Abstract
Fluorescent amplified fragment length polymorphism revealed that strains of Xanthomonas axonopodis pv. phaseoli and Xanthomonas fuscans subsp. fuscans are genetically distinct and can be grouped into four genetic lineages. Four suppression subtractive hybridizations were then performed to isolate DNA fragments present in these bean pathogens and absent from closely related xanthomonads. Virulence gene candidates were identified such as homologs of hemagglutinins, TonB-dependent receptors, zinc-dependent metalloproteases, type III effectors, and type IV secretion system components. Unexpectedly, homologs of the type III secretion apparatus components (SPI-1 family), usually reported in animal pathogens and insect symbionts, were also detected.
Understanding the molecular mechanisms used by plant pathogens to attack their hosts is central to the study of plant pathology. Such fundamental knowledge is essential for the development of new strategies for the control of the economically important diseases caused by these microorganisms. Xanthomonas axonopodis pv. phaseoli (44) and Xanthomonas fuscans subsp. fuscans (40) (also designated Xanthomonas axonopodis pv. phaseoli variant fuscans [44]) are the causative agents of common bacterial blight of bean (Phaseolus vulgaris L.), a disease that occurs worldwide and leads to important yield losses (5). Both pathogens have the same host range and epidemiological features (45), but it has been reported that the X. fuscans subsp. fuscans strains are generally more aggressive toward their hosts than X. axonopodis pv. phaseoli strains (31, 45). Both bacteria also have similar biochemical phenotypes, except that X. fuscans subsp. fuscans can produce a melanin-like pigment in culture (16). Currently, nothing is known about the virulence and host specificity determinants of these bean pathogens. To identify such determinants and to be as exhaustive as possible, we decided in this study to consider the genetic diversity of both bean pathogens as the basis for performing several suppression subtractive hybridizations (SSHs). We first report the determination of a large genetic diversity within X. axonopodis pv. phaseoli and X. fuscans subsp. fuscans strains by using fluorescent amplified fragment length polymorphism (F-AFLP), which has never been used with these bean pathogens. AFLP is known to be a very powerful DNA fingerprinting technique that allows very fine discrimination and reliable determination of taxonomic and phylogenetic relationships between strains (15, 20, 33, 39). Then, we describe the results of our SSHs. We used SSH, since it is reported to be an effective approach in the identification of virulence determinants and genetic diversity in bacteria (2, 6, 18, 34, 37, 42, 47).
F-AFLP revealed that strains of X. axonopodis pv. phaseoli and X. fuscans subsp. fuscans can be grouped into four genetic lineages.
In this study, we first assessed the genetic diversity of a worldwide collection of X. axonopodis pv. phaseoli and X. fuscans subsp. fuscans strains by F-AFLP (Table 1). We also worked with a set of selected strains with different host specificities (Table 1) in order to choose representative strains of phylogenetically closely related xanthomonads as the driver for our SSH approach. F-AFLP experiments were performed and analyzed as previously described (39).
TABLE 1.
Bacterial strains used in this study
| Species (genetic lineage no.) | Straina | Host | Geographic origin | Yr of isolation |
|---|---|---|---|---|
| X. fuscans subsp. fuscansb | CFBP4834d | Phaseolus vulgaris | France | 1998 |
| CFBP6165 | Phaseolus vulgaris | Canada | 1957 | |
| CFBP6166 | Phaseolus vulgaris | South Africa | 1963 | |
| CFBP6167 | Phaseolus sp. | United States | 1954 | |
| CFBP6960 | Phaseolus vulgaris | Reunion Island | 2000 | |
| CFBP6970 | Unknown | United States | 1990 | |
| CFBP6971 | Unknown | Tanzania | 1992 | |
| X. axonopodis pv. phaseoli (1)c,f | CFBP2534g | Phaseolus vulgaris | United States | Unknown |
| CFBP6164d | Phaseolus vulgaris | Romania | 1966 | |
| CFBP6982 | Phaseolus vulgaris | Reunion Island | 2000 | |
| CFBP6983 | Phaseolus vulgaris | Reunion Island | 2000 | |
| CFBP6984 | Phaseolus vulgaris | Reunion Island | 2000 | |
| CFBP6985 | Phaseolus vulgaris | Reunion Island | 2000 | |
| X. axonopodis pv. phaseoli (2)f | CFBP6989d | Phaseolus vulgaris | Reunion Island | 2000 |
| CFBP6990 | Phaseolus vulgaris | Reunion Island | 2000 | |
| X. axonopodis pv. phaseoli (3)f | CFBP6992 | Phaseolus vulgaris | Reunion Island | 2000 |
| CFBP6994d | Unknown | Tanzania | 1990 | |
| CFBP6996 | Phaseolus vulgaris | Reunion Island | 2000 | |
| JW162.16 | Phaseolus vulgaris | Reunion Island | 2000 | |
| JW351.4 | Phaseolus vulgaris | Reunion Island | 2000 | |
| JW352.2 | Phaseolus vulgaris | Reunion Island | 2000 | |
| X. alfalfae subsp. alfalfaeb | CFBP3836 | Medicago sativa | Sudan | Unknown |
| X. alfalfae subsp. citrumelonisb | CFBP3371 | Unknown | Unknown | 1989 |
| X. citri subsp. citrib | CFBP2866e | Citrus aurantiifolia | Brazil | 1982 |
| X. citri subsp. malvacearumb | CFBP2530 | Gossypium hirsutum | Sudan | 1958 |
| X. fuscans subsp. aurantifoliib | CFBP3528e | Citrus limon | Argentina | 1988 |
| X. axonopodis pv. alliic | CFBP6107 | Allium fistulosum | Japan | 1998 |
| CFBP6369e,g | Allium cepa | Reunion Island | 1996 | |
| X. axonopodis pv. begoniaec | CFBP2524g | Begonia sp. | New Zealand | 1962 |
| X. axonopodis pv. glycinesc | CFBP2526g | Glycine hispida | Sudan | 1956 |
| X. axonopodis pv. manihotisc | CFBP2603 | Manihot esculenta | Colombia | 1972 |
| X. axonopodis pv. vesicatoriac | CFBP1604 | Capsicum annuum | Guadeloupe | Unknown |
| CFBP5600 | Lycopersicon esculentum | Martinique | 1993 | |
| X. axonopodisc | CFBP4924h | Axonopus scoparius | Colombia | 1949 |
CFBP, Collection Française des Bactéries Phytopathogènes (INRA, Angers, France); JW, bacterial collection of the Pole de Protection des Plantes (CIRAD, Reunion Island, Saint-Pierre, France).
Taxonomy as proposed by Schaad et al. (40).
Taxonomy as proposed by Vauterin et al. (44).
Representative strains of the four genetic lineages of X. axonopodis pv. phaseoli and X. fuscans subsp. fuscans, as determined by our F-AFLP analyses (Fig. 1) and used separately as testers in our four SSHs.
Strains used as drivers in our four SSHs.
Genetic lineages 1, 2, and 3 represent the three genetic lineages of X. axonopodis pv. phaseoli, as revealed by our F-AFLP analyses (Fig. 1).
Pathotype strains.
Type strain.
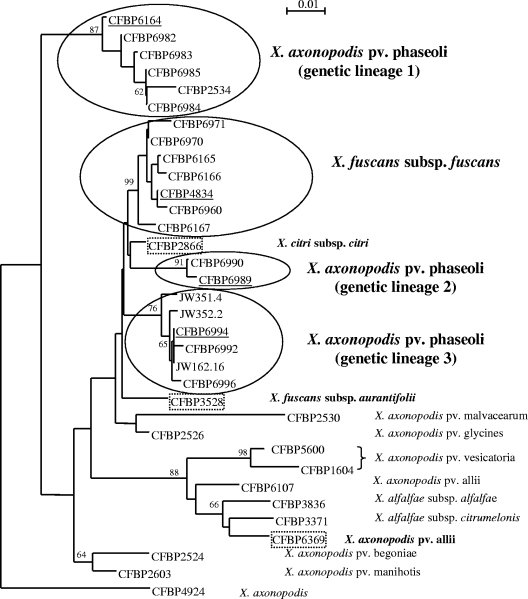
Interestingly, this study revealed that strains of X. axonopodis pv. phaseoli and X. fuscans subsp. fuscans are genetically different and can be grouped into four distinct genetic lineages (Fig. 1). The X. axonopodis pv. phaseoli strains are distributed within three lineages, and the X. fuscans subsp. fuscans strains formed the remaining lineage. High bootstrap values indicated that this clustering is well supported and that the dendrogram was robust (Fig. 1). Therefore, this result provides further data to show that X. axonopodis pv. phaseoli and X. fuscans subsp. fuscans strains are genetically distinct and that strains of X. axonopodis pv. phaseoli are more heterogeneous than those of X. fuscans subsp. fuscans (1, 3, 7, 19, 25, 26, 29, 40, 44). Moreover, F-AFLP provides new information, since it is the first technique that allows the identification of three distinct genetic lineages for X. axonopodis pv. phaseoli. Interestingly, genetic lineage 1 of X. axonopodis pv. phaseoli appears phylogenetically distant from genetic lineages 2 and 3, which are more closely related to X. fuscans subsp. fuscans. Furthermore, it is worth noting that this F-AFLP genetic clustering was supported by our SSH results, since strains belonging to genetic lineage 1 carry numerous DNA sequences that are not present in the other genetic lineages, such as those encoding a putative type III secretion system of the Salmonella pathogenicity island-1 (SPI-1) protein family (Table 2). Altogether, our investigations highlight the need for a novel taxonomic study including representative strains of the four newly identified genetic lineages, since it might lead to the reclassification of these strains into new species or subspecies.
FIG. 1.
A dendrogram constructed by using the neighbor-joining method shows the phylogenetic relationships of AFLP fingerprints of X. fuscans subsp. fuscans and X. axonopodis pv. phaseoli strains and related xanthomonads. The representative strains of the four genetic lineages of these bean pathogens (black circles), selected as testers for our four SSHs, are underlined. The three closely related xanthomonad (X. axonopodis pv. allii [44], X. fuscans subsp. aurantifolii [40], and Xanthomonas citri subsp. citri [40]) strains, selected as the drivers for our four SSHs, are indicated by dotted boxes. Numbers at the branch points represent the bootstrap values (1,000 replicates). Only high bootstrap values (>60) are displayed. Strain CFBP4924 is the type strain of the X. axonopodis species (44).
TABLE 2.
Sequence and distribution analyses of the 39 unique DNA fragments subtracted from the four tester strains of X. axonopodis pv. phaseoli and X. fuscans subsp. fuscans
| SSH clonea | Size (bp) | % G+C content | Southern blot hybridization result with DNA probes fromd:
|
BLASTX results
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Driver | X. fuscans CFBP 4834 | X. axonopodis CFBP 6164 | X. axonopodis CFBP 6989 | X. axonopodis CFBP 6994 | Predicted function | E value(s) | GenBank accession no. | Organism | |||
| 4834-1b,c | 713 | 47 | − | + | + | + | + | AvrBsT protein; ISXax1 transposase | 5 E−96; 2 E−37 | AAD39255; AY935340 | X. axonopodis pv. vesicatoria; X. axonopodis pv. phaseoli |
| 4834-2b | 745 | 57 | − | + | − | − | − | Type IV secretion system protein TraF (VirB5) | 5 E−56 | YP_199242 | X. oryzae pv. oryzae |
| 4834-3 | 1,081 | 48 | − | + | + | − | − | Xanthomonas outer protein C (XopC) | 8 E−154 | YP_364166 | X. axonopodis pv. vesicatoria |
| 4834-4b,c | 1,149 | 48 | − | + | + | − | − | Xanthomonas outer protein C (XopC); ISXcd1 transposase | 5 E−86; 2 E−4 | YP_364166; NP_637461 | X. axonopodis pv. vesicatoria; X. campestris pv. campestris |
| 4834-5 | 1,118 | 55 | − | + | − | − | − | Integral membrane protein (WxcO) | 8 E−10 | AAK53474 | X. campestris pv. campestris |
| 4834-6b | 779 | 62 | − | + | − | + | + | Conserved hypothetical protein | 2 E−45 | YP_342068 | Nitrosococcus oceani |
| 4834-7c | 1,102 | 50 | − | + | − | − | − | Conserved hypothetical protein; C-5 cytosine-specific DNA methylase | 4.9 E−41; 6 E−37 | ZP_00681364; ZP_00681363 | Xylella fastidiosa; Xylella fastidiosa |
| 6164-1b | 598 | 40 | − | − | + | − | − | Type III secretion system protein (PrgH/EprH) | 3 E−20 | YP_001063173 | Burkholderia pseudomallei |
| 6164-2b | 864 | 60 | − | − | + | − | − | Type III secretion system protein (YscJ/HrcJ) | 7 E−27 | ZP_01768663 | Burkholderia pseudomallei |
| 6164-3 | 602 | 52 | − | − | + | − | − | Type III secretion system protein (BsaQ) | 4 E−07 | YP_106131 | Burkholderia mallei |
| 6164-4c | 508 | 65 | − | + | + | + | + | ISXax1 transposase; l-threonine 3-dehydrogenase | 5 E−50; 2 E−17 | AY935340; YP_362783 | X. axonopodis pv. phaseoli; X. axonopodis pv. vesicatoria |
| 6164-5c | 548 | 65 | − | + | + | + | + | ISXax1 transposase; hypothetical protein | 4 E−39; 3 E−43 | AY935340; YP_364143 | X. axonopodis pv. phaseoli; X. axonopodis pv. vesicatoria |
| 6164-6 | 480 | 40 | − | − | + | − | − | Putative type I restriction modification system protein | 4 E−54 | NP_717074 | Shewanella oneidensis |
| 6164-7c | 659 | 659 | − | + | + | + | + | ISXac3 transposase; putative TraI protein (TrwC) | 4 E−19; 3 E−42 | NP_643556; ABA25997 | X. campestris pv. campestris; Pseudomonas sp. |
| 6164−8b | 640 | 58 | − | − | + | − | − | Hypothetical protein | 5 E−59 | ZP_01365986 | Pseudomonas aeruginosa |
| 6164-9 | 759 | 63 | − | − | + | − | − | Hypothetical protein | 4 E−47 | AAM37073 | X. axonopodis pv. citri |
| 6164-10b | 556 | 53 | − | − | + | − | − | Hypothetical protein | 1 E−11 | ZP_01642272 | Stenotrophomonas maltophilia |
| 6164-11 | 579 | 65 | − | − | + | − | − | Unknown function | |||
| 6164-12 | 586 | 47 | − | − | + | − | − | Unknown function | |||
| 6164-13 | 563 | 34 | − | − | + | − | − | Unknown function | |||
| 6164-14b | 553 | 34 | − | − | + | − | − | Unknown function | |||
| 6164-15 | 717 | 47 | − | − | + | − | − | Unknown function | |||
| 6164-16 | 666 | 50 | − | − | + | − | − | Unknown function | |||
| 6164-17b | 745 | 36 | − | − | + | − | − | Unknown function | |||
| 6989-1 | 688 | 41 | − | − | − | + | − | Filamentous hemagglutinin-related protein | 2 E−51 | NP_642141 | X. axonopodis pv. vesicatoria |
| 6989-2 | 1,074 | 41 | − | − | − | + | − | Putative surface adhesin | 4 E−40 | YP_001098807 | Herminiimonas arsenicoxydans |
| 6989-3c | 730 | 59 | − | − | − | + | + | Site-specific DNA methyltransferase; hypothetical protein | 2.1 E−14; 3 E−07 | NP_638315ZP_00415806 | X. campestris pv. campestris; Azotobacter vinelandii |
| 6989−4 | 824 | 55 | − | + | − | + | + | Hypothetical protein | 5 E−05 | YP_363001 | X. axonopodis pv. vesicatoria |
| 6989-5 | 587 | 54 | − | − | − | + | − | Hypothetical protein | 3 E−18 | NP_643587 | X. axonopodis pv. citri |
| 6989-6 | 1,087 | 42 | − | − | − | + | − | Hypothetical protein | 5 E−18 | ZP_01638616 | Pseudomonas putida |
| 6989-7 | 658 | 41 | − | − | − | + | + | Unknown function | |||
| 6989-8b | 650 | 50 | − | − | − | + | − | Unknown function | |||
| 6994-1c | 793 | 46 | − | + | + | + | + | AvrBsT protein; ISXac2 transposase | 7 E−97 0.063 | AAD39255YP_363027 | X. axonopodis pv. vesicatoria; X. axonopodis pv. vesicatoria |
| 6994-2b | 244 | 54 | − | + | + | − | + | TonB-dependent receptor | 9.6 E−20 | NP_779483 | Xylella fastidiosa |
| 6994-3 | 439 | 40 | − | + | + | + | + | Putative transposase | 8 E−67 | NP_943128 | Pseudomonas sp. |
| 6994-4b | 361 | 61 | − | + | + | + | + | Putative secreted protein (putative zinc-dependent metalloprotease) | 4 E−56 | YP_365137 | X. axonopodis pv. vesicatoria |
| 6994-5 | 297 | 57 | − | + | + | + | + | Conserved hypothetical protein | 3 E−14 | YP_452829 | X. oryzae pv. oryzae |
| 6994-6 | 347 | 56 | − | + | + | + | + | Conserved hypothetical protein | 4 E−11 | YP_342069 | Nitrosococcus oceani |
| 6994-7 | 166 | 64 | − | + | + | − | + | Unknown function | |||
The first number of each SSH clone indicates the tester strain (X. fuscans subsp. fuscans CFBP4834, X. axonopodis pv. phaseoli CFBP6164, X. axonopodis pv. phaseoli CFBP6989, or X. axonopodis pv. phaseoli CFBP6994) used in each SSH experiment.
The insert sequences from these clones were found in more than one clone.
Clones contained sequences from two genes (based on BLASTX results).
The distribution of each clone was analyzed by Southern blot hybridization experiments with genomic DNA from driver strains (CFBP3528, CFBP6369, and CFBP2866) or tester strains (X. fuscans subsp. fuscans CFBP4834, X. axonopodis pv. phaseoli CFBP6164, X. axonopodis pv. phaseoli CFBP6989, or X. axonopodis pv. phaseoli CFBP6994) as probes.
Strains of the four genetic lineages were all pathogenic in bean and appeared to be genetically distinct from strains of other xanthomonads that are pathogenic in different host plants. These results suggest not only that strains of the four genetic lineages share specific DNA sequences that may be involved in pathogenicity in bean but also that these strains possess different DNA sequences that could account for a distinct host range. Pathogenicity tests for a large host range are under way to determine whether this genetic diversity could be related to distinct host ranges. In these pathogenicity tests, it will be interesting to include strains that are closely related to bean pathogens based on F-AFLP, such as the X. citri subsp. citri CFBP2866 strain (Fig. 1), in order to determine whether these strains could also be pathogenic in bean.
SSHs confirmed the genetic heterogeneity of X. axonopodis pv. phaseoli and of X. fuscans subsp. fuscans and revealed DNA fragments likely acquired by horizontal gene transfers.
Four SSH experiments were then performed by selecting as the tester representative strains of each genetic lineage (X. fuscans subsp. fuscans CFBP4834, X. axonopodis pv. phaseoli CFBP6164, X. axonopodis pv. phaseoli CFBP6989, and X. axonopodis pv. phaseoli CFBP6994) and by selecting as the driver a mixture of strains belonging to three closely related xanthomonads, as revealed by F-AFLP: X. fuscans subsp. aurantifolii (strain CFBP3528) (40) and X. citri subsp. citri (strain CFBP2866) (40), the causal agents of citrus canker, and X. axonopodis pv. allii (strain CFBP6369) (44), the causal agent of bacterial blight of onion. These four subtractions were achieved according to the protocol of the PCR-Select bacterial genome kit (Clontech), except that the hybridization temperature was increased from 63°C to 73°C due to the high G+C content (∼65%) in the genomes of these Xanthomonas strains (44). For each subtractive library, the specificity of inserts from selected recombinant clones was checked by performing Southern blot hybridizations using tester and driver genomic DNAs separately as probes. Inserts from tester-specific clones were sequenced at the Station Biologique de Roscoff (France), and sequences were examined by using BLASTN and BLASTX programs (http://www.ncbi.nlm.nih.gov/BLAST/). Genomic signatures of subtracted DNA sequences were also analyzed by using Genstyle software (http://genstyle.imed.jussieu.fr/).
First, we analyzed 353 clones obtained from four subtractive libraries by Southern blot hybridizations, as described above. This analysis showed that only 75 out of the 353 subtracted DNA fragments could be considered specific to the tester strains, since they were not detected in the driver strains. Then, sequencing these 75 DNA fragments allowed the identification of 39 unique DNA sequences, as redundant sequences were revealed (Table 2). Therefore, our investigation revealed that X. axonopodis pv. phaseoli and X. fuscans subsp. fuscans tester strains contained at least 39 DNA sequences that are not shared by driver strains belonging to closely related xanthomonads (X. axonopodis pv. allii, X. citri subsp. citri, and X. fuscans subsp. aurantifolii). Interestingly, almost half (17) of these sequences were from the X. axonopodis CFBP6164 strain. This result is consistent with that of our F-AFLP analysis (Fig. 1), since the X. axonopodis CFBP6164 strain is less closely related to the driver strains than to the three other tester strains. Another point of interest is that by performing further Southern blot hybridizations using tester-genomic DNA as probes, only 9 out of the 39 subtracted DNA fragments were shown to be shared by the four tester strains (Table 2). This result demonstrates that these bean pathogens are genetically heterogeneous, further confirming our F-AFLP analysis. It would now be interesting to extend this study in order to know the distribution of the 39 subtracted DNA fragments, not only in the many strains belonging to the four newly identified genetic lineages of X. axonopodis pv. phaseoli and X. fuscans subsp. fuscans but also in other xanthomonads strains that are pathogenic for different host plants. Such a study would determine whether these DNA fragments could be considered bean-specific pathogens or lineage-specific pathogens. These distribution studies combined with pathogenicity tests, as described above, should impact future functional studies of genes that could play a role in the X. axonopodis pv. phaseoli-bean and X. fuscans subsp. fuscans-bean interactions.
Sequencing of the subtracted DNA fragments revealed that the G+C content of the majority of these sequences was considerably lower (average value, 51%) (Table 2) than the average value of total DNA for X. axonopodis pv. phaseoli (∼65%) (44). This result confirms the tendency of SSH in the identification of A+T-rich regions (47) and suggests that many of the subtracted DNA fragments may have been acquired from other organisms by horizontal gene transfers. Sequence analyses showed that the DNA fragments subtracted from X. axonopodis pv. phaseoli and from X. fuscans subsp. fuscans have high identities with sequences from other Gammaproteobacteria (Xanthomonas sp., Xylella sp., Pseudomonas sp., Azotobacter sp., Shewanella sp., Nitrosococcus sp., Stenotrophomonas sp.) and more strikingly from Betaproteobacteria (Burkholderia sp., Herminiimonas sp.) (Table 2). Altogether, these observations support the idea that Xanthomonas genomes have been subjected to numerous horizontal transfer events during evolution and sometimes from phylogenetically distant bacteria (9, 23).
Pathogenicity gene candidates of X. axonopodis pv. phaseoli and X. fuscans subsp. fuscans are identified by SSHs.
Sequence analyses revealed that the 39 different subtracted DNA fragments can be assigned to diverse functional classes: metabolism, transposase, membrane structure, adhesion, secretion, and unknown functions (Table 2). An interesting feature is that the ISXax1 element was identified. This result demonstrates the effectiveness of our SSHs, as we recently reported that this IS element is carried only by the X. axonopodis pv. phaseoli, X. fuscans subsp. fuscans and X. axonopodis pv. vesicatoria strains (1).
Another interesting feature of our SSHs is the identification of several homologs to known genes encoding proteins involved in the pathogenicity and/or host specificity of bacteria (13/39 subtracted DNA fragments). For instance, two subtracted DNA fragments have significant similarities to putative filamentous hemagglutinins and surface adhesins (Table 2). Such proteins have already been shown to contribute to the virulence of several proteobacteria, and they could be considered good candidates for identifying determinants that control host specificity, since bacterial attachment to host tissues by these proteins is a first step in pathogenesis (14, 24, 36). Another subtracted DNA fragment is a TonB-dependent receptor homolog (Table 2). TonB-dependent receptors are outer membrane proteins known mainly for the active transport of iron-siderophore complexes in gram-negative bacteria, but some of them have been shown to play a major role in plant-X. campestris pv. campestris interactions (4, 46). We also identified a homolog of a putative secreted protein harboring a putative zinc-dependent metalloprotease motif (Table 2). Similar metalloproteases have been reported to account for the virulence or the host specificity of several gram-negative bacteria (22, 28). The identification of TraF and TraI homologs (Table 2) strongly suggests that the X. axonopodis pv. phaseoli and X. fuscans subsp. fuscans strains harbor a putative type IV secretion system, like other Xanthomonas strains (13, 35, 41), even though its contribution to virulence was demonstrated only with the X. campestris pv. campestris strain 8004 (35). We are currently investigating the roles of all of these genes in the virulence or host specificity of X. fuscans subsp. fuscans and X. axonopodis pv. phaseoli strains.
Interestingly, we also isolated two homologs of genes encoding type III secretion system (T3SS) effector genes avrBsT and xopC (Table 2). Both genes have been reported for only a few strains of X. axonopodis pv. vesicatoria, the causal agent of the bacterial spot disease of pepper and tomato (8, 27, 30, 38; our unpublished data). The avrBsT homolog may play a significant role in the pathogenicity of X. axonopodis pv. phaseoli and X. fuscans subsp. fuscans, since further Southern blot hybridizations and PCRs showed that this gene is carried by all strains belonging to these bean pathogens (Fig. 1 and Table 2; data not shown). AvrBsT is a member of the YopJ/AvrRxv protein family that is widely distributed in proteobacteria and is predicted to encode a Cys protease that targets intracellular host proteins (11, 32). AvrBsT from X. axonopodis pv. vesicatoria triggers the hypersensitive response from pepper plants, but its virulence contribution was not demonstrated (11). Regarding the xopC homolog, it appeared to be harbored only by strains belonging to X. fuscans subsp. fuscans and to the genetic lineage 1 of X. axonopodis pv. phaseoli, based on Southern blot hybridizations and PCRs (Fig. 1, Table 2, data not shown). The biochemical function of XopC remains unknown (30, 38). Studies to determine the roles of avrBsT and xopC homologs in the interactions between the X. axonopodis pv. phaseoli and X. fuscans subsp. fuscans strains and bean plants are in progress.
A putative type III secretion system of the SPI-1 family is detected in one X. axonopodis pv. phaseoli genetic lineage.
Strikingly, we found significant similarities between putative proteins encoded by three subtracted DNA fragments and components of a type III secretion system belonging to the SPI-1 family (10) (Table 2). This T3SS family is usually found in animal pathogens and insect symbionts and is required for host cell invasion (10, 43). These subtracted sequences were detected only in the X. axonopodis pv. phaseoli CFBP6164 strain which belongs to the X. axonopodis pv. phaseoli genetic lineage 1 (Fig. 1). Further Southern blot hybridizations and PCRs revealed that only strains belonging to this genetic lineage carry these DNA sequences (Table 1 and Table 2; data not shown). Moreover, by using specific PCR primers (12), we tried to detect in strains belonging to the four genetic lineages the T3SS of the Hrp2 family that has been identified in many xanthomonads (10, 13, 17, 35, 41, 43). Our data strongly suggest that genetic lineage 1 strains of X. axonopodis pv. phaseoli have two T3SSs (types Hrp2 and SPI-1), whereas those of the three others genetic lineages have only one T3SS (the Hrp2 type). What could be the contribution of the putative SPI-1 T3SS in this particular group of X. axonopodis pv. phaseoli strains? It is worth noting that a T3SS of the SPI-1 family was recently disclosed by using SSH with Erwinia amylovora, the causal agent of fire blight of apple and pear (42). It has been speculated that this SPI-1 T3SS may allow interactions of E. amylovora and insects involved in the pathogen spread (42). Interestingly, the transmission of X. axonopodis pv. phaseoli by insects was reported, but the precise interaction between the bacterium and the insect host remains poorly understood (21). Does this T3SS play a role in the X. axonopodis pv. phaseoli life cycle? Our main objectives are now to get the complete sequence of this T3SS, to study its distribution and evolution in xanthomonads as well as in other plant pathogenic bacteria, and to study its functionality in interactions with plant and animal cells.
In conclusion, this study helped us to identify pathogenicity gene candidates for X. axonopodis pv. phaseoli and X. fuscans subsp. fuscans strains and a putative type III secretion apparatus that is usually not found in plant pathogenic bacteria. It also provides new insights into the diversity and evolution of these plant pathogenic bacteria. Finally, this work provides an excellent basis for further exploration of the specific interaction between the X. axonopodis pv. phaseoli or X. fuscans subsp. fuscans strain and bean and before the forthcoming genome sequencing of our model strain, X. fuscans subsp. fuscans CFBP4834.
Acknowledgments
This work was supported by INRA and Région Pays de la Loire.
S. M. Alavi is supported by a grant from NIGEB, Tehran, Iran.
We thank T. Boureau (Université d'Angers) and M. A. Jacques (INRA, Angers) for critical reading of the manuscript.
Footnotes
Published ahead of print on 21 March 2008.
REFERENCES
- 1.Alavi, S. M., S. Poussier, and C. Manceau. 2007. Characterization of ISXax1, a novel insertion sequence restricted to Xanthomonas axonopodis pv. phaseoli (variants fuscans and non-fuscans) and Xanthomonas axonopodis pv. vesicatoria. Appl. Environ. Microbiol. 73:1678-1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernier, S. P., and P. A. Sokol. 2005. Use of suppression-subtractive hybridization to identify genes in the Burkholderia cepacia complex that are unique to Burkholderia cenocepacia. J. Bacteriol. 187:5278-5291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birch, P. R. J., L. J. Hyman, R. Taylor, A. F. Opio, C. Bragard, and I. K. Toth. 1997. RAPD PCR-based differentiation of Xanthomonas campestris pv. phaseoli from Xanthomonas campestris pv. phaseoli var. fuscans. Eur. J. Plant Pathol. 103:809-814. [Google Scholar]
- 4.Blanvillain, S., D. Meyer, A. Boulanger, M. Lautier, C. Guynet, N. Denance, J. Vasse, E. Lauber, and M. Arlat. 2007. Plant carbohydrate scavenging through TonB-dependent receptors: a feature shared by phytopathogenic and aquatic bacteria. PLoS ONE 21:e224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Broughton, W. J., G. Hernandez, M. Blair, S. Beebe, P. Gepts, and J. Vanderleyen. 2003. Beans (Phaseolus spp.)-model food legumes. Plant Soil 252:55-128. [Google Scholar]
- 6.Castaneda, A., J. D. Reddy, B. El-Yacoubi, and D. W. Gabriel. 2005. Mutagenesis of all eight avr genes in Xanthomonas campestris pv. campestris had no detected effect on pathogenicity but one avr gene affected race specificity. Mol. Plant Microbe Interact. 18:1306-1317. [DOI] [PubMed] [Google Scholar]
- 7.Chan, J. W. Y. F., and P. H. Goodwin. 1999. Differentiation of Xanthomonas campestris pv. phaseoli from Xanthomonas campestris pv. phaseoli var. fuscans by PFGE and RFLP. Eur. J. Plant Pathol. 105:867-878. [Google Scholar]
- 8.Ciesiolka, L. D., T. Hwin, J. D. Gearlds, G. V. Minsavage, R. Saenz, M. Bravo, V. Handley, S. M. Conover, H. Zhang, J. Caporgno, N. B. Phengrasamy, A. O. Toms, R. E. Stall, and M. C. Whalen. 1999. Regulation of expression of avirulence gene avrRxv and identification of a family of host interaction factors by sequence analysis of avrBsT. Mol. Plant Microbe Interact. 12:35-44. [DOI] [PubMed] [Google Scholar]
- 9.Comas, I., A. Moya, R. K. Azad, J. G. Lawrence, and F. Gonzalez-Candelas. 2006. The evolutionary origin of Xanthomonadales genomes and the nature of the horizontal gene transfer process. Mol. Biol. Evol. 23:2049-2057. [DOI] [PubMed] [Google Scholar]
- 10.Cornelis, G. R. 2006. The type III secretion injectisome. Nat. Rev. Microbiol. 4:811-825. [DOI] [PubMed] [Google Scholar]
- 11.Cunnac, S., A. Wilson, J. Nuwer, A. Kirik, G. Baranage, and M. B. Mudgett. 2007. A conserved carboxylesterase is a suppressor of AvrBsT-elicited resistance in Arabidopsis. Plant Cell 19:688-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darsonval, A., A. Darrasse, D. Meyer, M. Demarty, K. Durand, C. Bureau, C. Manceau, and M.-A. Jacques. 2008. The type III secretion system of Xanthomonas fuscans subsp. fuscans is involved in the phyllosphere colonization process and in transmission to seeds of susceptible beans. Appl. Environ. Microbiol. 74:2669-2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.da Silva, A. C., J. A. Ferro, F. C. Reinach, C. S. Farah, L. R. Furlan, R. B. Quaggio, C. B. Monteiro-Vitorello, M. A. Van Sluys, N. F. Almeida, L. M. Alves, A. M. do Amaral, M. C. Bertolini, L. E. Camargo, G. Camarotte, F. Cannavan, J. Cardozo, F. Chambergo, L. P. Ciapina, R. M. Cicarelli, L. L. Coutinho, J. R. Cursino-Santos, H. El-Dorry, J. B. Faria, A. J. Ferreira, R. C. Ferreira, M. I. Ferro, E. F. Formighieri, M. C. Franco, C. C. Greggio, A. Gruber, A. M. Katsuyama, L. T. Kishi, R. P. Leite, E. G. Lemos, M. V. Lemos, E. C. Locali, M. A. Machado, A. M. Madeira, N. M. Martinez-Rossi, E. C. Martins, J. Meidanis, C. F. Menck, C. Y. Miyaki, D. H. Moon, L. M. Moreira, M. T. Novo, V. K. Okura, M. C. Oliveira, V. R. Oliveira, H. A. Pereira, A. Rossi, J. A. Sena, C. Silva, R. F. de Souza, L. A. Spinola, M. A. Takita, R. E. Tamura, E. C. Teixeira, R. I. Tezza, M. Trindade dos Santos, D. Truffi, S. M. Tsai, F. F. White, J. C. Setubal, and J. P. Kitajima. 2002. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 417:459-463. [DOI] [PubMed] [Google Scholar]
- 14.Feil, H., W. S. Feil, and S. E. Lindow. 2007. Contribution of fimbrial and afimbrial adhesins of Xylella fastidiosa to attachment to surfaces and virulence to grape. Phytopathology 97:318-324. [DOI] [PubMed] [Google Scholar]
- 15.Gonzalez, C., B. Szurek, C. Manceau, T. Mathieu, Y. Séré, and V. Verdier. 2007. Molecular and pathotypic characterization of new Xanthomonas oryzae strains from West Africa. Mol. Plant-Microbe Interact. 20:534-546. [DOI] [PubMed] [Google Scholar]
- 16.Goodwin, P. H., and C. R. Sopher. 1994. Brown pigmentation of Xanthomonas campestris pv. phaseoli associated with homogentistic acid. Can. J. Microbiol. 40:28-34. [Google Scholar]
- 17.Gürlebeck, D., F. Thieme, and U. Bonas. 2006. Type III effector proteins from the plant pathogen Xanthomonas and their role in the interaction with the host plant. J. Plant Physiol. 163:233-255. [DOI] [PubMed] [Google Scholar]
- 18.Harakava, R., and D. W. Gabriel. 2003. Genetic differences between two strains of Xylella fastidiosa revealed by suppression subtractive hybridization. Appl. Environ. Microbiol. 69:1315-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hildebrand, D. C., N. J. Palleroni, and M. N. Schroth. 1990. Deoxyribonucleic acid relatedness of 24 xanthomonad strains representing 23 Xanthomonas campestris pathovars and Xanthomonas fragariae. J. Appl. Bacteriol. 68:263-269. [Google Scholar]
- 20.Janssen, P., R. Coopman, G. Huys, J. Swings, M. Bleeker, P. Vos, M. Zabeau, and K. Kersters. 1996. Evaluation of the DNA fingerprinting method AFLP as a new tool in bacterial taxonomy. Microbiology 142:1881-1893. [DOI] [PubMed] [Google Scholar]
- 21.Kaiser, W. J., and N. G. Vakili. 1978. Insect transmission of pathogenic xanthomonads to bean and cowpea in Puerto Rico. Phytopathology 68:1057-1063. [Google Scholar]
- 22.Kooi, C., B. Subsin, R. Chen, B. Pohorelic, and P. A. Sokol. 2006. Burkholderia cenocepacia ZmpB is a broad-specificity zinc metalloprotease involved in virulence. Infect. Immun. 74:4083-4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lima, W., M. A. Van Sluys, and C. F. Menck. 2005. Non-gamma Proteobacteria gene islands contribute to the Xanthomonas genome. OMICS 9:160-172. [DOI] [PubMed] [Google Scholar]
- 24.Locht, C., R. Antoine, and F. Jacob-Dubuisson. 2001. Bordetella pertussis, molecular pathogenesis under multiple aspects. Curr. Opin. Microbiol. 4:82-89. [DOI] [PubMed] [Google Scholar]
- 25.López, R., C. Asensio, and R. L. Gilbertson. 2006. Phenotypic and genetic diversity in strains of common blight bacteria (Xanthomonas campestris pv. phaseoli and X. campestris pv. phaseoli var. fuscans) in a secondary center of diversity of the common bean host suggests multiple introduction events. Phytopathology 96:1204-1213. [DOI] [PubMed] [Google Scholar]
- 26.Mahuku, G. S., C. Jara, M. A. Henriquez, G. Castellanos, and J. Cuasquer. 2006. Genotypic characterization of the common bean bacterial blight pathogens, Xanthomonas axonopodis pv. phaseoli and Xanthomonas axonopodis pv. phaseoli var. fuscans by rep-PCR and PCR-RFLP of the ribosomal genes. J. Phytopathol. 154:35-44. [Google Scholar]
- 27.Minsavage, G. V., D. Dahlbeck, M. C. Whalen, B. Kearney, U. Bonas, B. J. Staskawicz, and R. E. Stall. 1990. Gene-for-gene relationship specifying disease resistance in Xanthomonas campestris pv. vesicatoria-pepper interaction. Mol. Plant-Microbe Interact. 3:41-47. [Google Scholar]
- 28.Miyoshi, S. I., and S. Shinoda. 2000. Microbial metalloproteases and pathogenesis. Microbes Infect. 2:91-98. [DOI] [PubMed] [Google Scholar]
- 29.Mkandawire, A. B. C., R. B. Mabagala, P. Guzman, P. Gepts, and R. L. Gilbertson. 2004. Genetic diversity and pathogenic variation of common blight bacteria (Xanthomonas campestris pv. phaseoli and X. campestris pv. phaseoli var. fuscans) suggests pathogen coevolution with the common bean. Phytopathology 94:593-603. [DOI] [PubMed] [Google Scholar]
- 30.Noël, L., F. Thieme, J. Gabler, D. Buttner, and U. Bonas. 2003. XopC and XopJ, two novel type III effector proteins from Xanthomonas campestris pv. vesicatoria. J. Bacteriol. 185:7092-7102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Opio, A. F., D. J. Allen, and J. M. Teri. 1996. Pathogenic variation in Xanthomonas campestris pv. phaseoli, the causal agent of common bacterial blight in Phaseolus beans. Plant Pathol. 45:1126-1133. [Google Scholar]
- 32.Orth, K., Z. Xu, M. B. Mudgett, Z. Q. Bao, L. E. Palmer, J. B. Bliska, W. F. Mangel, B. J. Staskawicz, and J. E. Dixon. 2000. Disruption of signaling by Yersinia effector YopJ, a ubiquitin-like protein protease. Science 290:1594-1597. [DOI] [PubMed] [Google Scholar]
- 33.Poussier, S., D. Trigalet-Demery, P. Vandewalle, B. Goffinet, J. Luisetti, and A. Trigalet. 2000. Genetic diversity of Ralstonia solanacearum as assessed by PCR-RFLP of the hrp gene region, AFLP and 16S rRNA sequence analysis, and identification of an African subdivision. Microbiology 146:1679-1692. [DOI] [PubMed] [Google Scholar]
- 34.Qi, M., K. E. Nelson, S. C. Daugherty, W. C. Nelson, I. R. Hance, M. Morrison, and C. W. Forsberg. 2005. Novel molecular features of the fibrolytic intestinal bacterium Fibrobacter intestinalis not shared with Fibrobacter succinogenes as determined by suppressive subtractive hybridization. J. Bacteriol. 187:3739-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qian, W., Y. Jia, S. X. Ren, Y. Q. He, J. X. Feng, L. F. Lu, Q. Sun, G. Ying, D. J. Tang, H. Tang, W. Wu, P. Hao, L. Wang, B. L. Jiang, S. Zeng, W. Y. Gu, G. Lu, L. Rong, Y. Tian, Z. Yao, G. Fu, B. Chen, R. Fang, B. Qiang, Z. Chen, G. P. Zhao, J. L. Tang, and C. He. 2005. Comparative and functional genomic analyses of the pathogenicity of phytopathogen Xanthomonas campestris pv. campestris. Genome Res. 15:757-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ray, S., R. Rajeshwari, Y. Sharmaand, and R. V. Sonti. 2002. A high-molecular-weight outer membrane protein of Xanthomonas oryzae pv. oryzae exhibits similarity to non-fimbrial adhesins of animal pathogenic bacteria and is required for optimum virulence. Mol. Microbiol. 46:637-647. [DOI] [PubMed] [Google Scholar]
- 37.Reickseidler, S. L., D. DeShazer, P. A. Sokol, and D. E. Woods. 2001. Detection of bacterial virulence genes by subtractive hybridization: identification of capsular polysaccharide of Burkholderia pseudomallei as a major virulence determinant. Infect. Immun. 69:34-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roden, J. A., B. Belt, J. B. Ross, T. Tachibana, J. Vargas, and M. B. Mudgett. 2004. A genetic screen to isolate type III effectors translocated into pepper cells during Xanthomonas infection. Proc. Natl. Acad. Sci. USA 101:16624-16629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roumagnac, P., L. Gagnevin, L. Gardan, L. Sutra, C. Manceau, E. R. Dickstein, J. B. Jones, P. Rott, and O. Pruvost. 2004. Polyphasic characterization of xanthomonads isolated from onion, garlic and Welsh onion (Allium spp.) and their relatedness to different Xanthomonas species. Int. J. Syst. Evol. Microbiol. 54:15-24. [DOI] [PubMed] [Google Scholar]
- 40.Schaad, N. W., E. Postnikova, G. H. Lacy, A. Sechler, I. Agarkova, P. E. Stromberg, V. K. Stromberg, and A. K. Vidaver. 2006. Emended classification of xanthomonad pathogens on citrus. Syst. Appl. Microbiol. 29:690-695. [DOI] [PubMed] [Google Scholar]
- 41.Thieme, F., R. Koebnik, T. Bekel, C. Berger, J. Boch, D. Büttner, C. Caldana, L. Gaigalat, A. Goesmann, S. Kay, O. Kirchner, C. Lanz, B. Linke, A. C. McHardy, F. Meyer, G. Mittenhuber, D. H. Nies, U. Niesbach-Klösgen, T. Patschkowski, C. Rückert, O. Rupp, S. Schneiker, S. C. Schuster, F. J. Vorhölter, E. Weber, A. Pühler, U. Bonas, D. Bartels, and O. Kaiser. 2005. Insights into genome plasticity and pathogenicity of the plant pathogenic bacterium Xanthomonas campestris pv. vesicatoria revealed by the complete genome sequence. J. Bacteriol. 187:7254-7266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Triplett, L. R., Y. Zhao, and G. W. Sundin. 2006. Genetic differences between blight-causing Erwinia species with differing host specificities, identified by suppression subtractive hybridization. Appl. Environ. Microbiol. 72:7359-7364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Troisfontaines, P., and G. R. Cornelis. 2005. Type III secretion: more systems than you think. Physiology 20:326-339. [DOI] [PubMed] [Google Scholar]
- 44.Vauterin, L., B. Hoste, K. Kersters, and J. Swings. 1995. Reclassification of Xanthomonas. Int. J. Syst. Bacteriol. 45:472-489. [Google Scholar]
- 45.Vidaver, A. K. 1993. Xanthomonas campestris pv. phaseoli: cause of common bacterial blight of bean, p. 40-44. In J. G. Swings and E. L. Civerolo (ed.), Xanthomonas. Chapman and Hall, London, United Kingdom.
- 46.Wiggerich, H. G., and A. Pühler. 2000. The exbD2 gene as well as the iron-uptake genes tonB, exbB and exbD1 of Xanthomonas campestris pv. campestris are essential for the induction of a hypersensitive response on pepper (Capsicum annuum). Microbiology 146:1053-1060. [DOI] [PubMed] [Google Scholar]
- 47.Winstanley, C. 2002. Spot the difference: applications of subtractive hybridization to the study of bacterial pathogens. J. Med. Microbiol. 51:459-467. [DOI] [PubMed] [Google Scholar]