Abstract
In this study, we cloned and sequenced a virulence-associated gene (vacB) from a clinical isolate SSU of Aeromonas hydrophila. We identified this gene based on our recently annotated genome sequence of the environmental isolate ATCC 7966T of A. hydrophila and the vacB gene of Shigella flexneri. The A. hydrophila VacB protein contained 798 amino acid residues, had a molecular mass of 90.5 kDa, and exhibited an exoribonuclease (RNase R) activity. The RNase R of A. hydrophila was a cold-shock protein and was required for bacterial growth at low temperature. The vacB isogenic mutant, which we developed by homologous recombination using marker exchange mutagenesis, was unable to grow at 4°C. In contrast, the wild-type (WT) A. hydrophila exhibited significant growth at this low temperature. Importantly, the vacB mutant was not defective in growth at 37°C. The vacB mutant also exhibited reduced motility, and these growth and motility phenotype defects were restored after complementation of the vacB mutant. The A. hydrophila RNase R-lacking strain was found to be less virulent in a mouse lethality model (70% survival) when given by the intraperitoneal route at as two 50% lethal doses (LD50). On the other hand, the WT and complemented strains of A. hydrophila caused 80 to 90% of the mice to succumb to infection at the same LD50 dose. Overall, this is the first report demonstrating the role of RNase R in modulating the expression of A. hydrophila virulence.
A human diarrheal isolate SSU of Aeromonas hydrophila possesses various pathogenic mechanisms that contribute to the overall virulence of this bacterium in the host. Most importantly, our laboratory has described type II, type III, and type VI secretion system effector proteins that are involved in the disease state (gastroenteritis or septicemia) in animal models (11, 15, 17, 21, 32, 44-46, 48). However, to better understand the full virulence potential of any pathogen, it is important to identify new pathogenic factors and/or mechanisms that could be involved in their virulence. This is crucial since the expression of different virulence genes could be contributing factors leading to disease depending upon the anatomical niche where the organisms colonize and the environment in those regions which dictates the differential expression of genes (18).
Studies by Tobe et al. (50) identified a novel chromosomal gene, vacB, which was required for the production of S. flexneri virulence factors (e.g., IpaB, IpaC, IpaD, and VirG) from the large plasmid (230 kb) of this microorganism. More recently, it was shown that the vacB gene in S. flexneri and Escherichia coli strains encoded the 3′-5′ exoribonuclease RNase R (10). The deduced size of VacB (virulence-associated) protein is approximately 92 kDa, and it is located at 95 min on the E. coli chromosome, a position consistent with the earlier mapping studies of the gene encoding RNase R (10). These investigators provided evidence that RNase R has an essential cell function, in addition to its role in bacterial virulence, and hence they renamed the vacB gene to rnr (RNase R) based on its sequence similarity to another exoribonuclease, RNase II (5). Exoribonucleases (RNase R, polynucleotide phosphorylase [PNPase], and others) belong to the RNR family that has homologs widespread in most sequenced prokaryotic and eukaryotic genomes (6, 49), and they mediate many aspects of RNA metabolism and, in particular, the quality control of rRNA (9). More recent studies revealed multifaceted roles of exoribonucleases in the regulation of gene expression in pathogenic bacteria (3, 39, 55). For example, in Streptococcus pyogenes it was found that exoribonuclease PNPase activity was rate limiting for the decay of sagA and sda transcripts that coded for streptolysin S and streptodornase in the late exponential growth phase of the bacterium (3). PNPase enhanced the ability of Yersinia pseudotuberculosis and Y. pestis to withstand the killing activities of murine macrophages and was required for the optimal functioning of the Yersinia T3SS (39). In Salmonella enterica PNPase, apart from being a regulator of the cold shock response, it also functioned in tuning the expression of virulence genes and bacterial fitness during infection (55).
The first report regarding a broad-specificity RNase from Aeromonas was published for a gene from A. hydrophila AH1133 strain that coded for an RNase I-like RNase (20). In the present study, we describe the identification, cloning, and sequencing of the vacB gene from A. hydrophila SSU. Further, we demonstrate that VacB indeed is RNase R, since it exhibited exoribonuclease activity, and showed both a cold-sensitive phenotype and that VacB modulated virulence of A. hydrophila in in vitro and in vivo models of Aeromonas infection.
MATERIALS AND METHODS
Strains, plasmids, and media.
Bacterial strains and plasmids used in the present study are listed in Table 1. Bacterial cultures were grown at 37°C in Luria-Bertani (LB) broth and LB agar plates (41). LBNS (LB with no NaCl) solid medium with 15% sucrose (Suc) for levansucrase (sacB) counterselection was used for the isolation of the vacB isogenic mutant. The antibiotics ampicillin (Ap), kanamycin (Km), gentamicin (Gm), streptomycin (Sm), spectinomycin (Sp), tetracycline (Tc), and rifampin (Rif) were used at concentrations of 100, 50, 20, 30, 30, 15, and 200 μg/ml, respectively.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| A. hydrophila | ||
| SSU | CDCa | |
| SSU-R | Rifr strain of A. hydrophila SSU | Laboratory stock |
| vacB-Ω mutant | Isogenic vacB gene mutant of A. hydrophila SSU-R strain, Rifr Smr Spr | This study |
| vacB-Ω/pBR322 | vacB-Ω mutant strain containing pBR322 vector, Rifr Smr Spr Apr Tcr | This study |
| vacB-Ω/pBR322-vacB | vacB-Ω mutant complemented with the vacB gene via pBR322, Rifr Smr Spr Apr | This study |
| E. coli | ||
| TOP10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74recA1 deoR araD139 Δ(ara-leu)7697galU galK rpsL (Smr) endA1 nupG | Invitrogen |
| JM109 | endA1recA1gyrA96thihsdR17relA1supE44 λ− Δ(lac-proAB) [F′ traD36proA+B+lacIZΔM15] | Promega |
| SM10 (λpir) | Kmrthi-1thrleutonAlacYsupErecA::RP4-2-Tc::Mu pir | 14 |
| K12 CAN20-12E | RNase I− II− D− BN−, Tcr | 7 |
| K12 CAN20-12ER− | RNase I− II− D− BN− R−, Tcr Kmr | 7 |
| K12 CAN20-12ER−(pBAD) | RNase I− II− D− BN− R−, Tcr Kmr Apr | This study |
| K12 CAN20-12ER−(pBAD/vacB) | RNase I− II− D− BN− R−, Tcr Kmr Apr | This study |
| Plasmids | ||
| pCR2.1 | TA cloning vector, Apr Kmr | Invitrogen |
| pCR2.1/vacB | TA cloning vector carrying the vacB gene, Apr Kmr | This study |
| pHP45Ω | pHP45 plasmid containing a 2-kb Ω element, Apr Smr Spr | 33 |
| pCR2.1/vacB-Ω | TA cloning vector harboring the vacB gene disrupted by Ω element, Apr Smr Spr | This study |
| pJQ200SK | Suicide vector, P15A origin, sacB Gmr | 36 |
| pJQ200SK/vacB-Ω | Suicide vector containing the vacB gene with Ω element, Gmr Smr Spr | This study |
| pBR322 | Apr Tcr | Amersham |
| pBR322/vacB | Contains the vacB gene, Apr | This study |
| pBAD/Thio-E | Arabinose-inducible araBAD promoter-based vector, Apr | Invitrogen |
| pBAD | Control derivative plasmid, Apr | 15 |
| pBAD/vacB | pBAD expression vector with the vacB gene, Apr | This study |
CDC, Centers for Disease Control and Prevention, Atlanta, GA.
DNA isolation and PCR assays.
Plasmid DNA was isolated by using a QIAprep spin miniprep kit from Qiagen, Valencia, CA. A. hydrophila genomic DNA (gDNA) for sequencing was isolated by using the published protocol with some modifications (35) or by utilizing a DNeasy tissue kit (Qiagen) for PCR assays. DNA fragments were purified by using a PCR purification kit or gel extraction kit (Qiagen). The primers (Table 2) were synthesized by Integrated DNA Technologies, Inc., Coralville, IA.
TABLE 2.
Primers used in this study
| Primer | Sequence (5′-3′)a | Purpose |
|---|---|---|
| vacB-N | ATGATCCTCGACCTCATCAAGGGCCATGAA | PCR amplification of a portion (2.0 kb) of the vacB gene |
| vacB-C | TCACGAAGTCGATCTTGCGATCGTCCAGAT | |
| vacB-N1 | TGCTGGTCTCGGCGAACAGGTGCCG | DNA sequencing of the PCR product to obtain middle part of the vacB gene |
| vacB-C1 | CACCAGACCGTCGATGTGGATCTCG | |
| vacB-N/TGA | CTCACCAACGACTACTACCAGTTCGACCCG | DNA sequencing of the gDNA to obtain the ORF of the vacB gene |
| vacB-C/ATG | CAGTTGACCGTCGCGCTCCATGGCGCGCAG | |
| vacB-N/up | GTCTCATCATGACTGCAAACTGTGCATAGTATGAC | Cloning of the vacB gene into pCR2.1 vector |
| vacB-C/down | GGGGCCATCAGCCCCTTACGACCTCAGACCGGGTC | |
| vacB-N/g | GAACGCCTGCTTGAGATAGGCGGCGGATTGGGCGA | PCR verification of the vacB mutant |
| vacB-C/g | AGTAACAGGGTTCGGATTTTTTTCGTTTTCAGGAC | |
| vacB/HindIIIN | CCCAAGCTTGTCTCATCATGACTGCAAACTGTGCA | Cloning of the vacB gene into pBR322 vector |
| vacB/SalIC | ACGCGTCGACGGGGCCATCAGCCCCTTACGACCTC | |
| vacB/AflIIIN | CCCACATGTCTCAAAAAGATCCTTTCCTCGAACGCGAGGC | Cloning of the vacB gene into pBAD/Thio-E vector |
| vacB/PmeIC | AGCTTTGTTTAAACTTACCCCTTGGCCTTGTCGCGCGCTT |
Underlining indicates restriction endonuclease site.
Transformation of E. coli and electroporation of A. hydrophila strains.
The preparation of competent cells for transformation and electroporation was described previously (15, 47). Competent E. coli cells for transformation were prepared by using a Z-Competent transformation kit from Zymo Research, Orange, CA. The competent A. hydrophila cells in 300 mM sucrose were electroporated in 0.2-cm gap cuvettes (Bio-Rad, Hercules, CA) by using a GenePulser Xcell.
Sequencing of the A. hydrophila chromosomal vacB gene and construction of its knockout mutant.
To sequence the vacB gene of A. hydrophila SSU strain, we first PCR amplified a portion (∼2.0 kb) of this gene using gDNA of the wild-type (WT) bacterium as the template and the vacB-N/vacB-C pair of primers (Table 2). To sequence the middle part of the vacB gene, the above PCR product was DNA sequenced with vacB-N1/vacB-C1 primers. To determine the open reading frame (ORF) of the A. hydrophila SSU vacB gene, gDNA was sequenced using primers vacB-N/TGA and vacB-C/ATG (Table 2).
To clone the vacB gene (2394-bp) into pCR2.1 vector (Invitrogen, Carlsbad, CA), the target gene with up- and downstream DNA flanking sequences was PCR amplified using gDNA and the vacB-N/up and vacB-C/down primers, and the ligation mixture was transformed into chemically competent E. coli TOP10 cells (Table 1). To generate the vacB knockout mutant, an Sm/Sp cassette (the Ω element) from a pHP45Ω plasmid (33) was introduced into a unique SmaI restriction enzyme site of the vacB gene in the pCR2.1/vacB recombinant plasmid. Then, the vacB-Ω fragment from a pCR2.1/vacB-Ω plasmid was cloned into a suicide pJQ200SK vector (36) at the XbaI and SpeI restriction enzymes sites and transformed into E. coli SM10 (with λpir) strain (14). After conjugation between A. hydrophila SSU and the E. coli SM10 strain harboring the pJQ200SK/vacB-Ω plasmid, the knockout vacB mutant was screened on LBNS plates containing Rif, Sp, Sm, and sucrose. From single A. hydrophila SSU colonies with a Rif-, Sp-, Sm-, and sucrose-resistant but Gm-sensitive-phenotype, we isolated gDNA and performed PCR to confirm the disruption of the vacB gene using vacB-N/g and vacB-C/g primers (Table 2). The mutant colonies were identified as Aeromonas by a positive oxidase test to differentiate them from E. coli, which is oxidase negative (26).
Complementation of the A. hydrophila vacB mutation.
The vacB gene was PCR amplified by using the vacB/HindIIIN and vacB/SalIC primers (Table 2) and A. hydrophila gDNA as the template. We included 125-bp of the upstream flanking sequence containing the potential promoter region of the vacB gene for complementation studies. The PCR product was purified by a PCR purification kit, digested with appropriate enzymes, and ligated to the pBR322 vector at the HindIII-SalI restriction-enzymes sites. This ligation mixture was then transformed into E. coli JM109 cells, and plasmid DNA was isolated from Apr and Tcs transformants. The presence of pBR322/vacB plasmid (Table 1) was confirmed by restriction endonuclease mapping of the recombinant plasmid. Then, pBR322/vacB plasmid DNA was electroporated into the vacB isogenic mutant of the A. hydrophila strain. As a negative control, the A. hydrophila vacB mutant strain, containing the pBR322 vector alone, was also generated (Table 1).
Cloning of the vacB gene under the araBAD promoter.
To regulate the expression of the vacB gene, the recombinant plasmid pBAD/vacB was generated by using the vacB/AflIIIN and vacB/PmeIC primers (Table 2) by replacing the NcoI-PmeI fragment of the pBAD/Thio-E vector (Invitrogen) under its arabinose PBAD promoter. To induce expression of the gene from the plasmid, arabinose (0.2%) was added to the medium (24). To construct the pBAD plasmid for use as a control, pBAD/Thio-E vector was digested with NcoI and PmeI restriction endonucleases, treated with DNA polymerase I (Klenow fragment), and ligated. Both plasmids (pBAD and pBAD/vacB) were then electroporated in E. coli K-12 strain CAN20-12ER− (RNases I−, II−, D−, BN−, and RNase R−) (Table 1) for measuring RNase R activity.
Preparation of cell extracts.
Overnight-grown E. coli K-12 strains (CAN20-12E [RNases I−, II−, D−, BN−, but RNase R+], CAN20-12ER− with pBAD and CAN20-12ER− with pBAD/vacB) were reinoculated in the morning, and at an optical density at 600 nm of 0.4 arabinose, at a final concentration of 0.2%, was added. After 4 h of growth, the pellets from 10-ml cultures were resuspended in 1 ml of solution containing 20 mM Tris-HCl (pH 7.5), 1 mM dithiothreitol, 300 mM KCl, and 0.1 mM phenylmethylsulfonyl fluoride (7). After incubation for 1 h on ice with lysozyme (300 μg/ml) and 0.1% Triton X-100, the cells were disrupted by sonication on ice by using two 10-s pulses. The cell extracts were recovered by centrifugation at 13,000 × g for 10 min at 4°C to remove cell debris, and the protein concentration of the extracts was determined by a Bradford assay (4). Bovine serum albumin was used as the standard. E. coli K-12 strain CAN20-12E was used as the positive control, and E. coli strain CAN20-12ER− containing pBAD vector alone was used as the negative control.
Assay for RNase R.
The assay for RNase R activity was conducted according to the described procedure (7). Reaction mixtures (100 μl) contained 20 mM Tris-HCl (pH 8.0), 0.25 mM MgCl2, 180 mM KCl, 40 μg of [3H]poly(A) (50 to 100 cpm/nmol; GE Healthcare Bio-Sciences Corp., Piscataway, NJ), and 30 μg of the indicated cell extracts. Reaction mixtures were incubated at 37°C for 1 h. After precipitation with 10% cold trichloroacetic acid, the release of acid-soluble radioactivity was determined by using a Beckman scintillation counter LS 6500.
Cold growth assay.
For the cold-growth experiment, A. hydrophila overnight cultures (WT, vacB mutant, and complemented strains) were reinoculated in LB broth with the appropriate antibiotics and grown at 37°C up to an optical density at 600 nm of 0.4, and then the diluted (10−5) strains were plated (50 μl) on LB agar plates with antibiotics. The plates were incubated for 7 days at 4°C. As a control, plates inoculated similarly were incubated at 37°C.
Motility assay.
The equal numbers of CFU of tested Aeromonas cultures were stabbed into 0.3% LB agar plates and incubated at 37°C overnight. Their motility was assayed by examining the migration of bacteria through the agar from the center toward the plate's periphery (15).
Cytotoxicity assay.
RAW 264.7 murine macrophages were seeded onto 96-well plates (105 cells/well) and infected with either the live WT A. hydrophila SSU strain or its vacB knockout mutant at an multiplicity of infection of 10 for 4 h (43). In another set of experiment, host cells were treated with 5 μl of filter-sterilized, overnight-grown (at 37°C with shaking [180 rpm] in selected LB medium) bacterial culture supernatants. After incubation at 37°C for 2 h, the tissue culture medium was examined for the release of lactate dehydrogenase enzyme by using a CytoTox96 kit (Promega, Madison, WI). The percentage of lactate dehydrogenase released by RAW 264.7 macrophages infected with bacterial cells was determined according to the manufacturer's instructions. In macrophages treated with culture supernatants, cytotoxicity was reported per milliliter of the culture filtrate per 108 CFU and was expressed as a fold change compared between WT A. hydrophila SSU and the vacB mutant strains. The purpose of these experiments was to show whether VacB directly or indirectly contributed to host cell toxicity by altering functionality of type III and type VI secretion systems (17, 44-46, 48). The culture filtrates were used to demonstrate whether the isogenic mutant of the vacB gene affected cytotoxic and hemolytic activities associated with a potent cytotoxic enterotoxin (Act) or other secreted cytotoxins or hemolysins produced by A. hydrophila SSU (17, 21, 44).
Hemolytic activity.
A. hydrophila SSU strains (WT, vacB knockout, and complemented) were grown at 37°C overnight with shaking (180 rpm) in selected LB medium. The cultures were spun down, and supernatants were added to the first well in each row of a 96-well microtiter plate, followed by serial twofold dilution (before 100 μl of 1× Dulbecco phosphate-buffered saline [DPBS] was added to each of the wells of the microtiter plate). Then, 100 μl of 3% rabbit erythrocytes (Colorado Serum Co., Denver, CO) was added, and the plate was incubated at 37°C for 1 h. A control included 1× DPBS alone. The supernatants were taken from wells that showed partial lysis of red blood cells, and the hemoglobin release was recorded at 540 nm by using a microplate reader. The hemolytic units were reported per ml/108 CFU of culture supernatants.
Animal experiments.
Eight-week-old female Swiss-Webster mice were inoculated intraperitoneally with a lethal dose (3 × 107 CFU, representing two 50% lethal doses) of WT, vacB knockout mutant, and vacB-complemented Aeromonas strains in groups of 10 mice each. One mouse group was inoculated with DPBS and served as a control. Mice were observed daily for signs of distress and mortality for up 2 weeks.
Statistics.
Wherever applicable, at least three independent experiments were performed. The data were analyzed by using Student t test or the Fisher exact test, and P values of ≤0.05 were considered significant.
Nucleotide sequence accession number.
The nucleic acid and translated amino acid sequences of the A. hydrophila SSU strain vacB gene were deposited in the GenBank database under accession number EU380595.
RESULTS
Identification and cloning of the A. hydrophila SSU vacB gene.
To define additional genetic determinants that could control virulence gene expression in a pathogenic diarrheal A. hydrophila SSU isolate, the nucleotide sequence of the vacB gene in S. flexneri (50) was compared to that of the genome sequence of A. hydrophila ATCC 7966T (42) from the TIGR database, which we recently annotated. We noted that the gDNA of A. hydrophila ATCC strain exhibited a 66% identity (from nucleotide position 84493 to 86780) with the S. flexneri vacB gene. Based upon the ORF sequence of the vacB gene of A. hydrophila ATCC 7966T, the vacB-N and vacB-C primers were designed (Table 2) to obtain a PCR product, using gDNA of A. hydrophila SSU strain as the template. One major band (∼2.0 kb) was PCR amplified and then sequenced using the “internal” primers vacB-N1 and vacB-C1 to obtain the complete sequence of this DNA product (a portion of vacB gene). Finally, to identify the coding region of the vacB gene of A. hydrophila SSU, we used chromosomal DNA sequencing with the vacB-N/TGA and vacB-C/ATG primers (Table 2). For further studies, the vacB gene was cloned into the EcoRI site of a pCR2.1 plasmid vector using PCR products from gDNA and primers without any sites for restriction endonucleases. The inserted vacB gene was verified by DNA sequencing of the recombinant plasmid pCR2.1/vacB (Table 1) with M13 forward and reverse primers.
The vacB gene encoded a protein (RNase R) of 798 amino acid residues with a molecular mass of 90.5 kDa. The overall identity of the A. hydrophila SSU vacB gene with that of A. hydrophila ATCC 7966T was 94%. At the protein level, RNase R (VacB) of the A. hydrophila SSU strain showed a 97% homology with A. hydrophila ATCC 7966T RNase R, 94% homology with A. salmonicida A449 RNase R (GenBank accession no. YP_001140610), 65% homology with S. flexneri VacB (GenBank accession no. D11024), 65% homology with E. coli CFT073 RNase R (GenBank accession no. NP_757111) (53), and 62% homology with RNase R of Y. pestis CO92 (GenBank accession no. NP_404028) (31).
VacB of A. hydrophila SSU strain possesses RNase R activity.
To assess the ability of A. hydrophila VacB to show RNase R activity, cell extracts were tested from the E. coli K-12 strain CAN20-12ER− that lacks RNases I, II, D, BN, and RNase R activities but carries the recombinant pBAD/vacB plasmid (Table 1). E. coli K-12 strain CAN20-12ER− containing pBAD vector alone was used as a negative control (RNase R− cell extract), and E. coli K-12 CAN20-12E (10) deficient in RNases I, II, D, and BN activities, but not in RNase R activity, was used as a positive control strain for this assay (RNase R+ cell extract). As noted from Table 3, cell extract from the E. coli K-12 strain CAN20-12ER− with the pBAD/vacB plasmid showed an RNase R activity comparable to that of the positive control. In contrast, the RNase R activity in the cell extracts of control E. coli CAN20-12ER− (with the pBAD vector alone) strain was 61% lower than that of the positive control. The E. coli K-12 strains indicated above were grown in the presence of 0.2% arabinose to regulate the vacB gene expression from the araBAD promoter and to allow similar growth conditions for all bacterial cultures. We could not directly measure RNase R activity in the cell extracts of the WT versus the vacB mutant of A. hydrophila SSU since other RNases masked the effect of RNase R.
TABLE 3.
RNase R activity of VacB from the A. hydrophila SSU straina
For the RNase R activity assay, we used the same total protein concentration (30 μg) for each indicated E. coli K-12 strain cell extract.
RNase R of A. hydrophila SSU is a cold-shock protein.
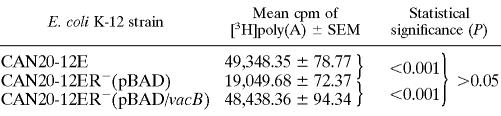
To address whether the A. hydrophila SSU RNase R is a cold-shock protein, A. hydrophila WT, vacB knockout, and complemented strains were tested for their ability to grow at low temperatures (Fig. 1). The vacB mutant strain was unable to grow at low temperatures (Fig. 1B), whereas the WT strain was able to grow at 4°C (Fig. 1A). The complemented strain harboring the recombinant plasmid that encoded RNase R showed the recovery of this strain to grow at a low temperature (Fig. 1C). In order to delineate whether the vacB mutant of A. hydrophila died at 4°C or was dormant, we shifted the temperature of growth from 4 to 37°C after an initial incubation period of 7 days. We observed no growth of the vacB mutant after the temperature shift for up to 48 h. The above-mentioned A. hydrophila WT, mutant, and the complemented cultures grew similarly at 37°C (data not shown). These results indicated that the vacB gene is required for the A. hydrophila SSU strain to grow at a low temperature.
FIG. 1.
Effect of RNase R on cold growth of the bacterium. A. hydrophila WT (A), vacB knockout mutant (B), and complemented (C) strains were plated (same CFU) on LB agar plates with appropriate antibiotics and incubated at 4°C for 7 days.
Effect of RNase R on A. hydrophila SSU virulence.
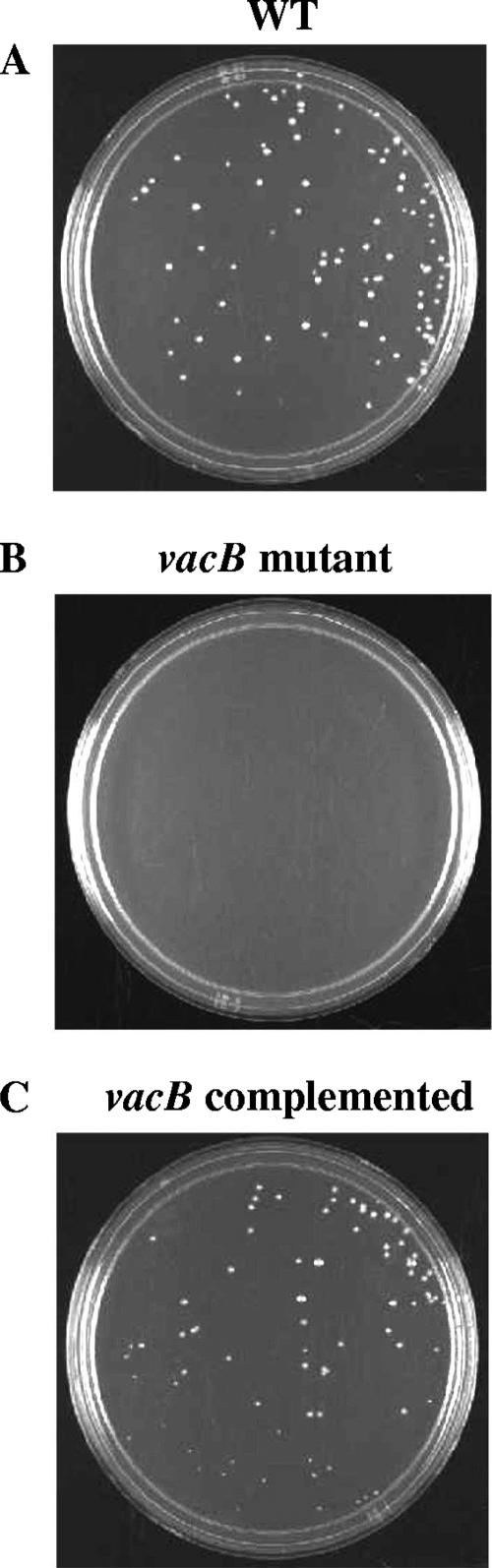
To evaluate the effect of RNase R activity associated with the vacB gene on the pathogenesis of the A. hydrophila SSU strain, we assessed the biological activities of various virulence factors of WT A. hydrophila and its isogenic vacB mutant. The motility of A. hydrophila is an important factor for bacteria to reach the host target tissue, to colonize, and then to cause disease (22). We noted that the vacB A. hydrophila mutant strain had 57% less motility (Fig. 2B) than the WT strain (Fig. 2A). The motility phenotype was restored in the strain that was complemented with the pBR322/vacB recombinant plasmid (Fig. 2C).
FIG. 2.
A. hydrophila SSU with knockout mutant of the vacB gene shows a reduced motility (B) compared to those of the WT and complemented A. hydrophila strains (A and C).
The cytotoxicity on macrophages essentially remained unaltered when infected with the WT and the vacB knockout mutant strain of A. hydrophila SSU, indicating that the RNase activity was not essential for modulating cytotoxicity associated with various effectors of type III and type VI secretion systems (data not shown). Also, we did not observe any differences in the hemolytic and/or cytotoxic activities in culture supernatants of Aeromonas WT and its vacB mutant strain, indicating that VacB did not modulate the activities of Act and the other hemolysins (e.g., HlyA) or cytotoxins that we recently described in A. hydrophila SSU (16).
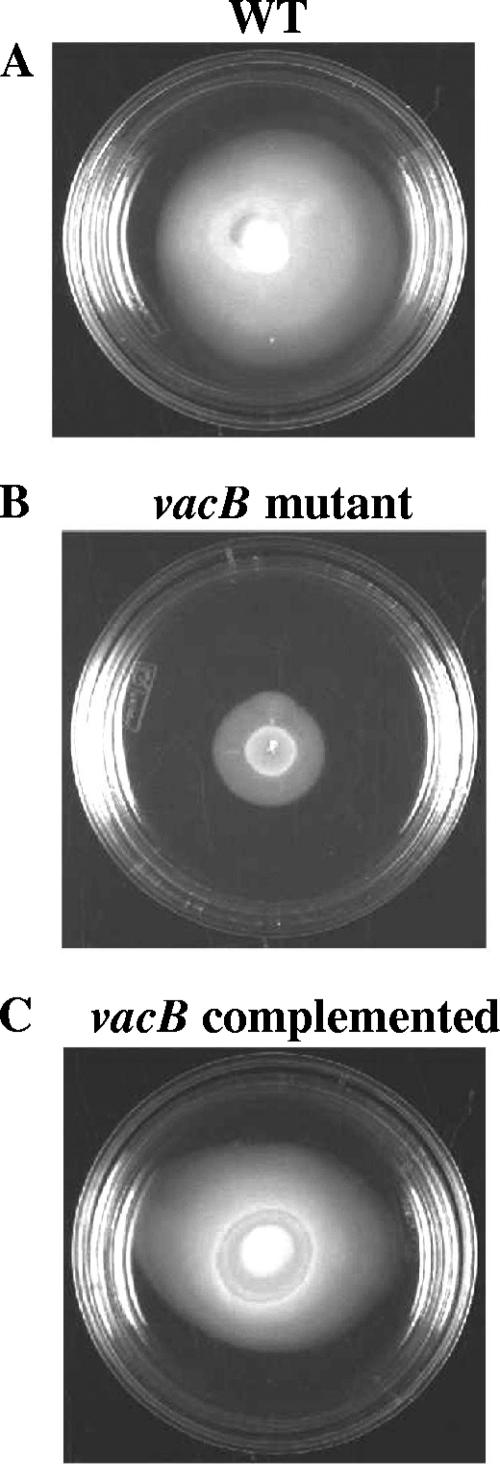
Subsequently, we examined the effect of RNase R of A. hydrophila in contributing to mouse mortality. We injected mice intraperitoneally with the isogenic vacB gene mutant strain, the vacB complemented strain (pBR322/vacB), and the WT control strain with pBR322 plasmid at a lethal dose of 3 × 107 CFU (Fig. 3). The animals infected with the A. hydrophila control strain or the complemented strain showed 80 to 90% mortality within 3 days. However, mice infected with the vacB mutant strain exhibited significant lower mortality (only 30%) over a tested period of 15 days, indicating bacterial attenuation as a result of a loss in the RNase R activity. We chose to use two 50% lethal doses for these studies since higher doses would not be appropriate because of other virulence factors (e.g., Act, AexU [T3SS effector], and hemolysin-coregulated protein [type VI secretion system effector]) (45, 46, 48) produced by A. hydrophila that also lead to mouse lethality.
FIG. 3.
The A. hydrophila vacB knockout mutant strain was less virulent in a mouse model. The complemented strain (pBR322/vacB) led to mouse mortality within 48 h, similar to the findings with the WT A. hydrophila strain harboring pBR322 plasmid. An asterisk denotes statistically significant values as determined by the Fisher exact test.
DISCUSSION
A majority of the studies concerned with exoribonucleases indicate their prominent role in quality control of rRNA (9), cell cycle-regulated degradation of tmRNA (small stable RNA) (25), and mRNA decay (8), which is a significant determinant of gene expression. Since publication of the report regarding the role of the vacB gene in the pathogenesis of serotypes of Shigella spp. and enteroinvasive E. coli strains (50), only recently did studies emerge that indicated a potential contribution of RNases, in particular PNPase, in regulating bacterial virulence (12, 39, 55). However, the role of RNase R in bacterial virulence is not fully explored. Interestingly, the vacB gene of Brucella abortus has no impact on bacterial virulence (29). Recent studies did indicate that the expression of RNase R is tightly regulated by temperature (6) and that it is essential for growth of the organism at low temperatures (e.g., 4°C) (34). These investigators demonstrated that the cold-sensitive phenotype of the rnr mutant was due to the cell death of the mutant strain and that defective protein synthesis was one of the important factors responsible for killing of Pseudomonas syringae rnr mutants at low temperature (34). These data confirmed our results indicating that RNase R from A. hydrophila SSU strain is a cold-shock protein. We did not observe growth of the vacB (rnr) mutant strain at a low temperature, in contrast to the findings with Aeromonas WT and complemented strains (Fig. 1). Our data also indicated that the vacB mutant died when grown at 4°C. It is plausible that VacB might have a regulatory role and could modulate the translation of some proteins in A. hydrophila SSU that are required for bacterial growth. We will investigate this possibility as well as the ability of the vacB mutant to inhibit protein synthesis in our future study.
As with RNase R, the exoribonuclease PNPase is also clearly required for Y. pseudotuberculosis and Y. pestis to grow at low temperatures (e.g., 5°C), and PNPase plays a multifaceted role in enhancing yersiniae survival in response to stressful conditions (39). Likewise, the role of PNPase in the proper configuration of the T3SS effector proteins was also reported (39). For the rnr mutant of E. coli, a growth defect characterized by the formation of colonies that were considerably smaller than that of the WT strain was noted (6); however, these investigators used 10°C temperature as the cold shock conditions. Interestingly, PNPase-deficient mutants of P. putida did not exhibit a cold-sensitive phenotype at 15 and 5°C (19). It is possible that the expression of the RNase gene and adaptation to the cold may be different in various bacteria.
Motility is an important virulence factor of gram-negative bacterial pathogens and is a very complex process that depends on the cooperation of many gene products (23, 28). We explored the role of RNase R in the pathogenesis of A. hydrophila and showed the decreased motility of the vacB mutant strain (Fig. 2). The experiments with some rnc (RNase III−) mutants supported the notion that RNase III is involved in the motility of E. coli (2). These researchers noted that the mutants of E. coli deficient in RNase III were nonmotile. Further, these investigators also showed that all transductants and revertants that regained RNase III showed motility-positive phenotypes and that all transductants that remained or became rnc− were nonmotile.
Based on our observations, RNase R from A. hydrophila SSU did not alter other biological activities associated with this pathogen, e.g., the ability to exhibit hemolytic and cytotoxic activities. However, in a future study, we will explore the possibility whether disruption of the vacB gene from A. hydrophila alters expression of the act and/or the hemolysin gene. Earlier it was shown that a cationic protein secreted by human eosinophils which exhibited structural homology to other RNases that had a RNase activity was not essential for cytotoxicity (38). On the contrary, a recent study showed that the exoribonuclease PNPase was necessary for Yersinia to affect the morphology of HeLa cells and that the PNPase-dependent cytotoxicity of Yersinia-infected host cells was delayed relative to the WT-infected HeLa cells (39). It was also reported that a Burkholderia strain living inside the arbuscular mycorrhizal fungus Gigaspora margarita possessed the vacB gene, which was involved in host cell colonization by this pathogenic bacteria (40).
Most significantly, we observed that RNase R mutant of the A. hydrophila strain led to the attenuation of virulence in a mouse lethality model. A similar pattern of mouse mortality was shown for the SmpB-SsrA system (small protein B-small stable RNA A, also known as tmRNA) in Y. pseudotuberculosis. Deletion of smpB-ssrA genes from Y. pseudotuberculosis rendered the bacterium avirulent in a mouse model of lethality (30). Previous studies also indicated that the ssrA gene played a role in the full virulence of S. enterica serovar Typhimurium. A >200-fold virulence defect was observed in BALB/c mice and was attributed to the altered expression of several genes induced during infection with the ssrA mutant (13, 27). Recently, it was reported that RNase R is required for the selective degradation of SsrA RNA in stalked cells and that SmpB binding controls the timing of SsrA RNA degradation by RNase R (25). E. coli has an enabling mechanism that requires RNase R activity and is dependent on the presence of SmpB protein and tmRNA, suggesting a requirement for active trans-translation in facilitating RNase R engagement and promoting nonstop mRNA decay (37). Most surprisingly, RNase II and PNPase do not play a significant role in the tmRNA-facilitated disposal of aberrant mRNAs (37). Whether VacB regulates RNA decay in A. hydrophila SSU is unknown and will be explored in a future study.
Thus, on one hand, vacB was first defined as a virulence factor (50), but on the other hand, VacB is known to be an exoribonuclease RNase R involved in mRNA posttranscriptional processing, ribosome rescue, and rRNA methylation (9, 10, 51). The process of mRNA decay is integral to the posttranscriptional control of gene expression, and mRNA turnover is a means of coordinating this process, first through integration with control of transcription and export and translation of mRNAs and second through enabling mRNAs involved in similar processes to decay at similar rates (54). Very recently, a study indicated that the Salmonella mutants deficient in several RNases (RNase E, RNase G, RNase III, and PNPase) that affected sRNA and mRNA turnover were very important for posttranscriptional studies in this bacterial model of pathogenesis (52).
In nature, bacteria remain mostly in the stationary phase of the life cycle, and RNase R can be a modulator of gene expression in this phase of cell growth (1). The impressive spectrum of bacterial diseases in the world probably can be explained by the ability of pathogens to modify adequately the gene expression in response to environmental stimuli and to control virulence factors by RNA-mediated transcriptional regulators. RNase R could be one such regulator that could modulate bacterial virulence and, hence, future studies will focus on a global assessment of host responses after infection of animals with the WT and vacB mutant of A. hydrophila.
Acknowledgments
We thank Murray Deutscher (University of Miami School of Medicine, Miami, FL) for providing E. coli K-12 CAN20-12E and CAN20-12ER− strains. We thank Mardelle Susman for editorial assistance.
This study was supported by a grant from the NIH/NIAID (AI41611) and from the Environmental Protection Agency. The NSF grant (EF-0334247) that was conduit for getting the ATCC 7966 A. hydrophila genome sequenced is also acknowledged.
Footnotes
Published ahead of print on 14 March 2008.
REFERENCES
- 1.Andrade, J. M., F. Cairrao, and C. M. Arraiano. 2006. RNase R affects gene expression in stationary phase: regulation of ompA. Mol. Microbiol. 60219-228. [DOI] [PubMed] [Google Scholar]
- 2.Apirion, D., and N. Watson. 1978. Ribonuclease III is involved in motility of Escherichia coli. J. Bacteriol. 1331543-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnett, T. C., J. V. Bugrysheva, and J. R. Scott. 2007. Role of mRNA stability in growth phase regulation of gene expression in the group a streptococcus. J. Bacteriol. 1891866-1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72248-254. [DOI] [PubMed] [Google Scholar]
- 5.Bycroft, M., T. J. Hubbard, M. Proctor, S. M. Freund, and A. G. Murzin. 1997. The solution structure of the S1 RNA binding domain: a member of an ancient nucleic acid-binding fold. Cell 88235-242. [DOI] [PubMed] [Google Scholar]
- 6.Cairrao, F., A. Cruz, H. Mori, and C. M. Arraiano. 2003. Cold shock induction of RNase R and its role in the maturation of the quality control mediator SsrA/tmRNA. Mol. Microbiol. 501349-1360. [DOI] [PubMed] [Google Scholar]
- 7.Chen, C., and M. P. Deutscher. 2005. Elevation of RNase R in response to multiple stress conditions. J. Biol. Chem. 28034393-34396. [DOI] [PubMed] [Google Scholar]
- 8.Cheng, Z. F., and M. P. Deutscher. 2005. An important role for RNase R in mRNA decay. Mol. Cell 17313-318. [DOI] [PubMed] [Google Scholar]
- 9.Cheng, Z. F., and M. P. Deutscher. 2003. Quality control of ribosomal RNA mediated by polynucleotide phosphorylase and RNase R. Proc. Natl. Acad. Sci. USA 1006388-6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng, Z. F., Y. Zuo, Z. Li, K. E. Rudd, and M. P. Deutscher. 1998. The vacB gene required for virulence in Shigella flexneri and Escherichia coli encodes the exoribonuclease RNase R. J. Biol. Chem. 27314077-14080. [DOI] [PubMed] [Google Scholar]
- 11.Chopra, A. K., and C. W. Houston. 1999. Enterotoxins in Aeromonas-associated gastroenteritis. Microbes Infect. 11129-1137. [DOI] [PubMed] [Google Scholar]
- 12.Clements, M. O., S. Eriksson, A. Thompson, S. Lucchini, J. C. Hinton, S. Normark, and M. Rhen. 2002. Polynucleotide phosphorylase is a global regulator of virulence and persistency in Salmonella enterica. Proc. Natl. Acad. Sci. USA 998784-8789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conner, C. P., D. M. Heithoff, S. M. Julio, R. L. Sinsheimer, and M. J. Mahan. 1998. Differential patterns of acquired virulence genes distinguish Salmonella strains. Proc. Natl. Acad. Sci. USA 954641-4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edwards, R. A., L. H. Keller, and D. M. Schifferli. 1998. Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene 207149-157. [DOI] [PubMed] [Google Scholar]
- 15.Erova, T. E., L. Pillai, A. A. Fadl, J. Sha, S. Wang, C. L. Galindo, and A. K. Chopra. 2006. DNA adenine methyltransferase influences the virulence of Aeromonas hydrophila. Infect. Immun. 74410-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Erova, T. E., J. Sha, A. J. Horneman, M. A. Borchardt, B. K. Khajanchi, A. A. Fadl, and A. K. Chopra. 2007. Identification of a new hemolysin from diarrheal isolate SSU of Aeromonas hydrophila. FEMS Microbiol. Lett. 275301-311. [DOI] [PubMed] [Google Scholar]
- 17.Fadl, A. A., C. L. Galindo, J. Sha, T. E. Erova, C. W. Houston, J. P. Olano, and A. K. Chopra. 2006. Deletion of the genes encoding the type III secretion system and cytotoxic enterotoxin alters host responses to Aeromonas hydrophila infection. Microb. Pathog. 40198-210. [DOI] [PubMed] [Google Scholar]
- 18.Faruque, S. M., G. B. Nair, and J. J. Mekalanos. 2004. Genetics of stress adaptation and virulence in toxigenic Vibrio cholerae. DNA Cell Biol. 23723-741. [DOI] [PubMed] [Google Scholar]
- 19.Favaro, R., and G. Deho. 2003. Polynucleotide phosphorylase-deficient mutants of Pseudomonas putida. J. Bacteriol. 1855279-5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Favre, D., P. K. Ngai, and K. N. Timmis. 1993. Relatedness of a periplasmic, broad-specificity RNase from Aeromonas hydrophila to RNase I of Escherichia coli and to a family of eukaryotic RNases. J. Bacteriol. 1753710-3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galindo, C. L., C. Gutierrez, Jr., and A. K. Chopra. 2006. Potential involvement of galectin-3 and SNAP23 in Aeromonas hydrophila cytotoxic enterotoxin-induced host cell apoptosis. Microb. Pathog. 4056-68. [DOI] [PubMed] [Google Scholar]
- 22.Galindo, C. L., J. Sha, A. A. Fadl, L. Pillai, and A. K. Chopra. 2006. Host immune responses to Aeromonas virulence factors. Curr. Immunol. Rev. 213-26. [Google Scholar]
- 23.Gardel, C. L., and J. J. Mekalanos. 1996. Alterations in Vibrio cholerae motility phenotypes correlate with changes in virulence factor expression. Infect. Immun. 642246-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1774121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hong, S. J., Q. A. Tran, and K. C. Keiler. 2005. Cell cycle-regulated degradation of tmRNA is controlled by RNase R and SmpB. Mol. Microbiol. 57565-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hunt, L. K., T. L. Overman, and R. B. Otero. 1981. Rapid oxidase method for testing oxidase-variable Aeromonas hydrophila strains. J. Clin. Microbiol. 131117-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Julio, S. M., D. M. Heithoff, and M. J. Mahan. 2000. ssrA (tmRNA) plays a role in Salmonella enterica serovar Typhimurium pathogenesis. J. Bacteriol. 1821558-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krukonis, E. S., and V. J. DiRita. 2003. From motility to virulence: sensing and responding to environmental signals in Vibrio cholerae. Curr. Opin. Microbiol. 6186-190. [DOI] [PubMed] [Google Scholar]
- 29.Miyoshi, A., G. M. Rosinha, I. L. Camargo, C. M. Trant, F. C. Cardoso, V. Azevedo, and S. C. Oliveira. 2007. The role of the vacB gene in the pathogenesis of Brucella abortus. Microbes Infect. 9375-381. [DOI] [PubMed] [Google Scholar]
- 30.Okan, N. A., J. B. Bliska, and A. W. Karzai. 2006. A Role for the SmpB-SsrA system in Yersinia pseudotuberculosis pathogenesis. PLoS Pathog. 2e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parkhill, J., B. W. Wren, N. R. Thomson, R. W. Titball, M. T. Holden, M. B. Prentice, M. Sebaihia, K. D. James, C. Churcher, K. L. Mungall, S. Baker, D. Basham, S. D. Bentley, K. Brooks, A. M. Cerdeno-Tarraga, T. Chillingworth, A. Cronin, R. M. Davies, P. Davis, G. Dougan, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Leather, S. Moule, P. C. Oyston, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413523-527. [DOI] [PubMed] [Google Scholar]
- 32.Pillai, L., J. Sha, T. E. Erova, A. A. Fadl, B. K. Khajanchi, and A. K. Chopra. 2006. Molecular and functional characterization of a ToxR-regulated lipoprotein from a clinical isolate of Aeromonas hydrophila. Infect. Immun. 743742-3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prentki, P., and H. M. Krisch. 1984. In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29303-313. [DOI] [PubMed] [Google Scholar]
- 34.Purusharth, R. I., B. Madhuri, and M. K. Ray. 2007. Exoribonuclease R in Pseudomonas syringae is essential for growth at low temperature and plays a novel role in the 3′ end processing of 16 and 5 S rRNA. J. Biol. Chem. 28216267-16277. [DOI] [PubMed] [Google Scholar]
- 35.Qimron, U., N. Madar, R. Ascarelli-Goell, M. Elgrably-Weiss, S. Altuvia, and A. Porgador. 2003. Reliable determination of transposon insertion site in prokaryotes by direct sequencing. J. Microbiol. Methods 54137-140. [DOI] [PubMed] [Google Scholar]
- 36.Quandt, J., and M. F. Hynes. 1993. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene 12715-21. [DOI] [PubMed] [Google Scholar]
- 37.Richards, J., P. Mehta, and A. W. Karzai. 2006. RNase R degrades non-stop mRNAs selectively in an SmpB-tmRNA-dependent manner. Mol. Microbiol. 621700-1712. [DOI] [PubMed] [Google Scholar]
- 38.Rosenberg, H. F. 1995. Recombinant human eosinophil cationic protein: ribonuclease activity is not essential for cytotoxicity. J. Biol. Chem. 2707876-7881. [DOI] [PubMed] [Google Scholar]
- 39.Rosenzweig, J. A., G. Weltman, G. V. Plano, and K. Schesser. 2005. Modulation of Yersinia type three secretion system by the S1 domain of polynucleotide phosphorylase. J. Biol. Chem. 280156-163. [DOI] [PubMed] [Google Scholar]
- 40.Ruiz-Lozano, J. M., and P. Bonfante. 2000. A Burkholderia strain living inside the arbuscular mycorrhizal fungus Gigaspora margarita possesses the vacB gene, which is involved in host cell colonization by bacteria. Microb. Ecol. 39137-144. [DOI] [PubMed] [Google Scholar]
- 41.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 42.Seshadri, R., S. W. Joseph, A. K. Chopra, J. Sha, J. Shaw, J. Graf, D. Haft, M. Wu, Q. Ren, M. J. Rosovitz, R. Madupu, L. Tallon, M. Kim, S. Jin, H. Vuong, C. O. Stine, A. Ali, A. J. Horneman, and J. F. Heidelberg. 2006. Genome sequence of Aeromonas hydrophila ATCC 7966T: the jack of all trades. J. Bacteriol. 1888272-8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sha, J., A. A. Fadl, G. R. Klimpel, D. W. Niesel, V. L. Popov, and A. K. Chopra. 2004. The two murein lipoproteins of Salmonella enterica serovar Typhimurium contribute to the virulence of the organism. Infect. Immun. 723987-4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sha, J., L. Pillai, A. A. Fadl, C. L. Galindo, T. E. Erova, and A. K. Chopra. 2005. The type III secretion system and cytotoxic enterotoxin alter the virulence of Aeromonas hydrophila. Infect. Immun. 736446-6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sha, J., S. F. Wang, G. Suarez, J. C. Sierra, A. A. Fadl, T. E. Erova, S. M. Foltz, B. K. Khajanchi, A. Silver, J. Graf, C. H. Schein, and A. K. Chopra. 2007. Further characterization of a type III secretion system (T3SS) and of a new effector protein from a clinical isolate of Aeromonas hydrophila—part I. Microb. Pathog. 43127-146. [DOI] [PubMed] [Google Scholar]
- 46.Sierra, J. C., G. Suarez, J. Sha, S. M. Foltz, V. L. Popov, C. L. Galindo, H. R. Garner, and A. K. Chopra. 2007. Biological characterization of a new type III secretion system effector from a clinical isolate of Aeromonas hydrophila—part II. Microb. Pathog. 43147-160. [DOI] [PubMed] [Google Scholar]
- 47.Smith, A. W., and B. H. Iglewski. 1989. Transformation of Pseudomonas aeruginosa by electroporation. Nucleic Acids Res. 1710509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suarez, G., J. C. Sierra, J. Sha, S. Wang, T. E. Erova, A. A. Fadl, S. M. Foltz, A. J. Horneman, and A. K. Chopra. 2008. Molecular characterization of a functional type VI secretion system from a clinical isolate of Aeromonas hydrophila. Microb. Pathog. 44344-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Szymanski, M., and J. Barciszewski. 2003. Regulation by RNA. Int. Rev. Cytol 231197-258. [DOI] [PubMed] [Google Scholar]
- 50.Tobe, T., C. Sasakawa, N. Okada, Y. Honma, and M. Yoshikawa. 1992. vacB, a novel chromosomal gene required for expression of virulence genes on the large plasmid of Shigella flexneri. J. Bacteriol. 1746359-6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Buul, C. P., and P. H. van Knippenberg. 1985. Nucleotide sequence of the ksgA gene of Escherichia coli: comparison of methyltransferases effecting dimethylation of adenosine in rRNA. Gene 3865-72. [DOI] [PubMed] [Google Scholar]
- 52.Viegas, S. C., V. Pfeiffer, A. Sittka, I. J. Silva, J. Vogel, and C. M. Arraiano. 2007. Characterization of the role of ribonucleases in Salmonella small RNA decay. Nucleic Acids Res. 357651-7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Welch, R. A., V. Burland, G. Plunkett III, P. Redford, P. Roesch, D. Rasko, E. L. Buckles, S. R. Liou, A. Boutin, J. Hackett, D. Stroud, G. F. Mayhew, D. J. Rose, S. Zhou, D. C. Schwartz, N. T. Perna, H. L. Mobley, M. S. Donnenberg, and F. R. Blattner. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 9917020-17024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wilusz, C. J., and J. Wilusz. 2004. Bringing the role of mRNA decay in the control of gene expression into focus. Trends Genet. 20491-497. [DOI] [PubMed] [Google Scholar]
- 55.Ygberg, S. E., M. O. Clements, A. Rytkonen, A. Thompson, D. W. Holden, J. C. Hinton, and M. Rhen. 2006. Polynucleotide phosphorylase negatively controls spv virulence gene expression in Salmonella enterica. Infect. Immun. 741243-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]