Abstract
The intensely studied MHC has become the paradigm for understanding the architectural evolution of vertebrate multigene families. The 4-Mb human MHC (also known as the HLA complex) encodes genes critically involved in the immune response, graft rejection, and disease susceptibility. Here we report the continuous 1,796,938-bp genomic sequence of the HLA class I region, linking genes between MICB and HLA-F. A total of 127 genes or potentially coding sequences were recognized within the analyzed sequence, establishing a high gene density of one per every 14.1 kb. The identification of 758 microsatellite provides tools for high-resolution mapping of HLA class I-associated disease genes. Most importantly, we establish that the repeated duplication and subsequent diversification of a minimal building block, MIC-HCGIX-3.8–1-P5-HCGIV-HLA class I-HCGII, engendered the present-day MHC. That the currently nonessential HLA-F and MICE genes have acted as progenitors to today’s immune-competent HLA-ABC and MICA/B genes provides experimental evidence for evolution by “birth and death,” which has general relevance to our understanding of the evolutionary forces driving vertebrate multigene families.
The chromosome 6p21.3-located human MHC is densely packed with genes functioning at key checkpoints in the adaptive immune system (1). Preeminent among these are the antigen-presenting HLA class I and II molecules, which initiate the cell-mediated immune response by displaying antigenic oligopeptides to the αβ T cell receptor (2). This interaction is central for restraining microbiological invasions, controlling malignant cell proliferation, and governing transplant success. The 4-Mb HLA segment is divided into three regions (from centromere to telomere): class II (1 Mb), class III (1 Mb), and class I (2 Mb) (1). They, by several criteria, occupy a unique position within the human genome, most notably an unusually high gene density of more than 180 genes per 4 Mb, the highest degree of genetic polymorphism ever encountered within the genome (with close to 900 alleles at the eight classical class I and II loci; see http://www.anthonynolan.com/HIG/nomenc.html for regular updates), and allelic and haplotypic association to more than 100 diseases (3).
The telomeric class I region spans 2 Mb from MICB to HLA-F and is known to contain six expressed HLA class I genes: the three classical (HLA-A, HLA-B, and HLA-C), the three nonclassical (HLA-E, HLA-F, and HLA-G) (1); and the two class I chain-related (MICA and MICB) (4–6) genes. This region also encompasses 12 HLA class I pseudogenes, truncated, or fragmental genes, (HLA-X, -17, -30, -L/92, -J/59, -80, -21, -K/70, -16, -H/54, -90, and -75) (7) and three class I chain-related pseudogenes (MICC, MICD, and MICE) (4).
Here we present the complete 1,796,938-bp HLA class I sequence solved by shotgun strategy. This continuum links the centromeric MICB gene to the telomeric HLA-F gene, allows a direct and in-depth analysis of the region with respect to overall structure, gene content, and microsatellite density, and permits us to grasp the complex development of this plastic segment of the vertebrate genome at the molecular level.
Materials and Methods
Yeast Artificial Chromosome (YAC), Bacterial Artificial Chromosome (BAC), P1-derived artificial chromosome (PAC), and Cosmid Clones.
Large-insert bacterial clones were identified by PCR-based screening of a human BAC library (Research Genetics, Huntsville, AL) constructed from the B cell line, 978SK (Fig. 3A, which is provided on the PNAS web site, www.pnas.org.), and two PAC libraries derived from human lymphocyte DNA (Genome Systems, St. Louis) (Fig. 3A), and human male lymphocyte DNA (supplied by Pieter J. de Jong, Roswell Park Cancer Institute, Buffalo, NY) (Fig. 3A) (8). PCR screening and physical mapping followed the protocol provided by Research Genetics and Osoegawa et al. (8). Two YAC clones, 745D12 with a 590-kb insert and 960H11 with a 1,600-kb insert (Fig. 3A) (6, 9), were obtained from the Centre d’Étude du Polymorphisme Humain (Paris) YAC library constructed from the HLA-homozygous B cell line BOLETH (HLA-A2, -B62, -Cw10, -DR4, -DQ8, and -DR53) (10), and one YAC clone (Y109) with a 240-kb insert (Fig. 3A) (5) was obtained from a YAC library constructed from the B cell line CGM1 harboring the following HLA haplotypes (HLA-A3, -B8, -Cw-, -DR3, -DQ2, -DR52, -A29, -B14, -Cw-, -DR7, -DQ2, and -DR53) (11). Construction, handling, screening, and mapping of cosmid libraries derived from YAC clones 745D12 and 960H11 in the Super Cos1 cosmid vector (Stratagene) and from YAC Y109 in the pWE15 cosmid vector (Stratagene) have been described (5, 6, 9). Chromosomal mapping and chimerism analysis of these BAC, PAC, and cosmid clones were analyzed by fluorescent in situ hybridization as described (9).
Sequencing Strategy, Assembly, and Analyses.
Three BACs, eight PACs, and 24 cosmid clones covering the 1.8-Mb segment from the MICB to HLA-F genes were subjected to nucleotide sequence determination by the shotgun strategy, as originally reported (12). Assembly and database analyses were performed following previously established procedures (6).
Results
Contig Construction and Sequencing.
To establish the genomic sequence of the entire HLA class I region between the MICB and HLA-F genes, a sequence-ready contig was first constructed by using one BAC-, two PAC-, and three YAC-derived cosmid libraries (5, 6, 9). By screening these libraries with human Alu-repeat probes, and gene-specific and sequence-tagged site-specific probes or PCR primers, seven BACs, 39 PACs, and 199 cosmid clones were isolated and assembled into a single contig after Southern hybridizations with clone-derived PCR products and EcoRI fragments (ref. 9 and unpublished work). In this manner, 245 densely overlapping clones spanning the HLA class I region between MICB and HLA-F were identified. Of these, three BACs, eight PACs, and 24 cosmids were selected for sequencing (Fig. 3A). Fluorescent in situ hybridization confirmed that all clone inserts were derived from chromosome 6, band p21.3 (data not shown). Furthermore, the physical map obtained by this BAC, PAC, and cosmid contig was consistent with that previously constructed on the basis of pulsed field gel electrophoresis analysis using independently isolated YAC clones (13). Altogether, these results suggested that these BAC, PAC, and cosmid clones used for sequencing were devoid of gross deletions, rearrangements, or chimerisms.
After shotgun sequencing, the total length of the contig linking MICB to HLA-F was established to be 1,796,938 bp (Fig. 3B). This result was obtained with a high redundancy of 7.14. All clone overlaps were ascertained at the nucleotide level. This defined length of roughly 1.8 Mb is slightly shorter than the previously predicted 2.0 Mb. The sequence contained precisely 483,365 A (adenine), 410,963 C (cytosine), 411,876 G (guanine), and 490,734 T (thymine), yielding an overall 45.8% G+C content, classifying this DNA segment within the relatively G+C-rich isochore H1 (14), putting it above the class II region that belongs to the G+C-poor isochore L (40%), but below the central class III segment, member of the most G+C-rich isochore H3 (around 53%) (15).
Gene Identification.
Homology searches with the entire sequence were carried out against the latest updates of DNA databases by using fasta, blastn, and blastx (http://www.ncbi.nlm.nih.gov/BLAST/). Searches for coding regions used the CRM/Grail Grail I, Ia, and II gene finding programs (genscan@gnomic.stanford.edu), the hexon exon finding program, the genscan gene prediction program (genscan@gnomic.stanford.edu), along with the SwissProt database and Smith-Waterman algorithm. As a result, the HLA class I region was found to include 23 known expressed genes, 12 new expressed genes (previously reported cDNA clones of unknown or ambiguous locations), three possibly expressed sequences (Fig. 3B and Table 1), and 22 potentially coding sequences (nearly 100% expressed sequence tag (EST)-matched sequences with exon-intron organization) (Table 2). Thus, a total of 37 new expressed genes or possibly coding sequences were identified. Our analysis also determined the precise location and structure of 30 known pseudogenes. Moreover, 37 new pseudogenes (Table 1) also were revealed during this study. These include one MIC (MICF), seven P5, six HCGII, seven HCGIV, three HCGIX, and 12 other pseudogenes. It must be noted that most of these pseudogenes are members of multigene families restricted to the HLA class I region. In sum, 127 genes were identified within this region, which corresponds to one gene per 14.1 kb. Among these, 60 are expressed genes or potentially coding sequences, corresponding to one expressed gene for every 29.9 kb.
Table 1.
Genes identified in the HLA class I region
Location is indicated by nucleotide positions numbered from the 5′ end of the MICB gene. The color code has been established as follows: orange indicates known expressed genes, green possibly expressed sequences, and red new expressed loci, i.e. for which cDNA clones were previously reported but where physical location was unknown or ambiguous. Regarding pseudogene, black depicts known pseudogenes and blue new ones. Gene orientation from centromere to telomere is shown by the letter C, and + depicts the opposite.
Table 2.
EST-matched sequences in the HLA class I region
Zooming In on the Segment Between the S and HLA-E Genes.
The 625-kb segment between the S and HLA-E genes is the least-characterized segment of the class I region because of previous difficulties in clone coverage (9); so far only three genes (TUBB, CAT56, and HSR1) have been mapped in this segment. By genomic sequencing, nine new expressed genes and seven EST-matched sequences were recognized in this segment. This makes the 250 kb between the PRG1 and HLA-E loci a gene-rich segment with 17 genes, including 15 expressed or potentially coding sequences (one expressed gene every 16.7 kb) (Fig. 3B and Tables 1 and 2). Novel genes noted in this segment are as follows (from centromere to telomere) (because of space restraints primary references for all these loci could be best accessed through their respective GenBank accession numbers). Transcription factor II H (TFIIH) is part of a protein complex involved in both transcription and DNA repair (Y07595); DDR encodes a receptor-tyrosine kinase up-regulated by the p53 (U48705) tumor suppressor gene; PRG1 (IEX-1) translates into an early response protein carrying functional binding sites for p53 and NF-κB (X96438); DBP2 gives rise to a putative nuclear ATP-dependent RNA helicase carrying a DEAH (Asp-Glu-Ala-His) box (AB001601); and ABC50 encodes an ATP binding cassette protein stimulated by tumor necrosis factor α (AF027302). Finally, the TC4 gene encoding a ras-like protein is juxtaposed (2.5 kb telomeric) to the HLA-E gene. It is noteworthy that all of these (six) newly mapped genes, packed around the PRG1–HLA-E segment, are likely to function in the process of DNA repair or cell proliferation, hence they may be possibly involved in the development of some cancers. The CAT56 (U63336) gene located just telomeric of ABC50 specifies a proline-rich protein homologous to the Wiskott-Aldrich syndrome gene located on Xp11.23-Xp11.22, mutation of which causes a rare immunodeficiency disorder affecting mainly platelets and lymphocytes. The HSR1 gene codes for a putative GTP-binding protein and is located 52 kb centromeric to HLA-E, not 2 kb as previously reported (indeed, a number of inaccuracies in previously published mapping as well as sequencing data have been discerned; these are available on request from the authors). Finally, no function has yet been ascribed to KIAA0170 (D79992), FB19 (Y13247), or GT260 and GT478 (X90535 and X90538, respectively).
Repetitive Elements.
Analysis of the complete sequence with the repeatmasker2 program unveiled the following numbers of repeats: 1,001 Alus, 105 MIRs, 411 LINEs (L1+L2), 290 LTRs, and 100 MERs. These collectively occupy 43.7% of the class I region, with Alus and LINEs representing 14.8% and 16.0% or one repeat per 1.8 kb and 3.6 kb, respectively. Although in case of Alus, this finding closely matches the theoretical value of 17.9% obtained for a genomic segment harboring a high G+C content of 45.8%, the LINE content is remarkably higher than the calculated value of 6.1% (16, 17).
Microsatellites.
A total of 758 microsatellite repeats were identified in the 1,796,938-bp genomic sequence. These consist of 203 di-, 139 tri-, 273 tetra-, and 143 penta-nucleotide repeats (Fig. 3C), yielding an overall density of one microsatellite per 2.3 kb, significantly higher than the one per 6 kb previously predicted by Beckman and Weber (18). Among the 758 microsatellite identified here, 70 already have been subjected to polymorphism analysis within the Japanese population. As expected, 38 of these 70 microsatellites are quite polymorphic with an average of 8.9 alleles and a 0.66 polymorphism content value (19). As these polymorphic microsatellites are evenly dispersed throughout the class I region, they should serve as much needed genetic markers in linkage and association analysis, enabling investigators to precisely map class I-associated disease susceptibility loci (3).
From the Original Building Block to Today’s MHC.
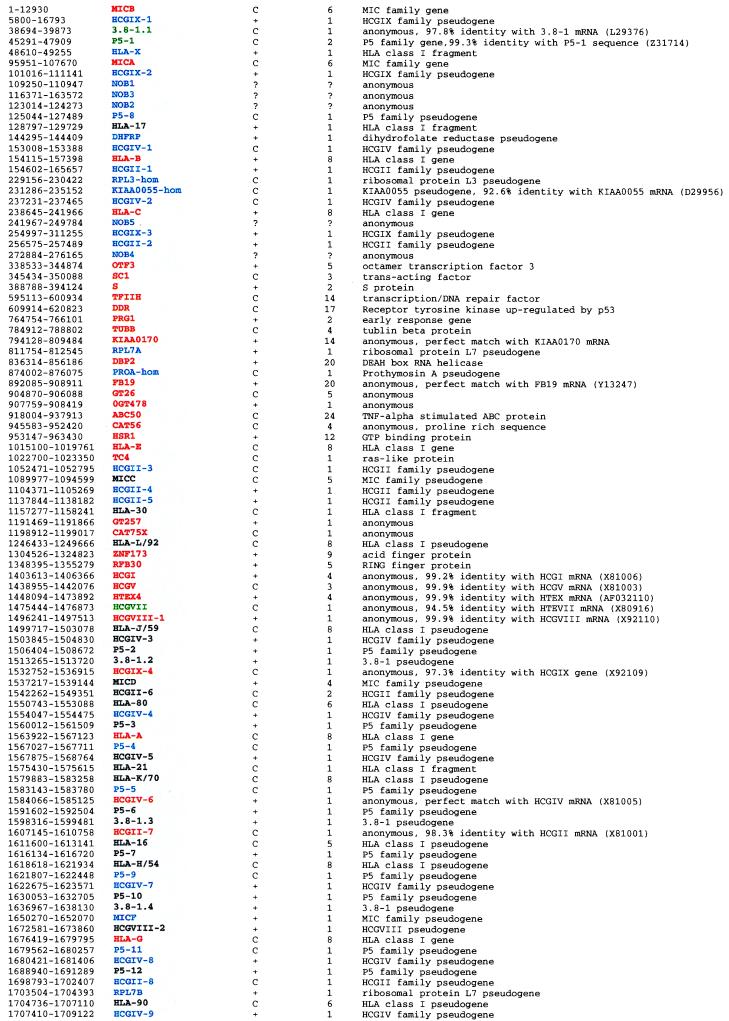
Dot matrix analysis using the entire 1,796,938-bp class I sequence versus itself revealed numerous large scale duplications (Fig. 1 A–C). These include several prominent homology sections: (a) 35-kb downstream segments of both HLA-B and HLA-C genes as well as 35-kb upstream regions of MICA and MICB genes display 80% and 85% nucleotide identity, respectively (5, 6), (b) around 39 kb upstream of MICD and 35 kb 5′ of MICE share significant nucleotide identity not only between themselves but also with (>50%) corresponding regions of MICA and MICB loci (Fig. 1A exemplifies the homology between MICA and MICD upstream segments), and (c) the 5′ segments of MICD and MICF are also homologous to each other (Fig. 1B). In sum, the upstream segments of all members of the MIC gene family (exception of MICC) display sequence homology to each other over distances longer than 15 kb. Interestingly all of these MIC-linked homologous segments share a unique mix of genes, all members of several multigene families. These are HCGIX, 3.8–1, P5, HCGIV, HLA class I, and HCGII in this order and within the same gene orientation in most cases. These facts strongly imply that successive segmental duplication of this basic unit gave rise to the present human MHC class I system. In this respect, the analysis of the 300-kb telomeric end of the HLA class I region, which links HLA-J/59 to HLA-F and includes MICD, MICE, and MICF, genes is staggering. Dot matrix analysis revealed this region to be filled with more than 30 pairs of homologous segments varying from 8 to 20 kb (Fig. 1C). This striking observation prompted us to look closely at each of these segments. Having done this, we realized that all 11 HLA class I genes (HLA-J/59, -80, -A, -21, -K/70, -16, -H/54, -G, -90, -75, and -F) are not only oriented in the same telomere to centromere orientation, but also display a high degree of homology within 8–20 kb of their upstream sequences. For example, the 20-kb upstream segment of the HLA-K/70 gene (HLA-70–P5–5–HCGIV-6–P5–6–3.8–1.3) shows significant (75.3%) homology to the 20-kb upstream segment of the HLA-H/54 gene (HLA-54–P5–9–HCGIV-7–P5–10–3.8–1.4).
Figure 1.
Dot matrix analysis (A) between the upstream segments of the MICA and MICD genes, (B) between the upstream segments of MICD and MICF genes, and (C) between the about 300-kb telomeric end segment of the HLA class I region from HCGVIII-1–HLA-F–P5–15 versus itself. Diagonals indicate the regions where contiguous sequences conform to the stated parameters.
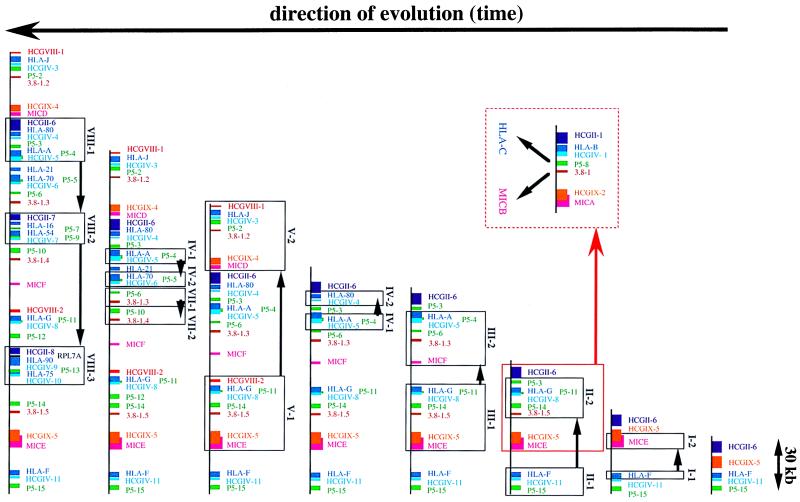
Based on these results, Fig. 2 depicts a model that explains how through seven rounds of successive segmental duplications of the above-mentioned elementary unit, the HLA class I region was shaped. Of the 18 HLA class I genes and six MIC genes localized, 15 HLA (except HLA-E, - 30, and -L/92) and five MIC (except MICC) loci are associated with these conserved shared segments. These generally consist of the HCGIX, 3.8–1, P5, HCGIV, and HCGII family members in this order and gene orientation. Furthermore, it must be emphasized that the conserved segments around the MIC genes always contain the HLA class I genes (MICB: HLA-X; MICA: HLA-17 and HLA-B; MICD: HLA-J/59; MICE: HLA-75 and HLA-90; and MICF: HLA-H/54 and HLA-16). Phylogenetic analysis of the HLA class I and MIC genes elegantly details the probable kinetics of these duplications. As shown in Fig. 4A (provided as supplemental data on the PNAS web site, www.pnas.org), the HLA-F and MICE genes first branched out from major clusters. Thus, HLA-F and MICE, which are in close proximity to each other at the telomeric end of the HLA class I region, possibly represent ancestral HLA class I and class I chain-related genes. According to this model, duplication of HLA-F gave birth to the MICE and HLA-G genes (from stage I-1 to I-2 in Fig. 2). The basic unit of MIC–HCGIX–3.8–1–P5–HCGIV–HLA-class I–HCGII therefore was created. Thereafter, two independent segmental duplications of this elementary unit simultaneously generated the HLA-A--MICF (from stage II-1 to II-2 in Fig. 2) as well as the MICA–HLA-B segments (from stage I-2 to II-3 in Fig. 2). The latter gave birth to both MICB and HLA-C genes after a single duplication (Fig. 2) (4, 5). The next partial segmental duplication gave rise to the HLA-80–HCGIV-4 segment from the HLA-A–HCGIV-5 segment (from stage III-1 to III-2 in Fig. 2). In a similar way, as illustrated in Fig. 2, four subsequent segmental duplications (stages IV to VII) including partial ones, led to the present-day gene organization of the HLA class I region. The order of the generation of each gene predicted by this model is supported by dendrograms of the HLA-class I, MIC, HCGIX, P5, 3.8–1, and HCGIV family members (Fig. 4 A–F). This model also is supported by the chronological order of the generation of the Alu, LINE, and LTR (long terminal repeat) subfamily members linked to respective repeat units (data not shown), which serve as a molecular clock to define the generation time of genomic segments of interest (for example, Alu J is older than Alu S).
Figure 2.
A model that explains how the HLA class I region was shaped by seven rounds of successive segmental duplications of a basic unit, MIC–HCGIX–3.8–1–P5–HCGIV–HLA class I. Arrows indicate the evolutionary path of the class I region paved by segmental duplications.
Discussion
The present work has established the nucleotide sequence of the human MHC class I region. Investigation of the obtained sequence unveiled the presence and exact location of a large number of genes, among which 37 were novel. This, combined with the identification of numerous highly polymorphic microsatellite repeats, will dramatically ease positional cloning approaches aimed at defining the molecular basis of HLA class I-associated diseases.
The molecular path taken across vertebrate evolution leading to the present-day MHC has been strongly debated for a number of years. Cloning and sequence analysis of MHC genes from various species, despite having brought a wealth of information, has failed to alleviate the confusion. The sequence data presented here resolves, at molecular level, this long controversy. It seems that HLA-F was the ancestor MHC class I gene that upon duplication gave rise to MICE and HLA-G. This contention is corroborated by substantial physical evidence: the phylogenetic tree analysis, the short genomic distance between HLA-G, MICE, and HLA-F, and the fact that the present-day MICE is the “least problematic” MIC pseudogene (in contrast to MICC, MICD, and MICF, which lack one or several exons/introns, MICE shows an intact genomic organization despite several nucleotidic defects). Moreover, no Alu, LINE, or LTR elements are linked to the MICE–HLA-F unit, whereas all the other MIC-HLA class I units share the oldest Alu, LINE, and LTR subfamily members. Therefore, these repetitive elements were inserted into these repeated basic units after duplication of MICE–HLA-F unit (after stage III-1 in Fig. 2). In fact, the ages of the Alu, LINE, and LTR subfamily members identified within the MIC–HLA class I basic units created after stage III-1 follow the order of the generation of these basic units predicted from our evolutionary model in Fig. 3. This primordial building block was already flanked by a number of structural units that upon a series of duplication and diversification gave birth to present-day MHC. This fact is evident in comparison of the upstream sequences of MIC and HLA genes. This scenario also explains the large number of pseudogenes present within the class I region. Indeed, among the 127 genes or gene candidates, more than half (67 genes) are pseudogenes. This accumulation of pseudogenes is unprecedented in the neighboring HLA class II and class III regions, paralleled by their respective “quiescent” evolutionary journey and is possibly reminiscent of the process of “birth and death process” (20) operational at the outset of MHC genesis. The large number of retrotransposon elements encountered throughout the class I region and some of the very structural genes part of the basic replicated unit (i.e., 3.8–1, P5, and HCGIV), which harbor LTR-like sequences, may have facilitated the entire process (21).
Among the 36 new expressed genes or potentially coding sequences around the S and HLA-E gene segment, six [TFIIH, DDR, PRG1 (IEX-1), DBP2, ABC50, and TC4] are involved in the process of DNA repair or cell proliferation. In this respect, the fact that numerous cancer cells exhibit decreased expression of HLA class I antigens as a result of deletion or loss of heterozygosity (LOH) of the HLA class I region is quite intriguing (22). This decline in the expression of HLA class I antigens is believed to provide an escape mechanism from the host immune system. Microsatellite alleles identified here provide the means to conclusively test this possibility, by narrowing candidate regions through the definition of LOH boundaries and subsequent investigation of tumor-associated mutations.
In sum, this highly accurate genomic sequence derived from a carefully tilled contig provides through a molecular blueprint of MHC structure, not only a platform for positional cloning experiments, but also a detailed case of evolution by “birth and death” relevant to our understanding of vertebrate genomics.
Supplementary Material
Acknowledgments
Grants from the Japan Science and Technology Corporation, an arm of the Science and Technology Agency; the Ministry of Education, Science, Sports and Culture, Japan, and the Tokai University School of Medicine supported this work. S.B. and H.I. are grateful for a French-Japanese collaborative grant awarded jointly by Institut National de la Santé et de la Recherche Médicale and Japan Society for the Promotion of Science. S.B. acknowledges the Fondation pour la Recherche Médicale (ARS2000) and the Association pour la Recherche sur le Cancer for additional support.
Abbreviations
- YAC
yeast artificial chromosome
- BAC
bacterial artificial chromosome
- PAC
P1-derived artificial chromosome
- LTR
long terminal repeat
- EST
expressed sequence tag
Footnotes
References
- 1.Campbell, R. D. & Trowsdale, J. (1997) Immunol. Today18, Suppl.
- 2.Bjorkman P J, Parham P. Annu Rev Biochem. 1990;59:253–288. doi: 10.1146/annurev.bi.59.070190.001345. [DOI] [PubMed] [Google Scholar]
- 3.Svejgaard A, Buus S, Fugger L, editors. HLA and Disease: The Molecular Basis (Alfred Benzon Symposium 40) Copenhagen: Munksgaard International Publishers; 1996. [Google Scholar]
- 4.Bahram S, Bresnahan M, Geraghty D E, Spies T. Proc Natl Acad Sci USA. 1994;91:6259–6263. doi: 10.1073/pnas.91.14.6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mizuki N, Ando H, Kimura M, Ohno S, Miyata S, Yamazaki M, Tashiro H, Watanabe K, Ono A, Taguchi S, et al. Genomics. 1997;42:55–66. doi: 10.1006/geno.1997.4708. [DOI] [PubMed] [Google Scholar]
- 6.Shiina T, Tamiya G, Oka A, Yamagata T, Yamagata N, Kikkawa E, Goto K, Mizuki N, Watanabe K, Fukuzumi Y, et al. Genomics. 1998;47:372–382. doi: 10.1006/geno.1997.5114. [DOI] [PubMed] [Google Scholar]
- 7.Geraghty D E, Koller B H, Pei J, Hansen J A. J Immunol. 1992;149:1947–1956. [PubMed] [Google Scholar]
- 8.Osoegawa K, Susukida R, Okano S, Kudoh J, Minoshima S, Shimizu N, de Jong P, Groet J, Ives J, Lehrach H, et al. Genomics. 1996;32:375–387. doi: 10.1006/geno.1996.0132. [DOI] [PubMed] [Google Scholar]
- 9.Shiina T, Kikkawa E, Yamagata T, Saito W, Tamiya G, Oka A, Watanabe K, Yamazaki M, Tashiro H, Okumura K, et al. Immunogenetics. 1998;48:402–407. doi: 10.1007/s002510050451. [DOI] [PubMed] [Google Scholar]
- 10.Albertsen H M, Abderrahim H, Cann H M, Dausset J, Paslier D L, Cohen D. Proc Natl Acad Sci USA. 1990;87:4256–4260. doi: 10.1073/pnas.87.11.4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Imai T, Olson M V. Genomics. 1990;8:297–303. doi: 10.1016/0888-7543(90)90285-3. [DOI] [PubMed] [Google Scholar]
- 12.Deininger P L. Anal Biochem. 1983;129:216–223. doi: 10.1016/0003-2697(83)90072-6. [DOI] [PubMed] [Google Scholar]
- 13.Abderrahim H, Sambucy J L, Iris F, Ougen P, Billault A, Chumakov I M, Dausset J, Cohen D, Le Paslier D. Genomics. 1994;23:520–527. doi: 10.1006/geno.1994.1538. [DOI] [PubMed] [Google Scholar]
- 14.Bernardi G. Annu Rev Genet. 1995;29:443–476. doi: 10.1146/annurev.ge.29.120195.002305. [DOI] [PubMed] [Google Scholar]
- 15.Tenzen T, Yamagata T, Fukagawa T, Sugaya K, Ando A, Inoko H, Gojobori T, Fujiyama A, Okumura K, Ikemura T, et al. Mol Cell Biol. 1997;17:4043–4050. doi: 10.1128/mcb.17.7.4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kazazian H H, Moran J V. Nat Genet. 1998;19:19–24. doi: 10.1038/ng0598-19. [DOI] [PubMed] [Google Scholar]
- 17.Schmid T. Prog Nucleic Acid Res Mol Biol. 1996;53:283–319. doi: 10.1016/s0079-6603(08)60148-8. [DOI] [PubMed] [Google Scholar]
- 18.Beckman J S, Weber J L. Genomics. 1992;12:627–631. doi: 10.1016/0888-7543(92)90285-z. [DOI] [PubMed] [Google Scholar]
- 19.Tamiya G, Shiina T, Oka A, Tomizawa M, Ota M, Katsuyama Y, Yoshitome M, Makino S, Kimura M, Inoko H, et al. Tissue Antigens. 1999;54:221–228. doi: 10.1034/j.1399-0039.1999.540302.x. [DOI] [PubMed] [Google Scholar]
- 20.Nei M. Nature (London) 1969;221:40–42. doi: 10.1038/221040a0. [DOI] [PubMed] [Google Scholar]
- 21.Van Arsdell S W, Denison R A, Bernstein L B, Weiner A M, Manser T, Gesteland R F. Cell. 1981;26:11–17. doi: 10.1016/0092-8674(81)90028-3. [DOI] [PubMed] [Google Scholar]
- 22.Garrido F, Ruiz-Cabello F, Cabrera T, Perez-Villar J J, Lopez-Botet M, Duggan-Keen M, Stern P L. Immunol Today. 1997;18:89–95. doi: 10.1016/s0167-5699(96)10075-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.