Abstract
Alveolar rhabdomyosarcoma is an aggressive pediatric cancer of striated muscle characterized in 60% of cases by a t(2;13)(q35;q14). This results in the fusion of PAX3, a developmental transcription factor required for limb myogenesis, with FKHR, a member of the forkhead family of transcription factors. The resultant PAX3-FKHR gene possesses transforming properties; however, the effects of this chimeric oncogene on gene expression are largely unknown. To investigate the actions of these transcription factors, both Pax3 and PAX3-FKHR were introduced into NIH 3T3 cells, and the resultant gene expression changes were analyzed with a murine cDNA microarray containing 2,225 elements. We found that PAX3-FKHR but not PAX3 activated a myogenic transcription program including the induction of transcription factors MyoD, Myogenin, Six1, and Slug as well as a battery of genes involved in several aspects of muscle function. Notable among this group were the growth factor gene Igf2 and its binding protein Igfbp5. Relevance of this model was suggested by verification that three of these genes (IGFBP5, HSIX1, and Slug) were also expressed in alveolar rhabdomyosarcoma cell lines. This study utilizes cDNA microarrays to elucidate the pattern of gene expression induced by an oncogenic transcription factor and demonstrates the profound myogenic properties of PAX3-FKHR in NIH 3T3 cells.
Tumor-specific chromosome translocations that encode chimeric transcription factors are thought to exert their oncogenic effects through the dysregulation of gene expression (1). New technologies for large-scale expression analysis, such as cDNA microarrays, provide the opportunity to observe the broad effects of oncogenic transcription factors on gene expression and potentially elucidate their role in oncogenesis. The PAX3-FKHR chimera found in alveolar rhabdomyosarcoma (ARMS) fuses the DNA binding domain of PAX3 with the trans-activation domain of FKHR (2, 3), thereby retaining the DNA binding specificity of PAX3 and potentially acting in ARMS to increase expression of genes containing PAX3 binding sites. Although PAX3-FKHR binds less avidly to its targets than PAX3, it is a much stronger transactivator than PAX3 (4, 5). In murine embryogenesis, PAX3 expression is temporally regulated to induce both myogenesis and neural crest development. PAX3 is necessary for limb muscle development as homozygous Splotch mice, which have PAX3 mutations, do not develop limb muscles and fail to express myogenic basic helix-loop-helix factors in these regions (6). PAX3 expression is found in the muscle precursor cells of the lateral dermomyotome; the cells then migrate to the limb buds, where PAX3 expression is later repressed upon the formation of the limb musculature. In ARMS, the chimeric PAX3-FKHR gene is often highly expressed (2, 7–9), and continuous expression of this gene may inhibit terminal differentiation and maintain the cells in a primitive myoblastic state that retains abnormal proliferative potential. In accord with this model, Epstein et al. (10) have demonstrated that expression of PAX3 or PAX3-FKHR can prevent terminal myogenic differentiation in C2C12 myoblasts. Additionally, PAX3-FKHR is able to transform chicken fibroblasts and NIH 3T3 cells (11, 12). Presumably, all of these effects result from alterations in the expression of PAX3 target genes by PAX3-FKHR.
To observe the effects of either Pax3 or PAX3-FKHR on gene expression, we introduced these genes into NIH 3T3 cells by retroviral transduction and monitored their effect on global gene expression by using a murine cDNA microarrray. Introduction of PAX3-FKHR resulted in increased expression of several genes including the transcription factors Myogenin, MyoD, and Six1, as well as numerous other muscle-specific genes. In sharp contrast, PAX3 failed to induce these genes but repressed several genes, some of which were also down-regulated by PAX3-FKHR. The relevance of these changes to ARMS was confirmed by Northern blot analysis of ARMS cell lines. Our observations are consistent with a model in which occurrence of the t(2;13)(q35;q14) in a tumor progenitor cell leads to a persistent myoblastic state followed by secondary genetic events leading eventually to ARMS.
Materials and Methods
Retroviral Transduction and Cell Cultures.
NIH 3T3 cells were grown in 90% DMEM and 10% FBS. The PAX3-FKHR and Pax3 cDNAs (the kind gift of Lee Helman, National Cancer Institute) were subcloned between the HpaI and ClaI sites of the retroviral vector pLNCX (13) and were sequenced to verify encoding of the correct proteins. Virus was generated from these plasmids and the empty vector by transient transfection of BING cells and was used to infect NIH 3T3 cells as per published protocols (14). The cells were incubated with viral supernatant for 6 hours and were split 48 hours later in medium containing LD100 levels of G418 (600 μg/ml). After 2 weeks of selection in this medium, polyclonal populations of G418 resistant cells (designated PAX3-C, containing Pax3 gene; PF-C, containing PAX3-FKHR; and NIL-C, containing empty vector) were derived. In addition, individual clones were derived by splitting cells at >1:100 dilution and picking individual colonies for expansion (PAX3–1, PF-1, and PF-3). Human ARMS cell lines (RH3, RH4, RH5, RH28, and RH30) containing the PAX3-FKHR gene and a sarcoma (A204) were cultured as described (15). All cell lines were harvested at 80–85% confluency.
Northern Blot Analysis.
RNA was extracted by using TRIzol (Life Technologies, Rockville, MD) followed by single round mRNA extraction (Amersham Pharmacia) for the human cell lines and RNeasy (Qiagen, Chatsworth, CA) for the NIH 3T3 cells. mRNA (6 μg) for human and15 μg of total RNA for murine lines were size fractionated, were transferred to nylon membranes, and were hybridized according to standard protocols. The 32P labeled probes were generated from DNA either by PCR (5′ Pax3, Myogenin, MyoD, Igf2, HSIX1, IGF2, Pdgfra), or restriction digest of insert (IGFBP5, Slug, MERTK) from sequence verified IMAGE consortium clones (Research Genetics, Huntsville, AL), or reverse transcription–PCR from RNA from clone PF-1 (Six1, Igfbp5, Slug, c-Met). Human and murine GAPDH probes were purchased from CLONTECH.
Western Blot Analysis.
Detergent lysates containing 60 μg of total protein from the cell lines NIL-C, PF-C, PF-1, PF-3, PAX3-C, and PAX3–1 were separated by using SDS/PAGE, were transferred to nylon membranes (Immobilon-P, Millipore), and were probed with the appropriate antibodies using standard protocols. Rabbit anti-PAX3 polyclonal antisera was a generous gift from Frank Rauscher III (Wistar Institute). Rabbit anti-tubulin IgG was from ICN. Rabbit anti-Myogenin and MyoD were from Santa Cruz Biotechnology, and horseradish peroxidase-conjugated goat anti-rabbit H&L was from Jackson ImmunoResearch.
Reporter Assays.
The PAX3 reporter plasmid, plucTKCD-19 (kindly provided by Frank Rauscher III, Wistar Institute), contains six e5 sequences adjacent to a minimal thymidine kinase promoter region. Transfection efficiency was controlled by using the plasmid pCMV-RL. plucTKCD-19 (3 μg) was co-transfected with 100 ng of the pCMV-RL into 5 × 105 cells for the lines NIL-C, PF-C, and PAX3-C, using lipofectamine (Life Technologies) for 4 hours. The cells were harvested at 48 hours and were analyzed for Renilla and firefly luciferase activity by using a dual luciferase kit (Promega).
cDNA Microarrays.
The cDNA microarray arrays consisted of 2,225 elements that included 2,092 known mouse genes that were identified by sequence comparison of the murine EST database with 3,282 mouse UniGene entries and obtained from Research Genetics. The remainder of the printed genes were conventional cDNA clones representing mouse developmental genes added to expand the utility of these arrays for developmental studies. PCR products from these clones were prepared and printed onto glass slides according to previously described protocols (16, 17). RNA was extracted from cells with two rounds of purification with the RNeasy kit, followed by TRIzol purification. Fluorescently labeled cDNA with either Cy3 or Cy5 (Amersham Pharmacia) was synthesized from 100–200 μg of total RNA by oligo(dT)-primed polymerization using SuperScript II reverse transcriptase (Life Technologies) as described (15). For each hybridization we used the same reference RNA, from NIL-C, labeled with one of the two fluorochromes, Cy3 or Cy5, and the test line (NIH 3T3, PF-C, PF-1, PF-3, PAX3-C, PAX3–1) with the other.
Imaging and Image Analysis.
Fluorescence intensities at the immobilized targets were measured by using a custom-designed laser confocal microscope as reported (18) with intensity data integrated over 15-micron square pixels and recorded at 16 bits. Image analysis was performed by using dearray software as described (19). All of the data, including fluorescent intensities of both channels and calibrated intensity ratios, were entered into a FileMaker Pro database. Results are available on the internet (http://www.nhgri.nih.gov/DIR/LCG/15K/HTML/).
Results
Retroviral Transfer of Pax3 and PAX3-FKHR into NIH 3T3 Cells.
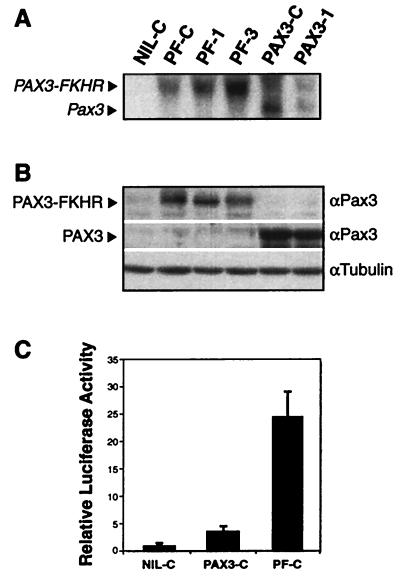
The expression levels of Pax3 and PAX3-FKHR in the pooled population and individual clones transduced with these genes were comparable as determined by both Northern blot and Western blot analysis (Fig. 1 A and B). The activity of the transduced genes was confirmed by transient transfection assays using a PAX3 responsive reporter. As expected from published reports (4), we observed that PAX3-FKHR was ≈6-fold more potent than PAX3 in this assay (Fig. 1C).
Figure 1.
(A) Northern blot analysis confirms expression of PAX3-FKHR in the lines PF-C, PF-1, and PF-3 and Pax3 in PAX3-C and PAX3–1. (B) Western blot analysis confirms comparable levels of these proteins in accord with the Northern blot data. (C) Relative luciferase activity of PAX3-FKHR (in PF-C) showing ≈6× the transcriptional activity of Pax3 (in PAX3-C), as described (4). The assay was performed according to published protocols (42) using the Pax3 reporter plasmid plucTKCD19.
Identification of Genes Responsive to Pax3 and PAX3-FKHR Transduction.
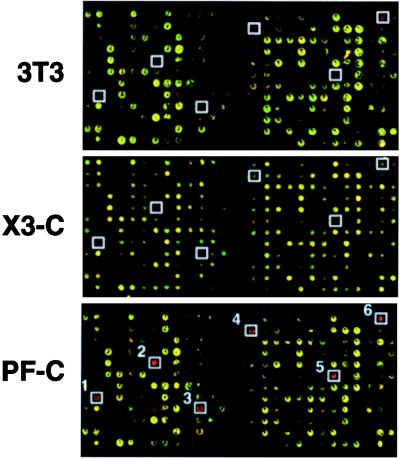
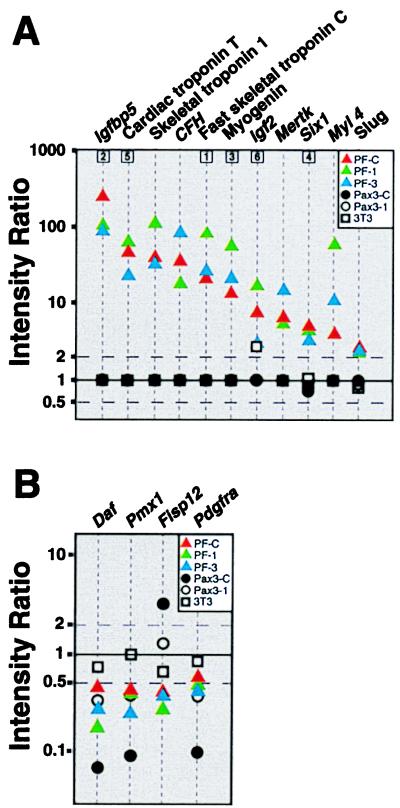
To determine the downstream effects of PAX3 and PAX3-FKHR, we performed cDNA micrroarray gene expression analysis on the pooled populations (PAX3-C and PF-C), their clonal derivatives (PAX3–1, PF-1 and PF-3), and the parental NIH 3T3 cells using 2,225 element murine cDNA microarrays. In each experiment, we used the empty vector transduced line (NIL-C) as the control. Fig. 2 illustrates corresponding portions of arrays in which NIL-C (labeled green) is compared with NIH 3T3 (red) and to the pooled populations PAX3-C and PF-C (both red). Expression levels of several genes were altered as a result of these manipulations in the experimental cells. To examine these effects, we entered the results into a database that was queried to identify the genes induced or repressed at the 99% confidence level as a result of PAX3-FKHR transduction in PF-C, PF-1, and PF-3. For this initial analysis, we used highly stringent filter, which included only genes with fluorescence signals ≥5,000 (on a scale of 0–65,535 fluorescent units) for at least one of the two channels. This process identified 11 genes that were induced in all three of PAX3-FKHR-transduced lines and 4 genes that were repressed. Fig. 3 summarizes the result of this inquiry. We confirmed that these changes were not attributable to the retrovirus or G418 treatment by comparing the gene expression of the parent NIH 3T3 cells to the NIL-C population. None of the 11 induced genes were induced to comparable levels by PAX3 in either the pooled population (PAX3-C) or in the clonal isolate (PAX3–1). Remarkably, of the 11 induced genes, 10 were muscle-related. These included Igfbp5 (>100-fold induction), Troponin T(49×), Troponin I (42×), Complement factor H (37×), Troponin C (22×), Myogenin (14×), Igf2 (8×), Six1 (5×), Myosin light chain 4 (4×), and Slug (3×). The final gene identified in this initial screen Mertk (6.8× induced) is an embryonic receptor tyrosine kinase protooncogene (20). Expression of several of these genes, including IGFBP5 and HSIX1, has not previously been reported in ARMS. At this level of stringency, there were also four genes repressed by PAX3-FKHR transduction (Fig. 3), including Pdgfra (0.6-fold), Fisp12 (0.4), Pmx1 (0.5), and Daf (0.5). Interestingly, Pax3 transduction also caused repression of three of these genes (Daf, Pmx1, and Pdgfra).
Figure 2.
Representative microarray hybridization of transduced lines. For each experiment, we used the same reference target prepared from cells transduced with empty vector (designated NIL-C). The pseudocolored images represent portions of a microarray with reference target, NIL-C, in green and parental NIH 3T3, PAX3-C, and PF-C in red. Up-regulation of several genes of interest by PAX3-FKHR are boxed (1, Fast skeletal troponin C; 2, Igfbp5; 3, Myogenin; 4, Six1; 5, Cardiac troponin T; 6, Igf2).
Figure 3.
(A) Eleven genes induced in PF-C, PF-1, and PF-3. The intensity ratio (log10 scale) is plotted against the 11 genes induced (at the 99% confidence level in each individual experiment) for all five microarray hybridizations. When the intensity values for both red and green channels for a probe/spot were <1,400, then the ratio of gene expression cannot be determined with precision, and the ratio value is plotted as 1. Boxed numbers correspond to genes from Fig. 2. (B) Four genes down-regulated by PAX3-FKHR, of which three (Daf, Pmx1, and Pdgfra) were also repressed by PAX3. A stringent filter, including only those genes with fluorescence ≥5,000 for either red or green channels, was applied for this analysis.
We then widened the search for genes induced by PAX3-FKHR transduction by using less stringent filters. We queried the database for all PAX3-FKHR-induced genes with ≥50% induction (ratio ≥1.5) in at least two of the three PAX3-FKHR-transduced cell lines by using an intensity filter of ≥1,400 for either channel. This identified 111 genes, of which a large proportion (28/111) were muscle related (Table 1). These 28 included genes involved in widely varied aspects of muscle function, including transcriptional regulation, signal transduction, contractility, energy metabolism, and synaptic transmission. Only one of these genes (Na+/H+-exchanger regulatory factor) was induced by Pax3 transduction in PAX3-C and PAX3–1. This observation suggests that PAX3-FKHR can function as a master regulator of muscle gene expression in NIH 3T3 cells.
Table 1.
Muscle-related genes induced by PAX3-FKHR transduction
| Gene symbol | PF-C | PF-1 | PF-3 | PAX3-C | PAX3-1 | |
|---|---|---|---|---|---|---|
| Transcription factors | ||||||
| Myogenin | Myog | 14 | 58.9 | 22.3 | NS | NS |
| Six1 | Six1 | 5.3 | 4.7 | 3.5 | 0.7 | 0.8 |
| Slug | Slug | 2.6 | 2.4 | 2.5 | 0.8 | 0.9 |
| Signal transduction/growth factors | ||||||
| Insulin-like growth factor binding protein 5 | Igfbp5 | >100 | >100 | 91.2 | NS | NS |
| Insulin-like growth factor 2 | Igf2 | 7.9 | 17.6 | 3.1 | 0.3 | NS |
| Transforming growth factor β2 | Tgfβ2 | 2.8 | 3.8 | 0.9 | NS | NS |
| Contractility/Cell structure | ||||||
| Cardiac troponin T | Tnnt2 | 49.1 | 65.9 | 23.9 | NS | NS |
| Fast skeletal muscle troponin 1 | Tnni2 | 41.8 | >100 | 34.3 | NS | NS |
| Fast skeletal troponin C | Tnnc2 | 22.4 | 81.0 | 27.7 | NS | NS |
| Skeletal muscle myosin light chain 1 isoform | Myl1 | 16.9 | 80.4 | 0.7 | NS | NS |
| Perinatal skeletal myosin heavy chain | Myh8 | 10.2 | 76.1 | 7.9 | NS | NS |
| Atrial/fetal myosin alkali light chain | Myl4 | 4.3 | 60.7 | 11.7 | NS | NS |
| Alpha cardiac actin | Actc | 2.9 | 7.0 | 2.1 | 1.9 | 1.3 |
| Beta-tubulin gene M-β-2 | Tubb2 | 1.8 | 2.1 | 2.5 | 1.3 | 0.7 |
| Myosin binding protein H | Mybph | 1.8 | 7.8 | 1.8 | NS | NS |
| Skeletal muscle α actin | Acta1 | 1.8 | 7.4 | 1.4 | NS | NS |
| Tropomyosin isoform 2 | Tmp2 | 1.6 | 4.2 | 0.8 | 0.8 | 1.2 |
| Energy metabolism | ||||||
| Creatinine kinase M chain | CkM | 5.2 | 27.6 | 8.5 | NS | NS |
| Acyl-CoA dehydrogenase short-chain | Acads | 1.5 | 1.5 | 1.5 | 0.6 | 0.9 |
| Fructose-biphosphate aldolase A | AldoA | 1.2 | 2.0 | 2.1 | 1.4 | 2.0 |
| Synaptic transmission/electrochemical function | ||||||
| Acetylcholine receptor, γ-chain | Achrg | 17.5 | 68.8 | NS | NS | NS |
| Acetylcholine receptor α-subunit | Achra | NS | 17.3 | 7.9 | NS | NS |
| Ryanodine receptor, type 1 | Ryr1 | 3.4 | 19.8 | NS | NS | NS |
| Na+/H+-exchanger regulatory factor | Nherf | 2.9 | 3.4 | 2.7 | 2.1 | 3.2 |
| Na+/K+-transporting ATPase β-chain | Atp1b1 | 2.1 | 1.8 | 3.4 | 0.8 | 0.6 |
| Other | ||||||
| Complement factor H | Hf1 | 37.0 | 18.9 | 87.3 | NS | NS |
| H19 | H19 | 18.2 | 36.5 | NS | NS | NS |
| Clusterin | Clu | 1.6 | 0.3 | 2.0 | NS | NS |
Listed are genes with ≥50% induction (ratio of ≥1.5) by PAX3-FKHR transduction in at least two of the PF-C, PF-1, and PF-3 clones. A filter was used to exclude all spots with the maximum intensity <1,400 (on a scale of 0–65,535 fluorescent units) for either fluorochrome (designated NS). Of the 111 genes retrieved by this enquiry, 28 were found to be muscle-related as determined by a search of Medline database. Shown here are the gene name, gene symbol, functional categories, and the induction ratio for each of the experiments.
Confirmation of Gene Induction.
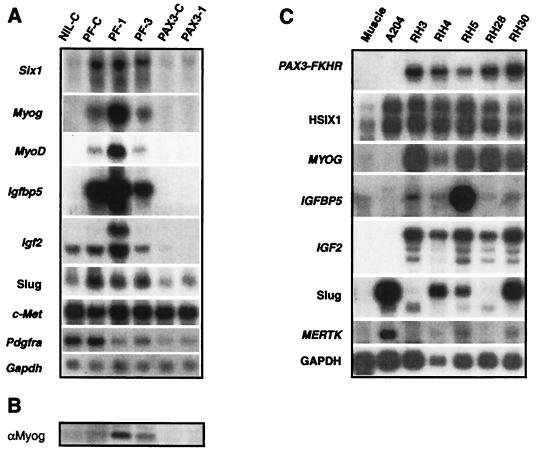
To verify the microarray results, we performed Northern blot hybridizations on RNA extracted from the transduced cells using probes for selected genes (Fig. 4A). Expression of Six1 in the NIL-C population (and NIH 3T3) (data not shown) was low, and induction was confirmed in the PF-C, PF-1, and PF-3 clones and not in the Pax3-transduced cells. Similarly, expression of the myogenic transcription factor Myogenin was undetectable in the control cells and was markedly induced after PAX3-FKHR introduction, particularly in PF-1. Because Myogenin is postulated to be downstream of MyoD and Myf5 (neither gene was represented on our cDNA microarray), we performed reverse transcription–PCR on PF-1 confirming the expression of MyoD but not Myf5 (data not shown). Northern blot analysis using the MyoD reverse transcription–PCR product as probe demonstrated (Fig. 4A) MyoD expression in all of the PAX3-FKHR but not in the control or Pax3 transduced clones. Myogenin expression was confirmed at the protein level by Western analysis (Fig. 4B), and expression of both Myogenin and MYOD also was observed by immunohistochemistry (data not shown). The expression and induction of Igfbp5 followed the same pattern, with highest expression found in PF-1. Interestingly PF-1 also had high levels of induction of Igf2, although Igf2 expression was detected at moderate levels in the NIL-C control cells. Similarly, Slug had a basal level of expression in the control cells but was further increased in the PAX3-FKHR transduced cells. c-Met has been implicated as a PAX3-FKHR target (21). In our system the basal level of c-Met expression was high in the control cells, and we did not observe induction of this gene by either PAX3-FKHR or PAX3. We also investigated three of the repressed genes by Northern blot hybridization, confirming repression of Pdgfra (Fig. 4A), Fisp12, and Pmx1 (data not shown).
Figure 4.
Northern blot analysis confirms array data. (A) Induction of Six1, Myogenin, MyoD, Igfbp5, Igf2, and Slug by PAX3-FKHR transduction is demonstrated, as is the failure to induce these genes by PAX3. Igf2 expression appears by Northern blotting to be suppressed by PAX3. Basal c-Met levels in NIH 3T3 cells transduced by the empty virus is high and remains high in the Pax3 and PAX3-FKHR transduced clones. Pdgfra expression is repressed in the clones PF-1, PF-3, PAX3-C, and PAX3–1. (B) Myogenin protein expression is confirmed in the PAX3-FKHR transduced clones PF-C, PF-1, and PF-3 by Western analysis. (C) Northern blot analysis reveals expression of HSIX1, MYOG, IGFBP5, IGF2, Slug, and MERTK in several of the ARMS cell lines (RH3, RH4, RH5, RH28, and RH30). The first lane is normal muscle. A204 is an embryonal rhabdomyosarcoma that lacks the PAX3-FKHR chimeric oncogene.
Expressions of Genes Predicted from the NIH 3T3 Model in ARMS Cell Lines.
To test whether these induced genes were also expressed in ARMS as predicted by our model system, we performed Northern blot analysis (Fig. 4C) on five ARMS cell lines (RH3, RH4, RH4, RH28, and RH 30) and compared them with normal muscle and a sarcoma (A204). Several of these genes (HSIX1, IGFBP5, Slug, and MERTK) have not previously been reported as expressed in ARMS. We confirmed the expression of the 7.3-kilobase PAX3-FKHR gene in all five ARMS and not in A204 or normal muscle. HSIX1 was expressed at high levels in all five ARMS and in A204 and, in accord with published reports (22), at low levels in normal muscle. As expected, expression of MYOG (23) and IGF2 (24) was high in all five ARMS but not A204. The expression of IGFBP5, not previously described in ARMS, evident in all five of the cell lines, was particularly high in RH5. In addition, Slug was expressed in all five ARMS and in A204. MERTK levels were relatively high in A204, and lower levels of expression were detected in RH3, RH4, and RH30.
Discussion
Induction of a Myogenic Transcription Program by PAX3-FKHR.
In this study, we have monitored the downstream effects of both PAX3 and PAX3-FKHR on NIH 3T3 cells with cDNA microarrays. We observed the remarkable ability of PAX3-FKHR to induce a myogenic transcription program, which includes induction of two important myogenic basic helix-loop-helix factors, Myogenin and MyoD, which are also expressed in ARMS (23). In addition, we observed induction of genes that represent several other key aspects of muscle function, including growth, energy metabolism, and contractility (Table 1). The observed induction of many of the genes associated with muscle differentiation is most likely a consequence of Myogenin and MyoD expression. Of interest, Myl4, a fetal muscle gene that is absent from adult muscle (25) was also identified in our previous study of ARMS using human cDNA microarrays in which we found MYL4 expression in all seven ARMS cell lines tested (15).
In addition to induction of genes previously associated with ARMS, we also observed induction by PAX3-FKHR of other potentially significant genes such as Six1, Slug, and Igfbp5, which has not previously related to the PAX3 pathway. Six1 is a homeobox gene, the murine ortholog of the Drosophila “sine oculis” (so) gene, which is expressed in the distal posterior and anterior limb regions of mouse embryos and is weakly expressed in skeletal muscles of the head and body during late development (22, 26). Of interest, Six1 has recently been shown to activate the Myogenin promoter (27). When we investigated ARMS cells for the presence of HSIX1, we found high expression in all five of the ARMS cell lines tested, as predicted by our results in the NIH 3T3 model. However, HSIX1 expression has also been noted in other human cancers, and, of interest, over-expression in MCF-7 cells has been shown to abrogate the G2 cell cycle check-point (28).
We also confirmed expression in ARMS of another gene, Slug, as predicted by the NIH 3T3 model. Slug is a zinc finger transcription factor that has been related to neural crest (29) and limb bud development, where it may be involved in mesenchymal differentiation (30, 31). A third gene not previously related to ARMS, IGFBP5, was highly induced in all PAX3-FKHR transduced lines. It belongs to a family of secreted proteins that bind IGF1 and 2, thereby modulating their biological actions. It is known to be expressed in embryogenesis during muscle development (32) and is rapidly induced during terminal muscle differentiation of C2l myoblasts coincident with the onset of Myogenin gene expression (33). Indeed, we found that PF-1, the clone that expressed the highest level of Myogenin, also showed high levels of Igfbp5. Additionally, IGFBP5 expression was observed in all five ARMS cell lines tested. Furthermore, Igf2 itself was induced in PAX3-FKHR transduced cells. This growth factor, which modulates muscle growth and differentiation, is thought to be an autocrine growth and motility factor in ARMS, and its induction links PAX3-FKHR directly to an important mechanism of dysregulated growth and impaired differentiation (24, 34). Of interest, expression of H19 (Table 1), which is closely linked to Igf2 in an imprinted region of mouse chromosome 7 (35), was also increased in the PAX3-FKHR-transduced cells.
Of the other possible PAX3 targets identified so far in the literature, including GFAP, NGFR, N-CAM, L1 (36), and c-Met (21, 37), only c-Met was on our array, and it was not induced by PAX3-FKHR. However, Northern blot analysis revealed an already high basal expression level in NIH 3T3 cells that did not increase in cells transduced with Pax3 or PAX3-FKHR (Fig. 4A), probably because of the high level of expression in the parental cells.
We noted that the genes that were strongly induced by PAX3-FKHR transduction were not induced or only weakly induced in Pax3 transduced cells, despite the similar DNA binding specificity of these proteins. Even with comparable levels of expression of PAX3 and PAX3-FKHR, only PAX3-FKHR was able to induce the expression of a myogenic program including MyoD and Myogenin. This is in accord with the stronger transcriptional activity of PAX3-FKHR relative to PAX3, despite its reduced affinity for PAX3 targets previously reported and confirmed in our system (4). The increased transcriptional activity is most likely essential to the oncogenic properties of PAX3-FKHR and may be related to both gain of FKHR sequences and unresponsiveness to the transcriptional inhibitors of PAX3 (38).
Repression was not as prominent as induction in our study, with only four genes identified as repressed. It is also more difficult to interpret repression because the ability to measure this effect depends on initial gene expression levels in NIH 3T3 cells. Interestingly, although gene induction by PAX3 was not observed in our study, Pax3-transduced cells did exhibit repression of three of the four repressed genes identified. Decay accelerating factor (Daf) shows precisely the same reciprocal relationship with another regulator of complement activity, Clusterin (Table 1), observed previously in myocytes (39). Another gene repressed in both Pax3 and PAX3-FKHR transduced cells, Pmx1, is a mesoderm-specific paired box type homeodomain protein related to Pax3, which is highly expressed in adult skeletal muscle (40). Repression of Pdgfra, which is normally expressed in mouse mesodermal and neural-crest derived tissue, contrasts with the study of Epstein et al., who demonstrated that PAX3-FKHR but not PAX3 can directly increase expression of PDGFRA (41). Their result was obtained in p19 embryonal carcinoma cells, which have an undetectable baseline level of PDGFRA expression. In contrast, NIH 3T3 cells have a high basal expression of Pdgfra and appear to respond to both PAX3 and PAX3-FKHR by repression of Pdgfra as confirmed by Northern blotting (Fig. 4A).
Although we have only sampled a portion of the murine genome in this study, the ability to assay parallel gene expression analysis provides a remarkable view of the myogenic gene expression program invoked by PAX3-FKHR in NIH 3T3 cells. In addition, our results correctly predicted the expression of HSIX1, IGFBP5, and Slug in ARMS. This study does not permit us to distinguish direct from indirect targets of PAX3-FKHR. Resolving this issue and the complex interrelationships among the induced genes will require both alternative gene transfer technologies and other approaches, which are complementary to large-scale gene expression analysis. Our results lead us to speculate on certain aspects of the pathogenesis of ARMS. First, it is critical that the as-yet unidentified tumor progenitor cell must be competent to respond to the effects of PAX3-FKHR. Second, considering the profound myogenic effects of PAX3-FKHR, this cell may have the properties of a primitive muscle precursor, which on transformation by PAX3-FKHR, exhibits the muscle markers (such as Myogenin and MYOD) characteristic of ARMS. Failure of these abnormal rhabdomyoblasts to terminally differentiate and exit the cell cycle would permit expansion of this population of cells. The subsequent acquisition of secondary genetic alterations would then lead to the development of fully malignant ARMS.
Acknowledgments
We thank Kimberly A. Gayton, Alicia J. Faller, and Robert L. Walker for their excellent technical assistance.
Abbreviation
- ARMS
alveolar rhabdomyosarcoma
References
- 1.Barr F G. Nat Genet. 1998;19:121–124. doi: 10.1038/475. [DOI] [PubMed] [Google Scholar]
- 2.Galili N, Davis R J, Fredericks W J, Mukhopadhyay S, Rauscher F J, III, Emanuel B S, Rovera G, Barr F G. Nat Genet. 1993;5:230–235. doi: 10.1038/ng1193-230. [DOI] [PubMed] [Google Scholar]
- 3.Shapiro D N, Sublett J E, Li B, Downing J R, Naeve C W. Cancer Res. 1993;53:5108–5112. [PubMed] [Google Scholar]
- 4.Fredericks W J, Galili N, Mukhopadhyay S, Rovera G, Bennicelli J, Barr F G, Rauscher F J., III Mol Cell Biol. 1995;15:1522–1535. doi: 10.1128/mcb.15.3.1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bennicelli J L, Fredericks W J, Wilson R B, Rauscher F J, III, Barr F G. Oncogene. 1995;11:119–130. [PubMed] [Google Scholar]
- 6.Bober E, Franz T, Arnold H H, Gruss P, Tremblay P. Development (Cambridge, UK) 1994;120:603–612. doi: 10.1242/dev.120.3.603. [DOI] [PubMed] [Google Scholar]
- 7.Bennicelli J L, Edwards R H, Barr F G. Proc Natl Acad Sci USA. 1996;93:5455–5459. doi: 10.1073/pnas.93.11.5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis R J, Barr F G. Proc Natl Acad Sci USA. 1997;94:8047–8051. doi: 10.1073/pnas.94.15.8047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barr F G, Nauta L E, Davis R J, Schafer B W, Nycum L M, Biegel J A. Hum Mol Genet. 1996;5:15–21. doi: 10.1093/hmg/5.1.15. [DOI] [PubMed] [Google Scholar]
- 10.Epstein J A, Lam P, Jepeal L, Maas R L, Shapiro D N. J Biol Chem. 1995;270:11719–11722. doi: 10.1074/jbc.270.20.11719. [DOI] [PubMed] [Google Scholar]
- 11.Scheidler S, Fredericks W J, Rauscher F J, III, Barr F G, Vogt P K. Proc Natl Acad Sci USA. 1996;93:9805–9809. doi: 10.1073/pnas.93.18.9805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lam P Y, Sublett J E, Hollenbach A D, Roussel M F. Mol Cell Biol. 1999;19:594–601. doi: 10.1128/mcb.19.1.594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller A D, Rosman G J. BioTechniques. 1989;7:980–2. , 984–6, 989–990. [PMC free article] [PubMed] [Google Scholar]
- 14.Pear W S, Nolan G P, Scott M L, Baltimore D. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khan J, Simon R, Bittner M, Chen Y, Leighton S B, Pohida T, Smith P D, Jiang Y, Gooden G C, Trent J M, Meltzer P S. Cancer Res. 1998;58:5009–5013. [PubMed] [Google Scholar]
- 16.DeRisi J, Penland L, Brown P O, Bittner M L, Meltzer P S, Ray M, Chen Y, Su Y A, Trent J M. Nat Genet. 1996;14:457–460. doi: 10.1038/ng1296-457. [DOI] [PubMed] [Google Scholar]
- 17.Shalon D, Smith S J, Brown P O. Genome Res. 1996;6:639–645. doi: 10.1101/gr.6.7.639. [DOI] [PubMed] [Google Scholar]
- 18.Khan J, Saal L H, Bittner M L, Chen Y, Trent J M, Meltzer P S. Electrophoresis. 1999;20:223–229. doi: 10.1002/(SICI)1522-2683(19990201)20:2<223::AID-ELPS223>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 19.Chen Y, Dougherty E R, Bittner M L. Biomed Optics. 1997;2:364–374. doi: 10.1117/12.281504. [DOI] [PubMed] [Google Scholar]
- 20.Graham D K, Bowman G W, Dawson T L, Stanford W L, Earp H S, Snodgrass H R. Oncogene. 1995;10:2349–2359. [PubMed] [Google Scholar]
- 21.Epstein J A, Shapiro D N, Cheng J, Lam P Y, Maas R L. Proc Natl Acad Sci USA. 1996;93:4213–4218. doi: 10.1073/pnas.93.9.4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boucher C A, Carey N, Edwards Y H, Siciliano M J, Johnson K J. Genomics. 1996;33:140–142. doi: 10.1006/geno.1996.0172. [DOI] [PubMed] [Google Scholar]
- 23.Clark J, Rocques P J, Braun T, Bober E, Arnold H H, Fisher C, Fletcher C, Brown K, Gusterson B A, Carter R L, et al. Br J Cancer. 1991;64:1039–1042. doi: 10.1038/bjc.1991.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.El-Badry O M, Minniti C, Kohn E C, Houghton P J, Daughaday W H, Helman L J. Cell Growth Differ. 1990;1:325–331. [PubMed] [Google Scholar]
- 25.Strohman R C, Micou-Eastwood J, Glass C A, Matsuda R. Science. 1983;221:955–957. doi: 10.1126/science.6879193. [DOI] [PubMed] [Google Scholar]
- 26.Oliver G, Wehr R, Jenkins N A, Copeland N G, Cheyette B N, Hartenstein V, Zipursky S L, Gruss P. Development (Cambridge, UK) 1995;121:693–705. doi: 10.1242/dev.121.3.693. [DOI] [PubMed] [Google Scholar]
- 27.Spitz F, Demignon J, Porteu A, Kahn A, Concordet J P, Daegelen D, Maire P. Proc Natl Acad Sci USA. 1998;95:14220–14225. doi: 10.1073/pnas.95.24.14220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ford H L, Kabingu E N, Bump E A, Mutter G L, Pardee A B. Proc Natl Acad Sci USA. 1998;95:12608–12613. doi: 10.1073/pnas.95.21.12608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nieto M A, Sargent M G, Wilkinson D G, Cooke J. Science. 1994;264:835–839. doi: 10.1126/science.7513443. [DOI] [PubMed] [Google Scholar]
- 30.Cohen M E, Yin M, Paznekas W A, Schertzer M, Wood S, Jabs E W. Genomics. 1998;51:468–471. doi: 10.1006/geno.1998.5367. [DOI] [PubMed] [Google Scholar]
- 31.Buxton P G, Kostakopoulou K, Brickell P, Thorogood P, Ferretti P. Int J Dev Biol. 1997;41:559–568. [PubMed] [Google Scholar]
- 32.Green B N, Jones S B, Streck R D, Wood T L, Rotwein P, Pintar J E. Endocrinology. 1994;134:954–962. doi: 10.1210/endo.134.2.7507840. [DOI] [PubMed] [Google Scholar]
- 33.Rotwein P, James P L, Kou K. Mol Endocrinol. 1995;9:913–923. doi: 10.1210/mend.9.7.7476973. [DOI] [PubMed] [Google Scholar]
- 34.Wang W, Kumar P, Epstein J, Helman L, Moore J V, Kumar S. Cancer Res. 1998;58:4426–4433. [PubMed] [Google Scholar]
- 35.Bartolomei M S, Zemel S, Tilghman S M. Nature (London) 1991;351:153–155. doi: 10.1038/351153a0. [DOI] [PubMed] [Google Scholar]
- 36.Kioussi C, Gross M K, Gruss P. Neuron. 1995;15:553–562. doi: 10.1016/0896-6273(95)90144-2. [DOI] [PubMed] [Google Scholar]
- 37.Ginsberg J P, Davis R J, Bennicelli J L, Nauta L E, Barr F G. Cancer Res. 1998;58:3542–3546. [PubMed] [Google Scholar]
- 38.Hollenbach A D, Sublett J E, McPherson C J, Grosveld G. EMBO J. 1999;18:3702–3711. doi: 10.1093/emboj/18.13.3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gasque P, Morgan B P, Legoedec J, Chan P, Fontaine M. J Immunol. 1996;156:3402–3411. [PubMed] [Google Scholar]
- 40.Cserjesi P, Lilly B, Bryson L, Wang Y, Sassoon D A, Olson E N. Development (Cambridge, UK) 1992;115:1087–1101. doi: 10.1242/dev.115.4.1087. [DOI] [PubMed] [Google Scholar]
- 41.Epstein J A, Song B, Lakkis M, Wang C. Mol Cell Biol. 1998;18:4118–4130. doi: 10.1128/mcb.18.7.4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schafer B W, Czerny T, Bernasconi M, Genini M, Busslinger M. Nucleic Acids Res. 1994;22:4574–4582. doi: 10.1093/nar/22.22.4574. [DOI] [PMC free article] [PubMed] [Google Scholar]