Abstract
Recently, Scm3p has been shown to be a nonhistone component of centromeric chromatin that binds stoichiometrically to CenH3–H4 histones, and to be required for the assembly of kinetochores in S. cerevisiae. Scm3p is conserved across fungi, and displays a remarkable variation in protein size, ranging from ~200 amino acids in Saccharomyces cerevisiae to ~1300 amino acids in Neurospora crassa. This is primarily due a variable C-terminal segment that is linked to a conserved N-terminal, CenH3-interacting domain. We have discovered that the extended C-terminal region is strikingly characterized by lineage-specific fusions of single or multiple DNA-binding domains—different versions of the MYB and C2H2 zinc finger domains, AT-hooks, and a novel cysteine-rich metal-chelating cluster—that are absent from the small versions of Scm3. Instead, S. cerevisiae point centromeres are recognized by components of the CBF3 DNA binding complex, which are conserved amongst close relatives of budding yeast, but are correspondingly absent from more distant fungi that possess regional centromeres. Hence, the C-terminal DNA binding motifs found in large Scm3p proteins may, along with CenH3, serve as a key epigenetic signal by recognizing and accommodating the lineage-specific diversity of centromere DNA in course of evolution.
Introduction
Despite being a universal feature of eukaryotic chromosomes, centromeres are characterized by enormous diversity. The structural spectrum of centromeres includes point centromeres that are short composite DNA elements like transcription regulatory elements or enhancers, extended regional centromeres that vary in length from 1kb to several megabases and holocentric elements that are distributed throughout the length of the chromosome 1–3. Within a monophyletic lineage, closely related organisms might exhibit diversity of centromere organization—for example, in fungi, Saccharomyces cerevisiae has point centromeres, whereas Candida albicans has regional centromeres 3. This diversity in form is accompanied by a concomitant diversity in the sequence of centromeric DNA (CEN) and in the proteins that assemble kinetochores on them 1–3 Yet, all these diverse centromeres perform a comparable set of functions across eukaryotes in chromosome segregation and spindle checkpoint control 1, 2. This conserved function has been attributed to an epigenetic role played by the centromeric chromatin proteins 1, 2. Hence, elucidating the assembly of the distinctive centromeric chromatin is necessary to understand the apparent paradox of centromere diversity form despite the presence of conserved functions.
Major advances in this direction have come from studies on the centromere specific histone H3 variant, CenH3 (Cse4p in yeast), which replaces the conventional histone H3 in centromeric nucleosomes. CenH3 is a unique and critical component of centromeric chromatin, and appears to have undergone adaptive evolution, probably in connection to the rapid sequence changes in CEN DNA 1, 2, 4. Recent discoveries of a CenH3 interacting protein, Scm3p (Suppressor of chromosome missegregation 3; a high-copy suppressor of mutations in cenH3) have further illuminated the architecture and assembly of centromeric chromatin 5–7. Scm3p was shown to specifically bind the cenH3-H4 complex, but not conventional histones H3-H4 and it is required for the assembly of CenH3 into the centromeric chromatin 6. Scm3p was also shown to selectively replace H2A-H2B from cenH3-H4-H2A-H2B octamers in vitro, consistent with the observed reduction of H2B, H2A and the H2A variant H2AZ at the centromeric nucleosomes in vivo. Furthermore, Scm3p is required for the centromeric localization of inner kinetochore proteins Cbf1p, Mif2p, Cep3p, and Ndc10p, suggesting that it functions at a key early step in the assembly of the unique centromeric chromatin structure 5–7.
Material and Methods
Sequence profile searches were conducted using the PSI-BLAST program 8 with either single sequences or multiple alignments as queries, with a profile inclusion expectation (e) value threshold of 0.01. For queries and searches containing computationally biased segments, the statistical correction option built into the BLAST program was used. Searches with conserved position-specific score matrices (PSSMs) and hidden Markov models (HMMs) were respectively conducted using the RPS-BLAST9 and HMMer package 10. Multiple alignments were constructed using the MUSCLE program 11, followed by manual adjustment based on PSI-BLAST hsp results and information provided by solved three-dimensional structures. Compositional bias and globular domains were identified using the SEG program 12 with parameters of 3.4 and 3.75 for complexity thresholds and window size of 45. Secondary structure was predicted using the combined information from residue frequencies, PSSMs and HMMs with the Jpred program 13. The non-redundant (NR) database of protein sequences and the Whole genome shotgun (WGS) (National Center for Biotechnology Information, NIH, Bethesda, MD) were the sequence databases used in this study. Translating searches of WGS was performed using TBLASTN.
Results and discussion
Sequence analysis of Scm3p shows that it is conserved across all fungi, irrespective of the type of centromere (point or regional) or CEN sequence they possess (Fig. 1). The conserved core of fungal Scm3p orthologs maps to a single globular domain that is predicted to adopt a predominantly α-helical fold and potentially chelate a metal ion. This conserved domain (hereinafter termed the Scm3p domain) corresponds to the region of Scm3, which is required for the cenH3-specific interaction 6, 7. The long predicted α-helix in this domain might be critical for the observed dimerization of Scm3, via a zipper-like pairing, as well as interaction with the corresponding helical segment in the histone fold. The incorporation of Scm3p into the nucleosome along with the histones also suggests that the Scm3p domain might itself have comparable non-specific DNA binding activity. Given the conservation of Scm3p across fungi, we investigated whether Scm3p has undergone evolutionary diversification that might correlate with diversity in CEN sequences and centromeric organization.
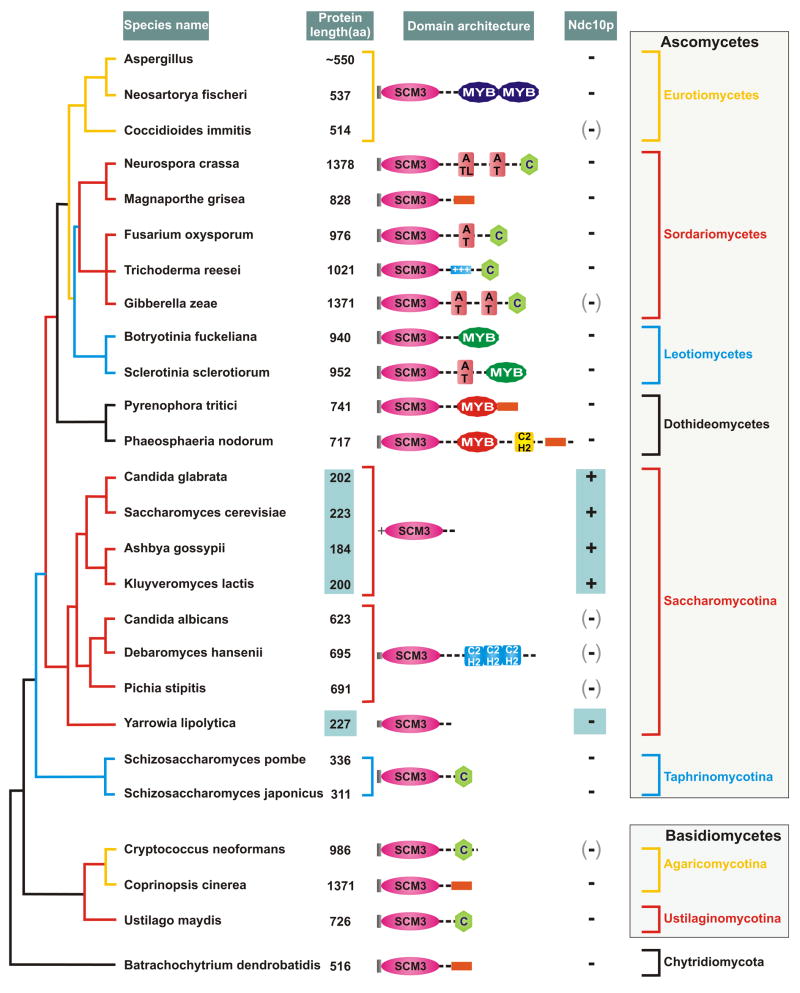
Figure 1. Diversity of domain architectures of Scm3p across fungal phylogeny.
The domain architectures of Scm3p proteins have been superimposed on a maximum-likelihood phylogenetic tree of fungi based on six conserved proteins. The major lineages shown in the figure are supported by 80% or greater relative logarithm likelihood bootstrap support. The proteins are not rendered to scale because of their enormous size difference, though the conserved domains are approximately rendered to size. The actual size of each protein is shown to its left and the small versions are boxed. The notations are: C- Cysteine cluster; the orange box represents positively charged segments. Notably, the N-termini of Scm3p orthologs across much of the fungal tree begin with a conserved block with the signature MxxP (shown as a grey block), but those of most members belonging to Saccharomycotina lack this signature. Instead they possess a distinctive positively charged helical segment indicated with a “+”. There is some uncertainty regarding the predicted gene structure, and hence the predicted protein of Cryptococcus neoformans. The different independently acquired versions of the MYB domain are shaded differently. The Ndc10 column reflects the presence or absence of orthologs of the Ndc10p protein, where +: presence, −: absence and (-): species with a homologous GCR1 domain, either in paralogous transcription factors or crypton family transposons, but no Ndc10 ortholog. All domains noted here were detected with e-values <.01 in profile and HMM searches. They were confirmed through reciprocal searches using PSI-BLAST that recovered known representatives of each of the depicted domains with e <.01 prior to convergence.
Examination of the fungal Scm3p orthologs indicates a remarkable variation in protein size. One group typified by S.cerevisiae, and closely related yeasts contain short Scm3p proteins (approximately 200 aa in size). The other group represented by yeasts such as Candida albicans, Pichia, Schizosaccharomyces, several diverse filamentous ascomycetes, basidiomycetes and chytrids are characterized by longer Scm3p proteins (300–1300 aa; see Fig. 1). Systematic analysis using the SEG program to measure compositional bias, and sensitive position-specific score matrices and hidden Markov models revealed striking features in the C-terminal extensions of the Scm3p orthologs. While the versions typified by S.cerevisiae had little more than a short tail of low complexity sequence, we discovered multiple, distinct DNA-binding modules in the C-terminal extensions of many of the longer orthologs (Fig. 1, all detected with e-value<.01 in sequence profile searches with the respective domain profiles. See supplementary material for further details). These include three distinct versions of the MYB domain with the helix-turn-helix fold10, two distinct versions of C2H2 zinc fingers 14 (C2H2-Znf), the AT-hook and related basic DNA-binding motifs 4, 15, and a novel potential DNA-binding Zn-cluster. Of the 3 versions of the MYB domain found in Scm3p tails one of them usually occurs as two tandem MYB domains resembles the classical Myb transcription factor, the second one is specifically related to the MYB domain in the RAP1p proteins, and the third a distinct highly divergent form (Fig. 1 and supplementary material). The C2H2-Znfs occur as either a single classical version, or else as a distinct module comprised of three specialized C2H2 zinc fingers (Fig. 1).
In addition to the classical AT-hook which binds minor grooves 15, typically of AT-rich DNA, some orthologs of Scm3p also possess a positively charged segment in the C-terminal tail with central motif of the form G[RK][RK]P (Fig. 1 and supplementary material). This charged motif is found in several DNA-binding proteins like Mif2p, Cbf1p, Cse4p, Sir4p, Orc1p, Dat1p, Arr1p and Yap7p (several of which are centromere or telomere proteins) and is comparable to the AT-hook and minor groove binding motifs in histone tails 4. Hence, in Scm3p orthologs this motif is likely to contact the minor groove of CEN DNA sequences, possibly specifically recognizing AT-rich DNA. Scm3p orthologs of several fungi, might also display a predicted Zn-cluster with the sequence signature [CH]x4-6Cx4Cx2C (where x is any residue) with an associated enrichment of positively charged and polar residues in their extreme C-terminal regions. Given these features, it is conceivable that this module has a distinctive DNA-binding or protein recruitment role. The C-terminal MYB domains and AT-hook motifs observed in Scm3p orthologs are reminiscent of homologous domains which are found in other key structural components of specialized chromatin at telomeres and centromeres, such as Rap1p 16 (MYB domain), Mif2p and Dat1p (both with AT-hooks) 15. These observations indicate that longer versions of Scm3p typically have a domain architecture of the form “Scm3+(DBD)n”, where (DBD)n represents one or more DNA-binding domains (Fig. 1) that might bind to CEN DNA sequences and locally alter DNA and chromatin structure at the centromere.
Superposing domain architectures of Scm3p on the phylogenetic tree of fungi 17 makes it clear that while different major lineages tend to have their own distinct architectures, these can dramatically vary even within closely related forms (Fig. 1). Thus, we have acquisition of the novel module with 3 C2H2-Znfs in one clade within Saccharomycotina while being absent in the sister clade. Likewise, Scm3p from the closely related Phaeosphaeria and Pyrenophora differ from each other in terms of the presence of a C2H2 Zn-finger. The Myb domains too appear to have been independently acquired of three different occasions in different orthologs of Scm3: versions from Aspergillus and related genera contain two tandem MYB domains, which are specifically related to the equivalent domains of classical Myb transcription factors. In contrast, those found in Sclerotinia and related species are specifically related to the version of the domain found in Rap1p, while the versions from Phaeosphaeria and relatives are highly divergent and unique (Fig. 1). Similarly, the presence of AT-hooks and related DNA-binding motifs in Scm3p tails show variability amongst related fungi, with these motifs being present only Gibberella, Neurospora, Fusarium and Sclerotinia, but not their close relatives (Fig. 1). The distribution of the novel Zn-cluster is similarly sporadic across fungi—it is found in both ascomycetes (e.g. Schizosaccharomyces) and basidiomycetes (e.g. Neurospora), with apparent losses in closely related sister taxa.
This variability of the C-terminal DNA-binding domains of Scm3p appears to have mirrored the diversification of the CEN sequences in the fungal centromeres. In this respect, Scm3p shows both certain parallels as well as differences to the concomitant diversification of cenH3 and CEN sequences in animals. In the case of animal cenH3, potential minor-groove-binding oligopeptide motifs in their N-terminal tails show notable diversity even between related species of the genus Drosophila 4. However, unlike Scm3, none of the cenH3 orthologs across eukaryotes show a comparable accretion of a diverse array of additional DNA-binding domains to their conserved core 4.
Members of the lineage within Saccharomycotina with the short Scm3p proteins, and lacking obvious DNA-binding motifs, (Fig. 1) are known or securely inferred to have point centromeres 3. Centromeres of Yarrowia lipolytica are more complex, but they possess CEN sequences that resemble S.cerevisiae sequences in size and structure. They appear to represent an intermediate condition between point and regional centromeres 18, which might be consistent with the short Scm3p ortholog in this yeast. In evolutionary terms, regional centromeres appear to be the primitive condition for fungi, with point centromeres being derived in a subgroup of Saccharomycotina 3. These yeasts possess a distinctive centromere-specific DNA-binding protein in the form of Ndc10p (Cbf2p), which along with the basic-helix-loop-helix transcription factor Cbf1p, is a component of the CBF3 inner kinetochore complex 3. Ndc10p, along with paralogous transcription factors like Hot1p, Msn1p, Gcr1p and Sum1p, contains a conserved DNA-binding domain that has been derived from a catalytically inactive version of the transposase of the crypton family of fungal mobile elements 19. This raises the possibility that recruitment of a degenerate transposable element a key CEN DNA-binding component played a role in the recent emergence of the point centromeres in the yeasts, and favored loss of the extended DNA-protein contact via the C-terminus of Scm3.
Though Scm3p has extensively diversified in course of fungal evolution to make different types of potential DNA contacts via its C-terminal regions, it is likely to mediate a conserved interaction with the CenH3-H4 complex via its N-terminal Scm3p domain. Hence, in functional terms, Scm3p is probably a key element that facilitates the conserved roles of centromeres by directing the assembly of an epigenetic signal in centromeric chromatin, while simultaneously recognizing and accommodating the lineage-specific diversity in centromere structure in course of speciation. Point centromeres in Saccharomycotina appear to be correlated on one hand with a short Scm3, without C-terminal DNA-binding extensions, while on the other with the presence of a distinctive sequence-specific CBF3 complex to bind CEN DNA. The regional centromeres, which apparently lack the precisely defined short sequence elements seen in the point chromosomes 1–3, might depend to a major extant on the Scm3p C-terminal modules to make DNA contacts, perhaps by recognizing structural features such as narrow minor groves. Evidence from the Scm3p architecture and the innovation of the CBF3 complex imply that the shift to point centromeres probably proceeded through origin or recruitment of a new set of proteins selected to mediate a qualitatively different kind of DNA contact, more akin to specific transcription factors.
CenH3 is a more universal component of eukaryotic chromatin, and is well-attested in terms of rapid sequence diversification 1, 2, but it shows less dramatic architectural variability than Scm3. Furthermore, the targeting of yeast CenH3 to centromeric chromatin itself seems to depend in the first place on Scm3. Hence, in more general terms, these observations raise the possibility that other eukaryotic lineages might similarly possess their own analogs of fungal Scm3. It is possible that these analogs might assist in assembly of cenH3 and provide additional moieties to recognize rapidly diversifying CEN sequences. Discovery and analysis of such proteins are likely to provide valuable insights into the role of centromere chromatin in diversification of organismal lineages.
Supplementary Material
Multiple alignments of conserved domains in fungal Scm3p proteins. The protein names and gis (genbank identifiers) are indicated to the right along with abbreviations for organism names.
Acknowledgments
We would like to thank J. Gerton and R. Baker for communication of results before publication, M. Smith and members of the Wu group for helpful comments. This work was supported by the Intramural Research Programs of National Center for Biotechnology Information, National Library of Medicine (L.A, L.M.I) and the Center for Cancer Research, National Cancer Institute (C.W.)
Footnotes
Note added in proof
After this paper was accepted for publication we became aware of recent publications identifying proteins involved in cenH3 in animals (Dev Cell. 2007 12(1):17–30. Priming of centromere for CENP-A recruitment by human hMis18alpha, hMis18beta, and M18BP1.Fujita Y, Hayashi T, Kiyomitsu T, Toyoda Y, Kokubu A, Obuse C, Yanagida M. and J Cell Biol. 2007 176(6):757–63. Maddox PS, Hyndman F, Monen J, Oegema K, Desai A.). These works implicates the orthologous MYB domain proteins Knl-2 (from Caenorhabditis elegans) and M18BP1 from vertebrates as potential functional analogs of Scm3p in fungi. Interestingly, these proteins contain a well-conserved N-terminal SANT-associated (SANTA) domain, while their C-terminal MYB domains are fast evolving. Their plant orthologs appear to entirely lack the MYB domains while conserving the SANTA domain. Furthermore, in plants the monocot and eudicot proteins show considerable differences in their C-terminal architectures. Based on the analogy to the situation in Scm3p, we suggest that the SANTA domain in these proteins might function as an analog of the Scm3p domain, with which it shares no apparent relationship. Similarly, the rapidly diverging C-termini might play a role in interacting rapidly diverging features of centromeric chromatin.
References
- 1.Henikoff S, Ahmad K, Malik HS. The centromere paradox: stable inheritance with rapidly evolving DNA. Science. 2001;293:1098–102. doi: 10.1126/science.1062939. [DOI] [PubMed] [Google Scholar]
- 2.Henikoff S, Dalal Y. Centromeric chromatin: what makes it unique? Curr Opin Genet Dev. 2005;15:177–84. doi: 10.1016/j.gde.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 3.Meraldi P, McAinsh AD, Rheinbay E, Sorger PK. Phylogenetic and structural analysis of centromeric DNA and kinetochore proteins. Genome Biol. 2006;7:R23. doi: 10.1186/gb-2006-7-3-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malik HS, Vermaak D, Henikoff S. Recurrent evolution of DNA-binding motifs in the Drosophila centromeric histone. Proc Natl Acad Sci U S A. 2002;99:1449–54. doi: 10.1073/pnas.032664299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Camahort R, Li B, Florens L, Swanson SK, Washburn MP, Gerton JL. Scm3 is essential to recruit the histone h3 variant cse4 to centromeres and to maintain a functional kinetochore. Mol Cell. 2007;26:853–65. doi: 10.1016/j.molcel.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 6.Mizuguchi G, Xiao H, Wisniewski J, Smith MM, Wu C. Nonhistone Scm3 and Histones CenH3-H4 Assemble the Core of Centromere-Specific Nucleosomes. Cell. 2007;129:1153–64. doi: 10.1016/j.cell.2007.04.026. [DOI] [PubMed] [Google Scholar]
- 7.Stoler S, Rogers K, Weitze S, Morey L, Fitzgerald-Hayes M, Baker RE. Scm3, an essential Saccharomyces cerevisiae centromere protein required for G2/M progression and Cse4 localization. Proc Natl Acad Sci U S A. 2007 doi: 10.1073/pnas.0703178104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schaffer AA, Aravind L, Madden TL, Shavirin S, Spouge JL, Wolf YI, Koonin EV, Altschul SF. Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res. 2001;29:2994–3005. doi: 10.1093/nar/29.14.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schaffer AA, Wolf YI, Ponting CP, Koonin EV, Aravind L, Altschul SF. IMPALA: matching a protein sequence against a collection of PSI-BLAST-constructed position-specific score matrices. Bioinformatics. 1999;15:1000–11. doi: 10.1093/bioinformatics/15.12.1000. [DOI] [PubMed] [Google Scholar]
- 10.Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–63. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 11.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wootton JC, Federhen S. Analysis of compositionally biased regions in sequence databases. Methods Enzymol. 1996;266:554–71. doi: 10.1016/s0076-6879(96)66035-2. [DOI] [PubMed] [Google Scholar]
- 13.Cuff JA, Barton GJ. Application of multiple sequence alignment profiles to improve protein secondary structure prediction. Proteins. 2000;40:502–11. doi: 10.1002/1097-0134(20000815)40:3<502::aid-prot170>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 14.Finn RD, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, Durbin R, Eddy SR, Sonnhammer EL, Bateman A. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–51. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aravind L, Landsman D. AT-hook motifs identified in a wide variety of DNA-binding proteins. Nucleic Acids Res. 1998;26:4413–21. doi: 10.1093/nar/26.19.4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Konig P, Rhodes D. Recognition of telomeric DNA. Trends Biochem Sci. 1997;22:43–7. doi: 10.1016/s0968-0004(97)01008-6. [DOI] [PubMed] [Google Scholar]
- 17.James TY, Kauff F, Schoch CL, Matheny PB, Hofstetter V, Cox CJ, Celio G, Gueidan C, Fraker E, Miadlikowska J, Lumbsch HT, Rauhut A, Reeb V, Arnold AE, Amtoft A, Stajich JE, Hosaka K, Sung GH, Johnson D, O’Rourke B, Crockett M, Binder M, Curtis JM, Slot JC, Wang Z, Wilson AW, Schussler A, Longcore JE, O’Donnell K, Mozley-Standridge S, Porter D, Letcher PM, Powell MJ, Taylor JW, White MM, Griffith GW, Davies DR, Humber RA, Morton JB, Sugiyama J, Rossman AY, Rogers JD, Pfister DH, Hewitt D, Hansen K, Hambleton S, Shoemaker RA, Kohlmeyer J, Volkmann-Kohlmeyer B, Spotts RA, Serdani M, Crous PW, Hughes KW, Matsuura K, Langer E, Langer G, Untereiner WA, Lucking R, Budel B, Geiser DM, Aptroot A, Diederich P, Schmitt I, Schultz M, Yahr R, Hibbett DS, Lutzoni F, McLaughlin DJ, Spatafora JW, Vilgalys R. Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature. 2006;443:818–22. doi: 10.1038/nature05110. [DOI] [PubMed] [Google Scholar]
- 18.Vernis L, Poljak L, Chasles M, Uchida K, Casaregola S, Kas E, Matsuoka M, Gaillardin C, Fournier P. Only centromeres can supply the partition system required for ARS function in the yeast Yarrowia lipolytica. J Mol Biol. 2001;305:203–17. doi: 10.1006/jmbi.2000.4300. [DOI] [PubMed] [Google Scholar]
- 19.Goodwin TJ, Butler MI, Poulter RT. Cryptons: a group of tyrosine-recombinase-encoding DNA transposons from pathogenic fungi. Microbiology. 2003;149:3099–109. doi: 10.1099/mic.0.26529-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Multiple alignments of conserved domains in fungal Scm3p proteins. The protein names and gis (genbank identifiers) are indicated to the right along with abbreviations for organism names.