD can be cross-linked to the polymerase (28), this was not the case for NtrC (29). In NifA (nitrogen fixation protein A), another activator of σ54-holoenzyme, the phenylalanine and threonine residues corresponding to F217 and T218 of NtrC, respectively, each were replaced by nine other amino acid residues (27). Only tyrosine and serine substitutions, respectively, yielded partially active forms of NifA (assessed in vivo). The strict requirement for an aromatic residue followed by a residue bearing a hydroxyl group at these two positions implicates these residues as having specific functional roles, again presumably in some aspect of the interaction with σ54-holoenzyme (14, 27).
Analogies and Differences Between the Switch I Regions of NtrC and p21ras.
The Switch I region of the G protein Ras serves as a contact interface with its target or effector proteins, such as the Raf kinase, which are responsible for biological output (30, 31). It is also involved in contact with G activating proteins (GAPS), such as p120 and the tumor suppressor protein neurofibromin, which stimulate GTP hydrolysis by Ras and appear to be regulatory (32). The target or effector protein for NtrC is σ54-holoenzyme. As discussed above, the Switch I region of NtrC may be involved in contact with σ54-holoenzyme because this region plays an essential role in transcriptional activation but not ATP hydrolysis. Interestingly, phosphorylation and the enhancer, though not proteins, appear to be analogous to GAPS. Phosphorylation is essential for detectable ATP hydrolysis, and binding to the enhancer greatly stimulates nucleotide hydrolysis by phosphorylated NtrC (4, 15, 33). The properties of NtrCA216C and its non-DNA-binding derivative NtrCA216C, 3ala indicate that substitutions in the Switch I region of NtrC can perturb the response to the enhancer and, hence, that this region is involved in mediating effects of the enhancer. Interestingly, the properties of NtrCG219K indicate directly that Switch I is near the DNA and, hence, presumably near the primary DNA-binding region of the protein (16). Some amino acid substitutions in the Switch I region of NtrC perturb the response of the protein to phosphorylation (unpublished data) and, hence, indicate that the Switch I region is also involved in mediating effects of phosphorylation. Although the Switch I region of NtrC appears to resemble that of Ras in mediating contact with its target and the response to “GAPS,” there are also differences in the behavior of the two proteins. Whereas Ras has high affinity for its targets in the nucleoside triphosphate-bound form (34), neither the triphosphate-bound form nor the diphosphate-bound form of NtrC appears to have high affinity for σ54-holoenzyme, and contact between the two proteins is transient (1, 29). GAPS decrease biological output by Ras because the GTP-bound form of Ras has much higher affinity for its target proteins than the GDP-bound form (34, 35). By contrast, the GAP analogs for NtrC increase biological output because ATP hydrolysis per se is required to change the conformation of σ54-holoenzyme. In the sense that energy coupling is required for the biological function of NtrC, analogies between NtrC and other members of the purine nucleotide-binding protein family such as the F1-ATPase and myosin are better than analogies to Ras. Despite intensive study, however, the roles of the Switch I regions of myosin and the F1-ATPase are less well defined than those of G proteins (36–38).
DNA Context Effects on Eukaryotic Enhancer-Binding Proteins.
DNA-binding context determines whether the glucocorticoid receptor (GR) and other intracellular receptors (IR) function as transcriptional activators or repressors (39). Base substitution mutations in one glucocorticoid-response element at which GR normally represses or is inert, plfG, enabled GR to activate transcription from this element (40). Analogously, change of one lysine residue (K461) in the zinc-binding region of GR enabled it to activate from wild-type plfG. We think it unlikely that specific alteration of the strong enhancer for NtrC would relieve inhibitory effects of the enhancer on the function of NtrCA216C because binding to “nonspecific” DNA also inhibits transcriptional activation by this protein (Fig. 5). However, second-site amino acid substitutions in NtrCA216C that relieve the inhibitory effects of the enhancer without eliminating DNA binding should be revealing with respect to a possible mechanism of communication between the C-terminal DNA-binding domain of NtrC and its Switch I region.
Acknowledgments
We thank Anne North for studies of DNA binding by NtrCG219K and Timothy Hoover, Michael Levine, and Boris Magasanik for valuable criticisms of the manuscript. This work was supported by National Institutes of Health Grant GM38361 to S.K.
Abbreviations
- GAPS
G activating proteins
- MBP
maltose-binding protein
- NtrC
nitrogen regulatory protein C
- NtrCWT
wild-type NtrC
References
- 1.Rombel I, North A, Hwang I, Wyman C, Kustu S. Cold Spring Harbor Symp Quant Biol. 1998;63:157–166. doi: 10.1101/sqb.1998.63.157. [DOI] [PubMed] [Google Scholar]
- 2.Porter S C, North A K, Kustu S. In: Two-Component Signal Transduction. Hoch J A, Silhavy T J, editors. Washington, D. C.: ASM Press; 1995. pp. 147–158. [Google Scholar]
- 3.Magasanik B. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt F C, editor. Vol. 1. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 1344–1356. [Google Scholar]
- 4.Austin S, Dixon R. EMBO J. 1992;11:2219–2228. doi: 10.1002/j.1460-2075.1992.tb05281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wyman C, Rombel I, North A K, Bustamante C, Kustu S. Science. 1997;275:1658–1661. doi: 10.1126/science.275.5306.1658. [DOI] [PubMed] [Google Scholar]
- 6.Sasse-Dwight S, Gralla J D. Proc Natl Acad Sci USA. 1988;85:8934–8938. doi: 10.1073/pnas.85.23.8934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morett E, Buck M. J Mol Biol. 1989;210:65–77. doi: 10.1016/0022-2836(89)90291-x. [DOI] [PubMed] [Google Scholar]
- 8.Popham D L, Szeto D, Keener J, Kustu S. Science. 1989;243:629–635. doi: 10.1126/science.2563595. [DOI] [PubMed] [Google Scholar]
- 9.Wedel A, Kustu S. Genes Dev. 1995;9:2042–2052. doi: 10.1101/gad.9.16.2042. [DOI] [PubMed] [Google Scholar]
- 10.Cannon W, Gallegos M T, Casaz P, Buck M. Genes Dev. 1999;13:357–370. doi: 10.1101/gad.13.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drummond M, Whitty P, Wootton J. EMBO J. 1986;5:441–447. doi: 10.1002/j.1460-2075.1986.tb04230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.North A K, Klose K E, Stedman K M, Kustu S. J Bacteriol. 1993;175:4267–4273. doi: 10.1128/jb.175.14.4267-4273.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morett E, Segovia L. J Bacteriol. 1993;175:6067–6074. doi: 10.1128/jb.175.19.6067-6074.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Osuna J, Soberon X, Morett E. Protein Sci. 1997;6:543–555. doi: 10.1002/pro.5560060304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weiss D S, Batut J, Klose K E, Keener J, Kustu S. Cell. 1991;67:155–167. doi: 10.1016/0092-8674(91)90579-n. [DOI] [PubMed] [Google Scholar]
- 16.North A K, Weiss D S, Suzuki H, Flashner Y, Kustu S. J Mol Biol. 1996;260:317–331. doi: 10.1006/jmbi.1996.0403. [DOI] [PubMed] [Google Scholar]
- 17.Li J, Passaglia L, Rombel I, Yan D, Kustu S. J Bacteriol. 1999;181:5443–5454. doi: 10.1128/jb.181.17.5443-5454.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y K, Lee J H, Brewer J M, Hoover T R. Mol Microbiol. 1997;26:373–386. doi: 10.1046/j.1365-2958.1997.5851955.x. [DOI] [PubMed] [Google Scholar]
- 19.Hwang I, Thorgeirsson T, Lee J, Kustu S, Shin Y-K. Proc Natl Acad Sci USA. 1999;96:4880–4885. doi: 10.1073/pnas.96.9.4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klose K E, North A K, Stedman K M, Kustu S. J Mol Biol. 1994;241:233–245. doi: 10.1006/jmbi.1994.1492. [DOI] [PubMed] [Google Scholar]
- 21.Porter S C, North A K, Wedel A B, Kustu S. Genes Dev. 1993;7:2258–2273. doi: 10.1101/gad.7.11.2258. [DOI] [PubMed] [Google Scholar]
- 22.Popham D, Keener J, Kustu S. J Biol Chem. 1991;266:19510–19518. [PubMed] [Google Scholar]
- 23.Wedel A, Weiss D S, Popham D, Droge P, Kustu S. Science. 1990;248:486–490. doi: 10.1126/science.1970441. [DOI] [PubMed] [Google Scholar]
- 24.Flashner Y, Weiss D S, Keener J, Kustu S. J Mol Biol. 1995;249:700–713. doi: 10.1006/jmbi.1995.0330. [DOI] [PubMed] [Google Scholar]
- 25.North A K, Kustu S. J Mol Biol. 1997;267:17–36. doi: 10.1006/jmbi.1996.0838. [DOI] [PubMed] [Google Scholar]
- 26.Wang Y K, Hoover T R. J Bacteriol. 1997;179:5812–5819. doi: 10.1128/jb.179.18.5812-5819.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gonzalez V, Olvera L, Soberon X, Morett E. Mol Microbiol. 1998;28:55–67. doi: 10.1046/j.1365-2958.1998.00772.x. [DOI] [PubMed] [Google Scholar]
- 28.Lee J H, Hoover T R. Proc Natl Acad Sci USA. 1995;92:9702–9706. doi: 10.1073/pnas.92.21.9702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stedman K. Ph.D. thesis. Berkeley: Univ. of California; 1996. [Google Scholar]
- 30.Wittinghofer A, Nassar N. Trends Biochem Sci. 1996;21:488–491. doi: 10.1016/s0968-0004(96)10064-5. [DOI] [PubMed] [Google Scholar]
- 31.Katz M E, McCormick F. Curr Opin Genet Dev. 1997;7:75–79. doi: 10.1016/s0959-437x(97)80112-8. [DOI] [PubMed] [Google Scholar]
- 32.McCormick F. Curr Biol. 1998;8:R673–R674. doi: 10.1016/s0960-9822(98)70431-2. [DOI] [PubMed] [Google Scholar]
- 33.Ninfa A J, Magasanik B. Proc Natl Acad Sci USA. 1986;83:5909–5913. doi: 10.1073/pnas.83.16.5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wittinghofer A. Biol Chem. 1998;379:933–937. [PubMed] [Google Scholar]
- 35.Wittinghofer A, Scheffzek K, Ahmadian M R. FEBS Lett. 1997;410:63–67. doi: 10.1016/s0014-5793(97)00321-9. [DOI] [PubMed] [Google Scholar]
- 36.Gulick A M, Rayment I. BioEssays. 1997;19:561–569. doi: 10.1002/bies.950190707. [DOI] [PubMed] [Google Scholar]
- 37.Li X, Rhodes T E, Ikebe R, Kambara T, White H, Ikebe M. J Biol Chem. 1998;273:27404–27411. doi: 10.1074/jbc.273.42.27404. [DOI] [PubMed] [Google Scholar]
- 38.Leslie A G, Abrahams J P, Braig K, Lutter R, Menz R I, Orriss G L, van Raaij M J, Walker J E. Biochem Soc Trans. 1999;27:37–42. doi: 10.1042/bst0270037. [DOI] [PubMed] [Google Scholar]
- 39.Lefstin J A, Yamamoto K R. Nature (London) 1998;392:885–888. doi: 10.1038/31860. [DOI] [PubMed] [Google Scholar]
- 40.Starr D B, Matsui W, Thomas J R, Yamamoto K R. Genes Dev. 1996;10:1271–1283. doi: 10.1101/gad.10.10.1271. [DOI] [PubMed] [Google Scholar]