Abstract
Single-strand conformation polymorphism (SSCP) analysis of ribosomal DNA (rDNA) was investigated for rapid differentiation of phenotypically similar yeast species. Sensitive tests indicated that some yeast strains with one, most strains with two, and all strains with three or more nucleotide differences in the internal transcribed spacer 1 (ITS1) or ITS2 region could be distinguished by PCR SSCP analysis. The discriminative power of SSCP in yeast species differentiation was demonstrated by comparative studies of representative groups of yeast species from ascomycetes and basidiomycetes, including Saccharomyces species, medically important Candida species, and phylloplane basidiomycetous yeast species. Though the species within each group selected are closely related and have relatively similar rDNA sequences, they were clearly differentiated by PCR-SSCP analysis of the ITS1 region, given the amplified fragments were less than 350 bp in sizes. By using SSCP analysis for rapid screening of yeast strains with different rDNA sequences, species diversity existing in a large collection of yeast strains from natural sources was effectively and thoroughly investigated with substantially reduced time and cost in subsequent DNA sequencing.
Yeasts are unicellular fungi and represent diversified microorganisms in the phyla of Ascomycota and Basidiomycota of the kingdom Fungi (30). They are widely distributed in natural environments and play an important role in the ecosystem (34). Recent studies have shown that a great number of yeast species, whether ascomycetes or basidiomycetes, remain to be discovered (6, 24, 47, 49, 59, 65). Estimates indicate that only 1% of yeast species that exist in nature have been described (6, 17).
The rapid and accurate identification of species is a common and basic need for yeast ecology and biodiversity studies. Traditional identification of yeasts relies on colony and cell morphology and distinctive reactions to a standardized set of fermentation and assimilation tests (68). These tests are laborious, time-consuming, and sometimes ambiguous because of strain variability. Consequently, molecular comparisons are increasingly used for yeast identification. A variety of molecular approaches for rapid identification of yeast species have been described. These methods include DNA probe hybridization (11, 15), multiplex PCR (42) and real-time PCR assays (25, 55), PCR-enzyme immunoassays (41), restriction fragment length polymorphism (RFLP) analysis of 5.8S internal transcribed spacer (ITS) ribosomal DNA (rDNA) (12, 16, 19, 45, 63), electrophoretic karyotyping by pulsed-field gel electrophoresis (2, 7), denaturing gradient gel electrophoresis (DGGE) (42, 44, 58), amplified fragment length polymorphism (20, 60), flow cytometry or Luminex (8, 13, 52), and DNA microarray (37, 38). These molecular approaches were usually developed for specific or limited groups of yeasts, e.g., medically important species.
The molecular studies aimed at general yeast species identification have emphasized either coding (D1/D2 variable domains of the large subunit rDNA) or noncoding (ITS) regions of the rDNA. As a result, databases of D1/D2 (17, 32) and ITS (33, 36, 56, 58) sequences are available for molecular classification and identification of yeasts. Taxonomic studies based on the molecular characters have resulted in the discovery of an unparalleled number of new yeast species in recent years and have greatly improved our understanding of yeast biodiversity (6).
The general molecular identification of yeasts relies on nucleotide sequence determination and comparison of the D1/D2 domain or ITS region of rDNA. Though DNA sequencing has become a common technique nowadays, the equipment is still unavailable in common laboratories and the experiment is still costly, especially when dealing with a large number of samples or strains in ecology and biodiversity studies. A practical approach commonly used to reduce cost and time is to pick only isolates with different colony morphologies at the isolation stage or select representative strains for further study after grouping morphologically similar strains that have been isolated. Since yeasts are usually single celled with very simple morphological characters, different species often have indistinguishable colony and cell appearances. Species and genetic diversity of yeasts in a sample or represented by the strains isolated may not be fully revealed by such practical methods.
We have investigated a simple and rapid approach to reveal the sequence polymorphism of rDNA by utilizing the technique of single-strand conformation polymorphism (SSCP). The SSCP technique was initially developed for point mutation detection in human DNA (50, 51). It has been demonstrated to be a powerful tool for gene mutation and variation analysis (14, 21) and has been used in species identification of human- and plant-pathogenic fungi (27, 28, 64). The present study shows that the SSCP technique is a powerful tool for rapid detection of rDNA sequence divergences among morphologically similar yeast strains and for rapid differentiation of phenotypically similar and phylogenetically closely related yeast species.
MATERIALS AND METHODS
Yeast strains.
Yeast strains with known rDNA ITS sequences were selected for the sensitivity test (Table 1). The other strains studied were indicated in the legends of appropriate figures.
TABLE 1.
Pairs of selected yeast strains with different nucleotide mismatches in the ITS1 or ITS2 region
| Strain pair | Species | Strain | Fragment amplified | Fragment size (bp) | Mismatch(es) |
|---|---|---|---|---|---|
| 1 | Bullera pseudoschimicola | W10.7 | ITS2 | 338 | 1 substitution |
| W21.8 | 338 | ||||
| 2 | Bullera oryzae | YLG10.1 | ITS1 | 212 | 1 substitution |
| YS2E1 | 212 | ||||
| 3 | Dioszegia zsoltii | AS 2.2089 | ITS1 | 229 | 1 substitution |
| AS 2.2231 | 229 | ||||
| 4 | Bullera variabilis | SM5.5 | ITS1 | 193 | 1 substitution and 1 indel |
| W15.5 | 194 | ||||
| 5 | Bullera oryzae | YLG10.1 | ITS2 | 311 | 2 substitutions |
| YS2E1 | 311 | ||||
| 6 | Bullera sinensis | JF23.4 | ITS2 | 307 | 2 substitutions |
| SY11.8 | 307 | ||||
| 7 | Bullera variabilis | CB165 | ITS2 | 319 | 2 substitutions |
| CB146 | 319 | ||||
| 8 | Bullera variabilis | SM5.5 | ITS2 | 320 | 3 substitutions |
| W15.5 | 320 | ||||
| 9 | Dioszegia zsoltii | H6C1 | ITS2 | 328 | 3 substitutions |
| ZXS31.1-2 | 328 | ||||
| 10 | Bullera variabilis | CB146 | ITS2 | 319 | 2 substitutions and 1 indel |
| CB149 | 318 | ||||
| 11 | Bulleromyces albus | CB14 | ITS2 | 366 | 2 substitutions and 1 indel |
| CB113 | 365 | ||||
| 12 | Dioszegia zsoltii | AS 2.2089 | ITS2 | 328 | 3 substitutions |
| AS 2.2091 | 328 | ||||
| 13 | Itersonilia perplexans | CB340 | ITS1 | 248 | 2 substitutions and 1 indel |
| HS19.9 | 247 | ||||
| 14 | Bulleromyces albus | H2C5 | ITS2 | 366 | 4 substitutions |
| D35.2 | 366 |
DNA extraction and PCR amplification.
Nuclear DNA was extracted by using the method of Makimura et al. (43). ITS1 or ITS2 regions of the yeasts studied were amplified by PCR. The universal primer pair ITS1 (5′-GTCGTAACAAGGTTTCCGTAGGTG-3′) and ITS2 (5′-GCTGCGTTCTTCATCGATGC-3′) was used for amplification of the ITS1 region, and primer pair ITS3 (5′-GCATCGATGAAGAACGCAGC-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′) was used for the ITS2 region. An alternative primer pair, ITS1 and ITS1R (5′-CTCCACAGTGTGTTGTATTG-3′), was used to amplify a part of the ITS1 region of Saccharomyces species. PCR amplification was performed in a thermocycler (ICycler; Bio-Rad, Hercules, CA) with a program consisting of an initial denaturing step at 94°C for 4 min; 36 cycles of denaturation at 94°C for 1 min, annealing at 52°C for 1 min, and extension at 72°C for 20 s; and a final extension step of 4 min at 72°C.
SSCP analysis.
The PCR products were first evaluated for purity and concentration by agarose gel electrophoresis. Then 2.5 to 5 μl of PCR products was mixed with the same volume of denaturing buffer (95% formamide, 20 mM EDTA, 0.05% bromophenol blue, 0.05% xylene cyanol). The mixtures were heated at 95°C for 10 min and then chilled on ice. Denatured PCR products were loaded on an 8% acrylamide-bisacrylamide (37.5:1) nondenaturing gel (200 by 200 by 0.75 mm) with 5% glycerol; the gel was cast using the gel sandwich set provided in the DCode Universal Mutation Detection System (Bio-Rad, Hercules, CA). Electrophoresis was performed in the same system in prechilled 1× TBE buffer (89 mM Tris-borate, 2 mM EDTA [pH 8.0]) at 220 V for 10 h at 10°C. A nondenatured double-stranded DNA (dsDNA) ladder of 2,000, 1,000, 750, 500, and 250 bp was used as the marker.
After electrophoresis, silver staining of the gel was carried out using the procedure reported by Beidler et al. (5) with minor modifications. Specifically, the polyacrylamide gel was soaked in 300 ml of 10% ethanol for 5 min and then in the same volume of 0.05% acetic acid solution for 5 min. After one brief wash with 300 ml of distilled water (dH2O), the gel was soaked in 300 ml of 0.1% (wt/vol) silver nitrate for 15 min and then washed with 300 ml of dH2O. The gel was developed by rinsing in 300 ml of 1.5 ppm formaldehyde in 1.5% sodium hydroxide solution. When the desired intensity was reached, the gel was washed twice with dH2O and then fixed in 0.75% sodium carbonate for 15 min.
DNA sequencing and molecular phylogenetic analysis.
When desired, ITS regions of yeast strains were directly sequenced using the ABI BigDye terminator cycle sequencing kit (Applied Biosystems, Foster City, CA). Sequences were aligned with the program ClustalX (61) and adjusted manually. Reference sequences were retrieved from GenBank. The phylogenetic tree was constructed from the evolutionary distance data calculated from Kimura's two-parameter model (26) using the neighbor-joining method (54). Bootstrap analysis (18) was performed on 1,000 random resamplings.
RESULTS
Sensitivity of SSCP in the detection of nucleotide differences in the ITS region of yeasts.
Extensive analyses of rDNA sequence differences between yeast species indicated that, for the majority groups of yeasts, interspecific sequence divergences in the ITS region are usually greater than those in the D1/D2 domain (17, 33, 56). Therefore, we selected the ITS region as the molecular marker in this study. The lengths of the whole ITS1-5.8S rDNA-ITS2 region of yeasts ranged approximately from 400 to 800 bp (27). Shorter fragments (less than 400 bp) are better suited for detection of mutations in the SSCP gel, and the optimal size of fragment for sensitive base substitution detection by SSCP is approximately 150 to 200 bp (23, 48, 57). Therefore, we amplified ITS1 or ITS2 region separately from yeast strains for SSCP analysis.
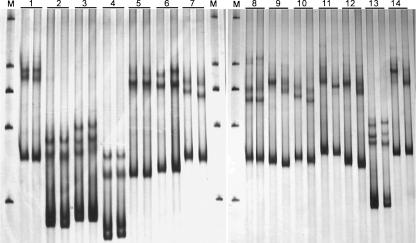
Yeast strains with known one to four nucleotide differences in the ITS1 or ITS2 region were selected for the sensitivity test of SSCP analysis (Table 1). Among the three pairs of strains with one base substitution each, slight and clear differences were shown between the SSCP patterns of the two strains in pairs 2 and 3, respectively (Fig. 1). The two strains in each of the four pairs with two base differences each were slightly or clearly differentiated, except for the strains in pair 5, which showed an indistinguishable SSCP pattern (Fig. 1). Clearly different SSCP patterns were observed for all strains with three or more mismatches in the ITS1 or ITS2 region (Fig. 1).
FIG. 1.
Comparison of SSCP patterns between yeast strains with 1 to 4 nucleotide differences in the ITS1 or ITS2 region in each of the pairs. The strain pairs employed are listed in Table 1. M, marker (a nondenatured dsDNA ladder of 2,000, 1,000, 750, 500, and 250 bp).
Differentiation of Saccharomyces species.
Seven species are currently included in the genus Saccharomyces Meyen ex Reess as redefined recently by Kurtzman (29) based on multigene sequence analysis (33). A new species of the genus was discovered from tree bark recently (67). The eight Saccharomyces species are phenotypically similar and phylogenetically closely related. They are difficult to distinguish by phenotypic criteria (62). The Saccharomyces species have longer ITS regions than most of the other groups of yeasts. When the universal primer pair ITS1 and ITS2 or ITS3 and ITS4 was used to amplify the ITS1 or ITS2 region, the lengths of the amplicons obtained were approximately 450 or 420 bp. The majority of the Saccharomyces species could not be clearly distinguished by SSCP analysis of the amplicons.
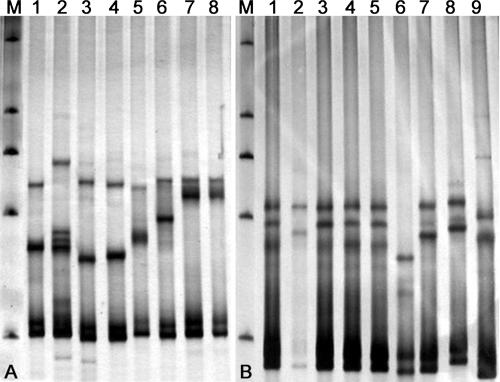
A new reverse primer, ITS1R, which is located near the 5′ end of the ITS1 region, was designed to obtain shorter fragments which covered the main part of ITS1 region. The lengths of the amplified fragments from the Saccharomyces species using primer pair ITS1 and ITS1R were 265 to 268 bp. SSCP analysis of the shortened fragments differentiated the Saccharomyces species clearly from each other, except for Saccharomyces bayanus and Saccharomyces pastorianus, which have identical ITS sequences (Fig. 2A). Even the two species Saccharomyces cariocanus and Saccharomyces paradoxus, which have only two substitutions in the amplified fragment, were distinguished clearly (Fig. 2A).
FIG. 2.
SSCP patterns of the ITS1 region from type or authentic strains of Saccharomyces (A) and Candida (B) species. (A) Lanes: 1, S. arboricolus AS 2.3317; 2, S. cariocanus AS 2.2374; 3, S. kudriavevii AS 2. 2408; 4, S. mikatae AS 2.2407; 5, S. paradoxus AS 2.2401; 6, S. cerevisiae AS 2.1882; 7, S. pastorianus AS 2.2402; 8, S. bayanus AS 2.1885. (B) Lanes: 1 to 5, C. albicans strains S6, 0034, S12, S0, and S22, respectively; 6, C. krusei AS 2.3194; 7, C. tropicalis AS 2.3195; 8, C. parapsilosis ATCC 22019; 9, C. dubliniensis CBS 7988. M, marker (a nondenatured dsDNA ladder of 2,000, 1,000, 750, 500, and 250 bp).
Differentiation of medically important Candida species.
The Candida albicans clade in the ascomycetous yeasts contains several clinically important species (31, 32). The opportunistically pathogenic Candida species other than C. albicans (the so called non-albicans Candida species) are more and more frequently isolated from clinical sources (53). Among the five medically important species compared, C. albicans, C. dubliniensis, C. parapsilosis, and C. tropicalis are closely related within the C. albicans clade, while C. krusei is located in a separate clade (32, 40). The sizes of ITS1 fragments amplified using the primer pair ITS1 and ITS2 from the former four species were 232 to 243 bp, and the size of the fragment from the last one was 196 bp. The five Candida species were clearly differentiated from each other by SSCP analysis of the amplified fragments (Fig. 2B).
The five C. albicans strains compared showed identical SSCP patterns, except strain 0034, which exhibited a unique SSCP pattern (lane 2 in Fig. 2B). Sequence analysis showed that strain 0034 differed from the other four strains compared in the ITS1 region by two mismatches. The identifications of all of the C. albicans strains studied were confirmed by D1/D2 domain sequence analysis (31).
Differentiation of closely related basidiomycetous yeast species.
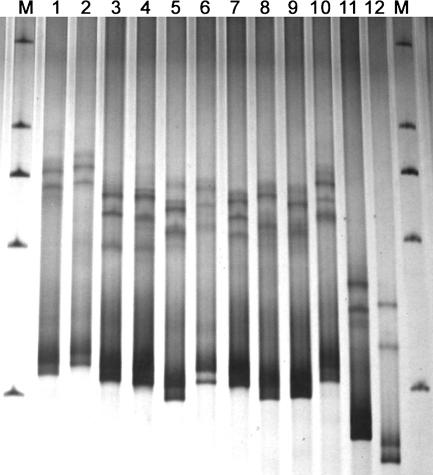
In basidiomycetous yeasts, the Bullera mrakii clade contains 11 closely related species which are also phenotypically similar (66). The lengths of the amplified fragments covering the whole ITS1 region from all 11 species except Bullera hubeiensis were 258 to 281 bp. The fragment length of B. hubeiensis, which was clearly separated from the other species of the clade in the phylogenetic tree (66), was much shorter (217 bp). SSCP analysis showed that the 11 closely related species were clearly differentiated from each other (Fig. 3). The SSCP pattern differences of the species were roughly parallel with their phylogenetic distances (66). B. hubeiensis differed from the other species of the same clade more obviously. Bullera anamola, which was located in a different clade (4), had a much more remarkable and different SSCP pattern (Fig. 3).
FIG. 3.
SSCP patterns of the ITS1 region from type strains of closely related Bullera species. Lanes: 1, B. pseudoschimicola AS 2.2201; 2, B. schimicola AS 2.2415; 3, B. boninensis AS 2.2413; 4, B. waltii AS 2.2414; 5, B. huiaensis JCM 8933; 6, B. komagatae AS 2.2202; 7, B. nakasei AS 2.2435; 8, B. cylindrica AS 2.2308; 9, B. pseudohuiaensis AS 2.2203; 10, B. mrakii JCM 8934; 11, B. hubeiensis AS 2.2466; 12, B. anamola AS 2.2094. M, marker (a nondenatured dsDNA ladder of 2,000, 1,000, 750, 500, and 250 bp).
Differentiation of unidentified yeast strains with similar colony morphology.
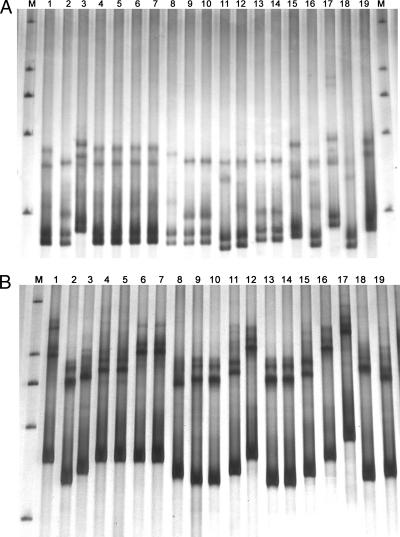
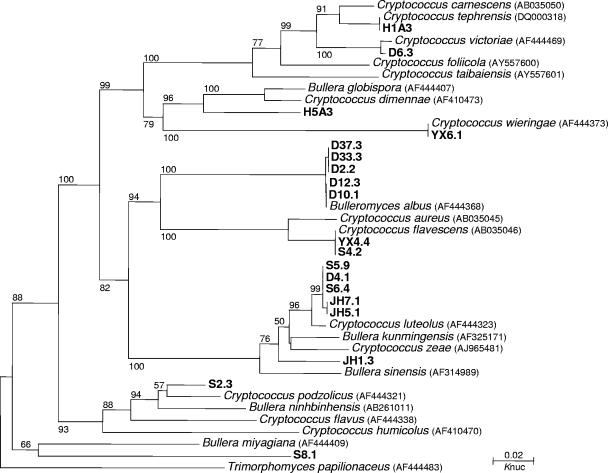
During the research on biodiversity of basidiomycetous yeasts in the phylloplane, we isolated a large number of strains forming similar pink or white colonies. PCR SSCP analysis of the ITS1 and ITS2 regions was used to screen the strains with different ITS sequences for further sequence analysis. The SSCP patterns of a part of the strains forming similar white colonies are shown in Fig. 4. Among the 19 strains compared, 10 ITS1 and 11 ITS2 SSCP patterns were recognized, respectively. Ten species, corresponding to the 10 ITS1 SSCP patterns, were identified from the strains compared by subsequent ITS sequence analysis (Fig. 5). Phylogenetic analysis showed that 5 of the 10 species identified might be new to science (Fig. 5). The Bulleromyces albus strains with the same ITS1 SSCP pattern were separated into two groups by ITS2 SSCP analysis (Fig. 4). Sequence comparison indicated that the two groups differed by one substitution in the ITS2 region. The sequence difference of the Bulleromyces albus strains was reflected in the tree (Fig. 5).
FIG. 4.
SSCP patterns of the ITS1 (A) and ITS2 (B) regions of basidiomycetous yeast strains isolated from phylloplane which formed similar white colonies. Lanes: 1, 4, 5, 6, and 7, Bulleromyces albus strains D2.2, D10.1, D12.3, D33.3, and D37.3, respectively; 2, 9, 10, 13, and 14, Cryptococcus sp. 1 strains D4.1, JH5.1, JH7.1, S5.9, and S6.4, respectively; 3, Cryptococcus victoriae D6.3; 8, Cryptococcus sp. 2 strain JH1.3; 11, Cryptococcus sp. 3 strain S2.3; 12 and 16, Cryptococcus flavescens strains S4.2 and YX4.4, respectively; 15, Cryptococcus sp. 4 strain S8.1; 17, Cryptococcus wieringae YX6.1; 18, Cryptococcus sp. 5 strain H5A3; 19, Cryptococcus tephrensis H1A3. M, marker (a nondenatured dsDNA ladder of 2,000, 1,000, 750, 500, and 250 bp).
FIG. 5.
Phylogenetic tree drawn from neighbor-joining analysis of the ITS (including 5.8S rDNA) sequences, depicting the relationships of the basidiomycetous yeast strains isolated from the phylloplane which form similar white colonies with closely related taxa. Bootstrap percentages over 50% from 1,000 bootstrap replicates are shown. Reference sequences were retrieved from GenBank under the accession numbers indicated.
DISCUSSION
Because of its technical simplicity and relatively high sensitivity for the detection of sequence variations, SSCP has become one of the most popular mutation detection strategies since its introduction in 1989 (50, 51). Previous studies have indicated that over 90% of single-base mutations can be detected for sequences in fragments up to 200 bp by SSCP analysis under optimized conditions. The detection rate falls to about 80% for fragments longer than 200 bp but shorter than 350 bp (22, 48).
For the majority of yeast species, the PCR-amplified rDNA fragments flanking the ITS1 or ITS2 region range in size from approximately 150 to 350 bp (9, 10), being suitable for detection of nucleotide differences in the SSCP gel. Although the amplified fragments of the yeasts compared were usually longer than the optimal size (<200 bp) for sensitive detection (Table 1), the majority of the strains differing by only one or two base mismatches in the target regions were differentiated (Fig. 1). All strains differing by three or more mismatches in the ITS1 or ITS2 region were readily and clearly distinguished by SSCP analysis (Fig. 1).
Extensive rDNA sequence comparisons have shown that separate yeast species usually differ by more than 1% substitution in the whole ITS region (3, 33, 56, 58). Considering the lengths of the ITS region of most yeast species, this means that yeast strains belonging to different species usually differed by at least three substitutions in the ITS1 or ITS2 region. Therefore, most yeast species are distinguishable by SSCP analysis using either of the ITS regions as the molecular marker.
The theoretically high discriminative power of PCR SSCP analysis in yeast species differentiation has been demonstrated in the present study. Representative groups of yeast species from ascomycetes and basidiomycetes were selected for comparative studies. The species within each group selected are closely related and have relatively similar rDNA sequences. All of them were clearly differentiated by SSCP analysis, given the amplified fragments were less than 350 bp in size. The yeast species in other groups or clades usually have more rDNA sequence divergence from each other (17, 32, 33, 56). Therefore, they should be differentiated by SSCP analysis more easily and clearly.
Among other molecular approaches developed for rapid identification of yeast species, RFLP analysis of 5.8S-ITS rDNA was more frequently used for a relatively broad range of yeast species because of the simplicity of the experiment (12, 16, 19, 45, 63). However, Villa-Carvajal et al. (63) found that, in general, groups of yeast species that are closely related on the basis of their 26S rDNA were difficult or impossible to differentiate on the basis of the RFLPs of their 5.8S ITS rDNA. The method is also subject to the use of multiple restriction enzymes selected based on known sequences of described species. Sequence differences occurring outside the restriction sites cannot be detected by RFLP analysis, and thus, fragments with identical sizes do not necessarily have identical sequences. Yeast species with different ITS sequences may have identical RFLP patterns (16, 63). Recently developed techniques employing DNA microarray (37, 38) or Luminex (8, 13, 52) can be used for high-throughput identification of yeasts. A specific DNA probe must be designed for a yeast species based on its known sequences, and specific equipment is required in the experiments. Therefore, these methods are only suitable for specific groups of yeasts with limited numbers of species. DGGE, another widely used mutation detection method, has been applied in differentiation of industrially and clinically important yeast species (44, 46, 60). However, the method has the following disadvantages. (i) The gradient gel casting process is complicated and needs experience. (ii) The necessary primer GC clamp decreases amplicon yield and favors primer dimers. (iii) It is almost impossible to achieve the same gradient in different gels. The last disadvantage makes the comparability of DGGE patterns between different gels difficult and limits its application to the comparison of a large number of strains or samples.
Compared to other molecular approaches, SSCP analysis is a rather competitive method for rapid species identification in specific groups with limited number of species (e.g., commercially and clinically important species, as shown in Fig. 2), by using the SSCP profiles of the target region of type or authentic strains of the species in the groups concerned as the standards. The more valuable application of SSCP analysis, however, is in the rapid molecular grouping of strains based on rDNA sequence differences in large-scale biodiversity and ecological studies of yeasts, as shown in Fig. 4. The advantages of SSCP analysis include the following. (i) SSCP has high discriminatory power, as shown in the present study. (ii) SSCP has simplicity of usage without the requirement for special equipment. (iii) Radioactive labeling is not necessary. (iv) Finally, SSCP is compatible with automated high-throughput analysis. In addition to the apparatus designed specially for mutation detection (e.g., the DCode system of Bio-Rad), SSCP analysis can be performed in the common apparatus used for polyacrylamide gel electrophoresis (27). Comparison of SSCP patterns between different gels can be achieved by employing a DNA ladder as shown in the present study and by Kong et al. (27) and by standardizing the parameters of the analysis procedure. By using automated capillary array sequencers (e.g., ABI Prism 310, 3100, and 3700), high-throughput SSCP analysis can be achieved (1, 35).
We have been using SSCP analysis for rapid screening of yeast strains with different rDNA sequences from a large number of strains isolated during our research on biodiversity of yeasts in natural environments. After the grouping of the strains based on colony morphology, the SSCP patterns of the strains within the same morphological groups were compared. The dsDNA ladder was used as the reference in comparisons among SSCP patterns of morphologically similar strains in different gels. As shown in Fig. 4 and 5, the strains with the same SSCP profiles belonged to the same species. Thus, only the strains with different SSCP patterns needed to be selected for sequencing. This procedure enabled us to effectively and thoroughly investigate the species diversity existing in a large collection of strains at substantially reduced time and cost and resulted in the discovery of many new taxa, as shown in Fig. 5, which included only a small part of the strains studied. The rapid SSCP screening method has also improved our understanding of species diversity of yeasts existing in small niches or microniches, such as, for example, a leaf. By analyzing as many isolates as from single plates, instead of only those with different colony morphologies, as usually done before, we found that a considerably higher diversity of yeast species—usually with close phylogenetic relationships—may exist in single leaves (data not shown).
Intraspecific variation can also be detected by SSCP analysis. In addition to the two SSCP patterns of ITS1 found in C. albicans strains as shown in Fig. 2B, one more SSCP pattern of the region has been found from strains of this species. The three SSCP patterns corresponded to three ITS types existing in C. albicans strains, which differed from each other by 1 or 2 nucleotides in the ITS region (data not shown). Bulleromyces albus strains differing by one base substitution in the ITS2 region were clearly differentiated, as shown in Fig. 4B. It will be adequate to analyze one region, either ITS1 or ITS2, for interspecific differentiation. However, it will be better to analyze both ITS1 and ITS2 regions for intraspecific variation detection. When more variable DNA markers, for example, microsatellites, are selected, SSCP can be used as a powerful tool for strain typing of yeasts (39).
Disadvantages of SSCP analysis should be considered when using the technique in yeast species or strain differentiation. First, the sizes of fragments analyzed are limited. Our experiences showed that if the fragments were longer than 400 bp, as in the case of Saccharomyces species, the discriminative power of SSCP analysis decreased remarkably. It is better to limit the sizes of the fragments to less than 350 bp for interspecific differentiation and 200 bp for intraspecific differentiation. Second, gels can sometimes be difficult to interpret by the principle of the SSCP technique (21). Theoretically, for a given homogenous DNA fragment, three bands, representing one double-stranded and two single-stranded bands, respectively, should appear in SSCP gel. However, extra bands usually appeared in SSCP gels when rDNA fragments or microsatellites were used as the markers, as shown in the present and previous studies (27, 39). The extra bands may come from the heterogeneous nature of the markers in the organisms studied or from the denatured fragments that refolded in different manners in the nondenaturing gel. Nevertheless, the extra bands may contribute to further and clearer differentiation of the strains compared.
Acknowledgments
This study was supported by grants 30470005 and 30628002 from the National Natural Science Foundation of China (NSFC).
Footnotes
Published ahead of print on 14 March 2008.
REFERENCES
- 1.Baba, S., Y. Kukita, K. Higasa, T. Tahira, and K. Hayashi. 2003. Single-stranded conformational polymorphism analysis using automated capillary array electrophoresis apparatuses. BioTechniques 34:746-750. [DOI] [PubMed] [Google Scholar]
- 2.Bai, F.-Y., H.-Y. Liang, and J.-H. Jia. 2000. Taxonomic relationships among the taxa in the Candida guilliermondii complex, as revealed by comparative electrophoretic karyotyping. Int. J. Syst. Evol. Microbiol. 50:417-422. [DOI] [PubMed] [Google Scholar]
- 3.Bai, F.-Y., M. Takashima, and T. Nakase. 2001. Phylogenetic analysis of strains originally assigned to Bullera variabilis: descriptions of Bullera pseudohuiaensis sp. nov., Bullera komagatae sp. nov. and Bullera pseudoschimicola sp. nov. Int. J. Syst. Evol. Microbiol. 51:2177-2187. [DOI] [PubMed] [Google Scholar]
- 4.Bai, F.-Y., M. Takashima, J.-H. Zhao, J.-H. Jia, and T. Nakase. 2003. Bullera anomala sp. nov. and Bullera pseudovariabilis sp. nov., two new ballistoconidium-forming yeast species from Yunnan, China. Antonie van Leeuwenhoek 83:257-263. [DOI] [PubMed] [Google Scholar]
- 5.Beidler, J. L., P. R. Hilliard, and R. L. Rill. 1982. Ultrasensitive staining of nucleic acids with silver. Anal. Biochem. 126:374-380. [DOI] [PubMed] [Google Scholar]
- 6.Boekhout, T. 2005. Gut feeling for yeasts. Nature 434:449-451. [DOI] [PubMed] [Google Scholar]
- 7.Boekhout, T., M. Renting, W. A. Scheffers, and R. Bosboom. 1993. The use of karyotyping in the systematics of yeasts. Antonie van Leeuwenhoek 63:157-163. [DOI] [PubMed] [Google Scholar]
- 8.Bovers, M., M. R. Diaz, F. Hagen, L. Spanjaard, B. Duim, C. E. Visser, H. L. Hoogveld, J. Scharringa, I. M. Hoepelman, J. W. Fell, and T. Boekhout. 2007. Identification of genotypically diverse Cryptococcus neoformans and Cryptococcus gattii isolates by Luminex xMAP technology. J. Clin. Microbiol. 45:1874-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, Y. C., J. D. Eisner, M. M. Kattar, S. L. Rassoulian-Barrett, K. Lafe, S. L. Yarfitz, A. P. Limaye, and B. T. Cookson. 2000. Identification of medically important yeasts using PCR-based detection of DNA sequence polymorphisms in the internal transcribed spacer 2 region of the rRNA genes. J. Clin. Microbiol. 38:2302-2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, Y.-C., J. D. Eisner, M. M. Kattar, S. L. Rassoulian-Barrett, K. Lafe, U. Bui, A. P. Limaye, and B. T. Cookson. 2001. Polymorphic internal transcribed spacer region 1 DNA sequences identify medically important yeasts. J. Clin. Microbiol. 39:4042-4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coignard, C., S. F. Hurst, L. E. Benjamin, M. E. Brandt, D. W. Warnock, and C. J. Morrison. 2004. Resolution of discrepant results for Candida species identification by using DNA probes. J. Clin. Microbiol. 42:858-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Llanos Frutos, R., M. T. Fernandez-Espinar, and A. Querol. 2004. Identification of species of the genus Candida by analysis of the 5.8S rRNA gene and the two ribosomal internal transcribed spacers. Antonie van Leeuwenhoek 85:175-185. [DOI] [PubMed] [Google Scholar]
- 13.Diaz, M. R., T. Boekhout, B. Theelen, M. Bovers, F. J. Cabañes, and J. W. Fell. 2006. Microcoding and flow cytometry as a high-throughput fungal identification system for Malassezia species. J. Med. Microbiol. 55:1197-1209. [DOI] [PubMed] [Google Scholar]
- 14.Dockhorn-Dworniczak, B., B. Dworniczak, L. Brömmelkamp, J. Bülles, J. Horst, and W. W. Böcker. 1991. Non-isotopic detection of single strand conformation polymorphism (PCR-SSCP): a rapid and sensitive technique in diagnosis of phenylketonuria. Nucleic Acids Res. 19:2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elie, C. M., T. J. Lott, E. Reiss, and C. J. Morrison. 1998. Rapid identification of Candida species with species-specific DNA probes. J. Clin. Microbiol. 36:3260-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Esteve-Zarzoso, B., C. Belloch, F. Uruburu, and A. Querol. 1999. Identification of yeasts by RFLP analysis of the 5.8S rRNA gene and the two ribosomal internal transcribed spacers. Int. J. Syst. Bacteriol. 49:329-337. [DOI] [PubMed] [Google Scholar]
- 17.Fell, J. W., T. Boekhout, A. Fonseca, G. Scorzetti, and A. Statzell-Tallman. 2000. Biodiversity and systematics of basidiomycetous yeasts as determined by large-subunit rDNA D1/D2 domain sequence analysis. Int. J. Syst. Evol. Microbiol. 50:1351-1371. [DOI] [PubMed] [Google Scholar]
- 18.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 19.Fernádez-Espinar, M. T., B. Esteve-Zarzoso, A. Querol, and E. Barrio. 2000. RFLP analysis of the ribosomal internal transcribed spacers and the 5.8S rRNA gene region of the genus Saccharomyces: a fast method for species identification and the differentiation of flor yeasts. Antonie van Leeuwenhoek 78:87-97. [DOI] [PubMed] [Google Scholar]
- 20.Gupta, A. K., T. Boekhout, B. Theelen, R. Summerbell, and R. Batra. 2004. Identification and typing of Malassezia species by amplified fragment length polymorphism and sequence analyses of the internal transcribed spacer and large-subunit regions of ribosomal DNA. J. Clin. Microbiol. 42:4253-4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hayashi, K. 1991. PCR-SSCP: a simple and sensitive method for detection of mutations in the genomic DNA. PCR Methods Appl. 1:34-38. [DOI] [PubMed] [Google Scholar]
- 22.Hayashi, K., and D. W. Yandell. 1993. How sensitive is PCR-SSCP? Hum. Mutat. 2:338-346. [DOI] [PubMed] [Google Scholar]
- 23.Highsmith, W. E., Jr., A. J. Nataraj, Q. Jin, J. M. O'Connor, S. H. El-Nabi, N. Kusukawa, and M. M. Garner. 1999. Use of DNA toolbox for the characterization of mutation scanning methods. II. Evaluation of single-strand conformation polymorphism analysis. Electrophoresis 20:1195-1203. [DOI] [PubMed] [Google Scholar]
- 24.Inácio, J., L. Portugal, I. Spencer-Martins, and A. Fonseca. 2005. Phylloplane yeasts from Portugal: seven novel anamorphic species in the Tremellales lineage of the Hymenomycetes (Basidiomycota) producing orange-coloured colonies. FEMS Yeast Res. 5:1167-1183. [DOI] [PubMed] [Google Scholar]
- 25.Innings, Å., M. Ullberg, A. Johansson, C. J. Rubin, N. Noreus, M. Isaksson, and B. Herrmann. 2007. Multiplex real-time PCR targeting the RNase P RNA gene for detection and identification of Candida species in blood. J. Clin. Microbiol. 45:874-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kimura, M. 1980. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 27.Kong, P., C. Hong, P. A. Richardson, and M. E. Gallegly. 2003. Single-strand-conformation polymorphism of ribosomal DNA for rapid species differentiation in genus Phytophthora. Fungal Genet. Biol. 39:238-249. [DOI] [PubMed] [Google Scholar]
- 28.Kumeda, Y., and T. Asao. 1996. Single-strand conformation polymorphism analysis of PCR-amplified ribosomal DNA internal transcribed spacers to differentiate species of Aspergillus section Flavi. Appl. Environ. Microbiol. 62:2947-2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kurtzman, C. P. 2003. Phylogenetic circumscription of Saccharomyces, Kluyveromyces and other members of the Saccharomycetaceae, and the proposal of the new genera Lachancea, Nakaseomyces, Naumovia, Vanderwaltozyma and Zygotorulaspora. FEMS Yeast Res. 4:233-245. [DOI] [PubMed] [Google Scholar]
- 30.Kurtzman, C. P., and J. W. Fell. 1998. Definition, classification and nomenclature of the yeasts, p. 3-5. In C. P. Kurtzman and J. W. Fell (ed.), The yeasts, a taxonomic study, 4th ed. Elsevier, Amsterdam, The Netherlands.
- 31.Kurtzman, C. P., and C. J. Robnett. 1997. Identification of clinically important ascomycetous yeasts based on nucleotide divergence in the 5′ end of the large-subunit (26S) ribosomal DNA gene. J. Clin. Microbiol. 35:1216-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kurtzman, C. P., and C. J. Robnett. 1998. Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie van Leeuwenhoek 73:331-371. [DOI] [PubMed] [Google Scholar]
- 33.Kurtzman, C. P., and C. J. Robnett. 2003. Phylogenetic relationships among yeasts of the 'Saccharomyces complex’ determined from multigene sequence analyses. FEMS Yeast Res. 3:417-432. [DOI] [PubMed] [Google Scholar]
- 34.Lachance, M.-A., and W. T. Starmer. 1998. Ecology and yeasts, p. 21-30. In C. P. Kurtzman and J. W. Fell (ed.), The yeasts, a taxonomic study, 4th ed. Elsevier, Amsterdam, The Netherlands.
- 35.Larsen, L. A., M. Christiansen, J. Vuust, and P. S. Andersen. 1999. High-throughput single-strand conformation polymorphism analysis by automated capillary electrophoresis: robust multiplex analysis and pattern-based identification of allelic variants. Hum. Mutat. 13:318-327. [DOI] [PubMed] [Google Scholar]
- 36.Leaw, S. N., H. C. Chang, H. F. Sun, R. Barton, J.-P. Bouchara, and T. C. Chang. 2006. Identification of medically important yeast species by sequence analysis of the internal transcribed spacer regions. J. Clin. Microbiol. 44:693-699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leaw, S. N., H. C. Chang, R. Barton, J.-P. Bouchara, and T. C. Chang. 2007. Identification of medically important Candida and non-Candida yeast species by an oligonucleotide array. J. Clin. Microbiol. 45:2220-2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leinberger, D. M., U. Schumacher, I. B. Autenrieth, and T. T. Bachmann. 2005. Development of a DNA microarray for detection and identification of fungal pathogens involved in invasive mycoses. J. Clin. Microbiol. 43:4943-4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li, J., and F.-Y. Bai. 2007. Single-strand conformation polymorphism of microsatellite for rapid strain typing of Candida albicans. Med. Mycol. 45:629-635. [DOI] [PubMed] [Google Scholar]
- 40.Li, J., Y.-C. Xu, and F.-Y. Bai. 2006. Candida pseudorugosa sp. nov., a novel yeast species from sputum. J. Clin. Microbiol. 44:4486-4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Loeffler, J., H. Hebart, S. Sepe, U. Schumacher, T. Klingebiel, and H. Einsele. 1998. Detection of PCR-amplified fungal DNA by using a PCR-ELISA system. Med. Mycol. 36:275-279. [DOI] [PubMed] [Google Scholar]
- 42.Luo, G., and T. G. Mitchell. 2002. Rapid identification of pathogenic fungi directly from cultures by using multiplex PCR. J. Clin. Microbiol. 40:2860-2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Makimura, K., Y. S. Murayama, and H. Yamaguchi. 1994. Detection of a wide range of medically important fungi by the polymerase chain reaction. J. Med. Microbiol. 40:358-364. [DOI] [PubMed] [Google Scholar]
- 44.Manzano, M., L. Cocolin, B. Longo, and G. Comi. 2004. PCR-DGGE differentiation of strains of Saccharomyces sensu stricto. Antonie van Leeuwenhoek 85:23-27. [DOI] [PubMed] [Google Scholar]
- 45.McCullough, M. J., K. V. Clemons, J. H. McCusker, and D. A. Stevens. 1998. Intergenic transcribed spacer PCR ribotyping for differentiation of Saccharomyces species and interspecific hybrids. J. Clin. Microbiol. 36:1035-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meroth, C. B., W. P. Hammes, and C. Hertel. 2003. Identification and population dynamics of yeasts in sourdough fermentation processes by PCR-denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 69:7453-7461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakase, T. 2000. Expanding world of ballistosporous yeasts: distribution in the phyllosphere, systematics and phylogeny. J. Gen. Appl. Microbiol. 46:189-216. [DOI] [PubMed] [Google Scholar]
- 48.Nataraj, A. J., I. Olivos-Glander, N. Kusukawa, and W. E. Highsmith, Jr. 1999. Single-strand conformation polymorphism and heteroduplex analysis for gel-based mutation detection. Electrophoresis 20:1177-1185. [DOI] [PubMed] [Google Scholar]
- 49.Naumov, G. I., S. A. James, E. S. Naumova, E. J. Louis, and I. N. Roberts. 2000. Three new species in the Saccharomyces sensu stricto complex: Saccharomyces cariocanus, Saccharomyces kudriavzevii and Saccharomyces mikatae. Int. J. Syst. Evol. Microbiol. 50:1931-1942. [DOI] [PubMed] [Google Scholar]
- 50.Orita, M., H. Iwahana, H. Kanazawa, K. Hayashi, and T. Sekiya. 1989. Detection of polymorphisms of human DNA by gel electrophoresis as single strand conformation polymorphisms. Proc. Natl. Acad. Sci. USA 86:2766-2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Orita, M., Y. Suzuki, T. Sekiya, and K. Hayashi. 1989. Rapid and sensitive detection of point mutations and DNA polymorphisms using the polymerase chain reaction. Genomics 5:874-879. [DOI] [PubMed] [Google Scholar]
- 52.Page, B. T., C. E. Shields, W. G. Merz, and C. P. Kurtzman. 2006. Rapid identification of ascomycetous yeasts from clinical specimens by a molecular method based on flow cytometry and comparison with identifications from phenotypic assays. J. Clin. Microbiol. 44:3167-3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pfaller, M. A., and D. J. Diekema. 2004. Rare and emerging opportunistic fungal pathogens: concern for resistance beyond Candida albicans and Aspergillus fumigatus. J. Clin. Microbiol. 42:4419-4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 55.Schabereiter-Gurtner, C., B. Selitsch, M. L. Rotter, A. M. Hirschl, and B. Willinger. 2007. Development of novel real-time PCR assays for detection and differentiation of eleven medically important Aspergillus and Candida species in clinical specimens. J. Clin. Microbiol. 45:906-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Scorzetti, G., J. W. Fell, A. Fonseca, and A. Statzell-Tallman. 2002. Systematics of basidiomycetous yeasts: a comparison of large subunit D1/D2 and internal transcribed spacer rDNA regions. FEMS Yeast Res. 2:495-517. [DOI] [PubMed] [Google Scholar]
- 57.Sheffield, V. C., J. S. Beck, A. E. Kwitek, D. W. Sandstrom, and E. M. Stone. 1993. The sensitivity of single-strand conformation polymorphism analysis for the detection of single base substitutions. Genomics 16:325-332. [DOI] [PubMed] [Google Scholar]
- 58.Sugita, T., A. Nishikawa, R. Ikeda, and T. Shinoda. 1999. Identification of medically relevant Trichosporon species based on sequences of internal transcribed spacer regions and construction of a database for Trichosporon identification. J. Clin. Microbiol. 37:1985-1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Suh, S.-O., J. V. McHugh, D. D. Pollock, and M. Blackwell. 2005. The beetle gut: a hyperdiverse source of novel yeasts. Mycol. Res. 109:261-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Theelen, B., M. Silvestri, E. Guého, A. van Belkum, and T. Boekhout. 2001. Identification and typing of Malassezia yeasts using amplified fragment length polymorphism (AFLP), random amplified polymorphic DNA (RAPD) and denaturing gradient gel electrophoresis (DGGE). FEMS Yeast Res. 1:79-86. [DOI] [PubMed] [Google Scholar]
- 61.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vaughan-Martini, A., and A. Martini. 1998. Saccharomyces Meyen ex Reess, p. 358-371. In C. P. Kurtzman and J. W. Fell (ed.), The yeasts, a taxonomic study, 4th ed. Elsevier, Amsterdam, The Netherlands.
- 63.Villa-Carvajal, M., A. Querol, and C. Belloch. 2006. Identification of species in the genus Pichia by restriction of the internal transcribed spacers (ITS1 and ITS2) and the 5.8S ribosomal DNA gene. Antonie van Leeuwenhoek 90:171-181. [DOI] [PubMed] [Google Scholar]
- 64.Walsh, T. J., A. Francesconi, M. Kasai, and S. J. Chanock. 1995. PCR and single-strand conformational polymorphism for recognition of medically important opportunistic fungi. J. Clin. Microbiol. 33:3216-3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang, Q.-M., and F.-Y. Bai. 2004. Four new yeast species of the genus Sporobolomyces from plant leaves. FEMS Yeast Res. 4:579-586. [DOI] [PubMed] [Google Scholar]
- 66.Wang, Q.-M., F.-Y. Bai, H.-Z. Lu, J.-H. Jia, and M. Takashima. 2004. Bullera cylindrica, Bullera hubeiensis and Bullera nakasei, ballistoconidium-forming yeast species isolated from plant leaves. Int. J. Syst. Evol. Microbiol. 54:1877-1882. [DOI] [PubMed] [Google Scholar]
- 67.Wang, S.-A., and F.-Y. Bai. 2008. Saccharomyces arboricolus sp. nov., a yeast species from tree bark. Int. J. Syst. Evol. Microbiol. 58:510-514. [DOI] [PubMed] [Google Scholar]
- 68.Yarrow, D. 1998. Methods for the isolation, maintenance and identification of yeasts, p. 77-100. In C. P. Kurtzman and J. W. Fell (ed.), The yeasts, a taxonomic study, 4th ed. Elsevier, Amsterdam, The Netherlands.