Abstract
Interactions involving genetically distinct bacteria, for example, between oral streptococci and actinomyces, are central to dental plaque development. A DNA microarray identified Streptococcus gordonii genes regulated in response to coaggregation with Actinomyces naeslundii. The expression of 23 genes changed >3-fold in coaggregates, including that of 9 genes involved in arginine biosynthesis and transport. The capacity of S. gordonii to synthesize arginine was assessed using a chemically defined growth medium. In monoculture, streptococcal arginine biosynthesis was inefficient and streptococci could not grow aerobically at low arginine concentrations. In dual-species cultures containing coaggregates, however, S. gordonii grew to high cell density at low arginine concentrations. Equivalent cocultures without coaggregates showed no growth until coaggregation was evident (9 h). An argH mutant was unable to grow at low arginine concentrations with or without A. naeslundii, indicating that arginine biosynthesis was essential for coaggregation-induced streptococcal growth. Using quantitative reverse transcriptase PCR, the expression of argC, argG, and pyrAb was strongly (10- to 100-fold) up-regulated in S. gordonii monocultures after 3 h of growth when exogenous arginine was depleted. Cocultures without induced coaggregation showed similar regulation. However, within 1 h after coaggregation with A. naeslundii, the expression of argC, argG, and pyrAb in S. gordonii was partially up-regulated although arginine was plentiful, and mRNA levels did not increase further when arginine was diminished. Thus, A. naeslundii stabilizes S. gordonii expression of arginine biosynthesis genes in coaggregates but not cocultures and enables aerobic growth when exogenous arginine is limited.
It is now widely accepted that most bacteria in nature exist in multispecies biofilm communities (48), where intergeneric interactions are commonplace (24). The outcome of these interactions plays a critical role in determining the success of an individual species in a mixed population. Dental plaque biofilms are particularly complex, harboring combinations of >700 species or phylotypes (1). From the earliest stages of plaque formation, microcolonies containing two or more genera of bacteria can be detected (24, 38), and therefore, significant potential for competitive or cooperative interactions exists through all phases in the development of dental biofilms. Specific cell-cell recognition, known as coaggregation, is thought to be fundamental for bringing together genetically distinct bacteria in dental plaque (21, 22).
Streptococcus spp. are key mediators of interactions in dental plaque since they produce multiple cell surface adhesins and receptors that bind both the salivary pellicle and numerous oral bacteria (17, 19). Streptococcus spp. are consistently identified as the predominant microorganisms in nascent dental plaque, comprising 60 to 80% of the total bacterial population on tooth surfaces for at least 24 h after tooth cleaning (8, 30, 36, 37). Actinomyces spp. constitute about one-third of nonstreptococcal cells in early plaque biofilms (37) and are often juxtaposed to Streptococcus spp. (38). A number of coaggregation interactions between Actinomyces spp. and Streptococcus spp. have previously been described. For example, type 2 fimbriae of Actinomyces naeslundii genospecies 2 are necessary for the recognition of streptococci that bear receptor polysaccharides (3, 34). Strong evidence that coaggregation between type 2 fimbriated actinomyces and receptor polysaccharide-bearing streptococci occurs in natural oral biofilms has been obtained with the use of specific antibodies against a known adhesin-receptor pair of antigens (38). These antigens were shown to be colocalized in plaque formed on enamel surfaces held in the mouths of human volunteers, indicating that the antigens interact in vivo (38). Such interactions are likely to initiate as well as stabilize biofilm communities in vivo and in vitro. Hence, it has been shown that the presence of Streptococcus gordonii DL1 on a saliva-conditioned surface in vitro significantly enhances the retention of a coaggregating partner, Actinomyces naeslundii T14V, in a biofilm under flowing saliva (39).
By promoting contact between distinct species of bacteria, coaggregation enhances the exchange of signals and metabolites between cells. Communication between Streptococcus oralis and A. naeslundii mediated by signaling molecule autoinducer 2 leads to the mutualistic growth of each species in a flow cell biofilm with saliva as the sole nutrient source, conditions under which neither species can grow alone (41). Other studies have identified specific changes in gene expression that occur in response to cell-cell communication between oral bacteria. For example, the sensing of arginine deiminase (ArcA) on the surface of Streptococcus cristatus leads to the down-regulation of the Porphyromonas gingivalis fimbrial gene fimA (56, 57). In a different system, a diffusible signal from Veillonella atypica mediates the up-regulation of S. gordonii amyB, encoding α-amylase (12). Coaggregation per se is not essential for signaling in a closed vessel since a coaggregation-deficient V. atypica mutant is able to elicit a response from S. gordonii. However, in biofilms formed under flowing saliva with wild-type (coaggregating) strains, only S. gordonii cells juxtaposed to V. atypica cells switch on the transcription of amyB (12). Therefore, in an open flowing system, akin to the human mouth, coaggregation is important for minimizing the dilution of diffusible signals and maximizing the productive exchange of signals between senders and responders.
Despite the clear evidence that cell-cell communication occurs between oral bacteria, no reports of systematic searches for genes regulated in response to coaggregation exist. DNA microarrays are an extremely powerful tool for investigating transcriptional changes in response to stimuli, and it has been demonstrated that microarrays can be employed to investigate gene expression in one bacterial species in the presence of another (33, 46). Here, we describe the application of a recently designed DNA microarray (55) to assess changes in S. gordonii gene expression in response to coaggregation with A. naeslundii. From this analysis, we show that coaggregation has a major impact on the expression of genes encoding enzymes for arginine biosynthesis. We therefore undertook an investigation into arginine biosynthesis in S. gordonii. Our data indicate that S. gordonii synthesizes arginine inefficiently and that coaggregation with A. naeslundii enables arginine biosynthesis under conditions that prohibit the growth of S. gordonii in monoculture. We propose that coaggregation enables S. gordonii cells to switch from quiescence to active growth when arginine is scarce, as it is in human saliva.
MATERIALS AND METHODS
Strains and growth media.
Streptococcus gordonii DL1 (Challis) and Actinomyces naeslundii MG1 (ATCC 43146) were routinely cultured in Todd-Hewitt broth (THB; Difco, Detroit, MI) or on THB medium solidified with 1.5% (wt/vol) Bacto agar, aerobically at 37°C and 5% CO2. The construction of the S. gordonii PK3337 argH::aphA3 strain is described below. This strain was subcultured in the presence of 250 μg/ml kanamycin. Chemically defined medium (CDM) was based on FMC (52) with the following modifications: l-leucine and l-isoleucine were each at a final concentration of 40 mg/liter rather than 100 mg/liter in FMC; l-arginine and l-histidine were at 100 mg/liter rather than 200 mg/liter in FMC; 0.1 mM CaCl2 (not present in FMC) was included, and the medium was adjusted to pH 7.3. All reagents for CDM were purchased from Sigma-Aldrich (St. Louis, MO). For some experiments, arginine was omitted or included at a different concentration. TYEG medium contained 1% (wt/vol) Bacto tryptone, 0.5% (wt/vol) yeast extract, 0.3% (wt/vol) K2HPO4, and 0.2% (wt/vol) d-glucose, adjusted to pH 7.5 before autoclaving.
Single- and mixed-species cultures.
For experiments involving growth in CDM, cells were first cultured in TYEG medium for 16 h anaerobically under a 90% N2-5% H2-5% CO2 atmosphere. Cells were harvested, resuspended in CDM, and adjusted to ∼5 × 109 CFU/ml. For monocultures, 300 μl of cell suspension was added to 14.7 ml CDM in a capped 15-ml glass tube (final concentration, 1 × 108 CFU/ml). To induce coaggregation in dual-species cultures, 300 μl of S. gordonii cells was combined with 300 μl of A. naeslundii cells, vortex mixed for 10 s, and adjusted to 15 ml with CDM. Vortex mixing dense suspensions of cells is a routine protocol for inducing coaggregation of organisms with complementary adhesins and receptors (23). For mixed-species cultures without induced coaggregation, 300 μl of S. gordonii cells and 300 μl of A. naeslundii cells were added to 14.4 ml CDM and mixed by gentle inversion. Cultures were incubated aerobically at 37°C.
Numbers of CFU were determined using 0.5-ml culture samples. Chains of streptococci, clumps of actinomyces, and coaggregates were disrupted by sonication in a Sonopuls ultrasonic homogenizer (Bandelin Electric, Berlin, Germany) equipped with a BR30 cup booster on 50% power for 1 min. Samples were serially diluted, and triplicate 20-μl portions of each dilution were dropped onto solid THB medium. Plates were incubated at 37°C and 5% CO2 for 24 h to enumerate S. gordonii CFU or for 48 h to quantify A. naeslundii CFU. Since A. naeslundii colonies were not visible after incubation for 24 h, this procedure was suitable for the specific enumeration of S. gordonii CFU from mixed-genus cultures. For the determination of A. naeslundii CFU in dual-species cultures, serial dilutions of the culture were dropped onto solidified THB medium supplemented with 128 mg/liter mupirocin and 2.5 mg/liter metronidazole (29), and plates were incubated at 37°C and 5% CO2 for 48 h. Turbidities of S. gordonii monocultures were measured using a Klett-Summerson colorimeter (Klett Manufacturing Co., Inc., New York, NY) with a 660-nm filter.
Fluorescence microscopy.
S. gordonii and A. naeslundii cells were visualized using Alexa Fluor-conjugated immunoglobulin G. Antibodies against S. gordonii DL1 (38) were labeled with Alexa 568, and anti-A. naeslundii T14V antibodies (5) were conjugated with Alexa 633 using Alexa Fluor labeling kits (Molecular Probes, Eugene, OR) according to the manufacturer's instructions. Cells were harvested, washed in phosphate-buffered saline (PBS), and resuspended in PBS containing 1% (wt/vol) bovine serum albumin and 10 μg of each antibody per ml. Cells were incubated at 25°C for 20 min, harvested, resuspended in PBS, and examined by epifluorescence microscopy.
Effects of arginine concentration on S. gordonii gene expression.
S. gordonii monocultures were prepared in CDM supplemented with arginine to a final concentration of 5 mM as described above and incubated anaerobically at 37°C to mid-exponential phase (140 to 160 Klett units). Cultures were divided into two aliquots, and cells were harvested by centrifugation at 3,500 × g for 7 min. One aliquot was resuspended in CDM containing 5 mM arginine, and the other was resuspended in CDM without arginine. Tubes were incubated anaerobically at 37°C for a further 30 min before RNA extraction. Results are shown as the geometric means and standard deviations for three independent cultures.
RNA extraction and purification.
Prior to the extraction of RNA from bacterial cells, intracellular RNA was stabilized by the addition of 2 volumes of RNAprotect (Qiagen, Valencia, CA) to 1 volume of sample from an exponentially growing culture, vortex mixing for 5 s, and incubation for 5 min at 20°C. Cells were harvested, the supernatant was discarded, and the pellet was stored at −70°C for up to 48 h. Total RNA was extracted using Trizol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions, with an extra step to ensure efficient disruption of gram-positive cells: following resuspension of the pellet in 1 ml Trizol, cells were mixed with lysing matrix B (Qbiogene, Morgan Irvine, CA), homogenized in a FastPrep bead beater (Qbiogene), and incubated at 20°C for 10 min. Extracted RNA was treated with RQ1 DNase I (Promega, Madison, WI) at 37°C for 1 h and repurified on Qiagen RNeasy MinElute columns. To ensure that RNA had not degraded during extraction and to determine the concentration of RNA, an aliquot of each sample was analyzed by capillary electrophoresis, using an RNA 6000 Nano chip in an Agilent 2100 bioanalyzer system (Agilent Technologies, Santa Clara, CA). Purified RNA was stored at −70°C.
Determination of extracellular arginine in CDM cultures.
Cell- and protein-free extracts were prepared from CDM cultures of S. gordonii or A. naeslundii incubated aerobically at 37°C for 3 h. Cells were pelleted by centrifugation at 15,000 × g for 10 min at 25°C. An aliquot (500 μl) of the supernatant was transferred to a Microcon YM-3 centrifugal filter device (Millipore, Billerica, MA) and centrifuged at 14,000 × g for 100 min at 25°C. Arginine concentrations were determined by Scientific Research Consortium (St. Paul, MN). Briefly, this process involved deproteinization in 13.5% (wt/vol) 5-sulfosalicylic acid hydrate and filtration through a 0.2-μm-pore-size membrane. Arginine was separated from the mixture on a dedicated high-performance liquid chromatography amino acid analyzer and detected by a colorimetric assay using ninhydrin reagent at 131°C. Three independent extracts of each monoculture and a coaggregate culture were analyzed, and arginine in uninoculated CDM was measured as a control.
Microarray hybridization and data analysis.
Labeling of RNA and microarray hybridization were performed according to the standardized protocols available at http://pfgrc.tigr.org/protocols.shtml. Briefly, aminoallyl-labeled cDNA was synthesized using Powerscript reverse transcriptase (Clontech, Mountain View, CA) and random hexamer primers (Invitrogen), with 5-(3-aminoallyl)-dUTP (Sigma) included in the reaction. Following the removal of unincorporated nucleotides on Qiagen MinElute columns, the cDNAs were labeled with Cy3 or Cy5 dye (Amersham Biosciences, Piscataway, NJ) and repurified on Qiagen MinElute columns. Labeling efficiencies were quantified using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE). Labeled cDNAs were denatured at 95°C for 5 min prior to use in hybridization reactions. Glass microarray slides containing 70-mer oligonucleotide probes for 2,195 S. gordonii open reading frames (ORFs) or loci (55) were prehybridized at 42°C for 1 h in 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.1% (wt/vol) sodium dodecyl sulfate (SDS), and 1% (wt/vol) bovine serum albumin. Slides were washed extensively in deionized water, washed once in isopropyl alcohol, and dried by centrifugation. Slides were hybridized with mixtures of Cy3- and Cy5-labeled cDNAs from two different populations (i.e., monocultured S. gordonii and S. gordonii-A. naeslundii coaggregate cultures) in hybridization buffer (40% formamide, 5× SSC, 0.1% [wt/vol] SDS, 0.6 mg/ml sheared salmon sperm DNA) at 42°C for 16 h. Following hybridization, slides were washed twice for 5 min at 25°C in each of three wash buffers: low-stringency buffer (2× SSC, 0.1% [wt/vol] SDS, 0.1 mM dithiothreitol [DTT] preheated to 55°C), medium-stringency buffer (0.1× SSC, 0.1% [wt/vol] SDS, 0.1 mM DTT), and high-stringency buffer (0.1× SSC, 0.1 mM DTT). Slides were rinsed several times in deionized water and air dried.
Images of microarray slides were produced using an Axon Genepix 4000B scanner (Molecular Devices, Sunnyvale, CA), and normalized spot intensities were determined with the associated Genepix Pro software. Data analysis was performed using the TM4 suite of applications (43). For statistical rigor, RNA was extracted from three independent sets of cultures and flip-dye replicates were performed for two of the three pairs of RNA preparations. Data from flip-dye replicate pairs were compared, and spots with inconsistent expression levels (varying by >2 standard deviations from the average intensity) were excluded from further analysis. For other spots, the geometric means of the fluorescence intensities were calculated. Data from six in-slide replicates per ORF/locus were combined, and the expression of each gene across three independent experiments was analyzed by significance analysis of microarrays (53), using a Δ value of 0.50.
Prediction of operon structure and regulatory motifs.
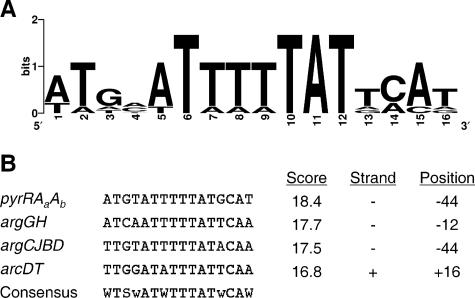
Promoters and terminators in the S. gordonii genome sequence were detected using the BProm and FindTerm modules of the fgenesB gene prediction program in Molquest software (Softberry Inc., Mount Kisco, NY). This software gives output scores from −1 to ∼25 to estimate the likelihood that a predicted promoter or terminator is functional; a higher score indicates that the prediction is more likely to be correct. The distribution of prediction scores was skewed, and >90% of putative promoters and terminators in the S. gordonii genome were predicted with scores below 10. We used an arbitrary range of values from 2.5 to 3.9 to indicate a promoter or terminator predicted with low confidence, and elements with scores of ≥4.0 were considered likely to be functional. Promoter/operator elements containing ARG box motifs were identified by searching the S. gordonii genome with a position weight matrix derived from known Bacillus subtilis AhrC recognition elements (see Fig. 6A), using the Virtual Footprint software program (http://prodoric.tu-bs.de/).
FIG. 6.
Identification of ARG box motifs in the promoter regions of arginine metabolism-related operons. (A) Sequence logo representing the B. subtilis ARG box consensus matrix that was used to search promoter regions of S. gordonii genes. (B) High-scoring matches upstream of S. gordonii operons involved in arginine biosynthesis and transport, identified using Virtual Footprint software. The similarity score is the sum of the weighted probabilities of finding the given base at each position. The maximum possible score for the ARG box element is 18.9, and the scores of known B. subtilis ARG box elements range from 15.4 to 18.4. The ARG box motifs upstream of pyrRAaAb, argGH, and argCJBD are on the negative strand relative to transcription, as indicated. The distances between the proximal base of the ARG box and the predicted transcription start sites, based on computational identification of σ70 promoters, are shown.
Disruption of argH in S. gordonii PK3337.
Routine cloning procedures were performed as described by Sambrook et al. (44). The argH gene, encoding arginosuccinate lyase, was replaced with the aphA3 kanamycin resistance determinant by allelic exchange mutagenesis. Sequences of primers used for mutagenesis are given in Table 1. Primers argHF1 and argHR1 were employed to amplify a 579-bp fragment comprising 30 bp of the 5′ end of argH and 549 bp upstream. A 606-bp fragment including 148 bp of the 3′ end of argH and 458 bp downstream was PCR amplified using primers argHF2 and argHR2. A second PCR was performed using equal amounts of the amplified products as templates and argHF1 and argHR2 primers and resulted in a 1,168-bp product comprising the ends of the argH gene with the surrounding sequence and a central EcoRI site. This fragment was cloned in pGEM-T, generating pGEM-argH. A 937-bp region harboring the aphA3 kanamycin resistance determinant was amplified from streptococcal integration plasmid pSF151 (50) by using primers aphA3F2 and aphA3R2 and ligated into the unique EcoRI site in pGEM-argH. The 2,086-bp argH::aphA3 insert was amplified from the vector by using primers argHF1 and argHR2 and used for the transformation of S. gordonii DL1 to generate the PK3337 ΔargH strain. The correct allelic exchange was verified by PCR amplification and DNA sequencing. To ensure that phenotypic effects of the gene disruption were not due to the presence of the aphA3 kanamycin resistance gene, a similar ΔargH mutant was constructed by allelic exchange with the ermAM erythromycin resistance determinant. This strain behaved similarly to the PK3337 argH::aphA3 strain in all assays (data not shown).
TABLE 1.
Primers used in this study
| Target genea | Primer | Primer sequence (5′ to 3′)b |
|---|---|---|
| argH | argHF1 | AGGTGTGCCGGTTGCTTTAGATG |
| argHR1 | ACGTCCGCCCCACAGTTTATG | |
| argH | argHF2 | AACTGTGGGGCGGACGTAGGAATTCTCGTCGCTGATTG |
| argHR2 | TTTCGCTCCGTCTCCTTGTAAT | |
| aphA3c | aphA3F2 | TTAGAATTCAAGGAACAGTGAATTGGAG |
| aphA3R2 | CGACGAATTCGATAAGCTTTTTAGACATCTAAATC | |
| 16S rRNA | 16SSgF1 | AGACACGGCCCAGACTCCTAC |
| gened | 16SSgR1 | CTCACACCCGTTCTTCTCTTACAA |
| 16S rRNA | 16SAnF1 | CGGGGTTGTGGGGCTGTCCTG |
| genee | 16SAnR1 | CACCCACTACGCCACGCCTTCC |
| spxB | 0292F | AGCACAAGGAGCTGTTGGAT |
| 0292R | GGAAGTGGACGGTGTTGAGT | |
| argG | 0175F | AAACGATCAGGTCCGTTTTG |
| 0175R | GATTTCTTCCTCCCGAGACC | |
| arcD | 1590F | GCGCGTGGTATCCAAGTTAT |
| 1590R | AGTCCTTGTTCACCCCAGTG | |
| bfbF | 1582F | TATCCGGCTACTTGCAATCC |
| 1582R | GCTCGCTAAAGTCCACCTTG | |
| bfbC | 1576F | ATTTTGGCGCCTATGACATC |
| 1576R | CCCAAGAAGGCTCCTATTCC | |
| argC | 1569F | AAAGAGCCTGCTGAAGACCA |
| 1569R | AGGGAATCAAGGCCAACTCT | |
| argD | 1566F | GCTTTTCAGGACCATCCAAA |
| 1566R | AATCCCCTGCTGAATTTCCT | |
| pyrAb | 1104F | CGCTAAGATTCCACGCTTTC |
| 1104R | TAGCCATGACTTCCCCTGTC | |
| amyB | 1075F | GACAGCGAAAACGGAAACTATGAC |
| 1075R | CCAATCGGAAGCCCTGTAT |
Genes are from S. gordonii unless otherwise indicated.
EcoRI sites, included in some primers to facilitate cloning, are underlined.
The aphA3 kanamycin resistance gene is carried on chimeric vector pSF151 (50).
The structural gene for S. gordonii 16S rRNA. There are four copies of this gene on the S. gordonii chromosome that are identical throughout the region encompassed by primers 16SSgF1 and 16SSgR1.
The structural gene for A. naeslundii 16S rRNA. The A. naeslundii chromosome harbors three copies of the template for 16S rRNA with identical sequences over the region amplified by primers 16SAnF1 and 16SAnR1.
Q-RT-PCR.
For the determination of relative mRNA concentrations by quantitative reverse transcription-PCR (Q-RT-PCR), RNA extracted from bacterial cells was reverse transcribed with Superscript II reverse transcriptase (Invitrogen) in accordance with the manufacturer's instructions. The reaction was terminated by heating to 70°C for 15 min, and RNA was degraded from RNA-DNA hybrid molecules by incubation with 2 U RNase H (Invitrogen) for 15 min at 37°C. cDNA was cleaned using MinElute columns (Qiagen). Primers for Q-RT-PCR were designed using Primer3 (42) and are listed in Table 1. Reaction mixtures contained 0 to 10 ng cDNA template, 12.5 μl Power Sybr green PCR master mix (Applied Biosystems, Foster City, CA), and forward and reverse primers, each at 300 nM, in a final reaction volume of 25 μl. Q-RT-PCR was performed in an MX3005P thermocycler (Stratagene, La Jolla, CA), using the following thermocycle program: 95°C for 10 min, 40 cycles of 30 s at 95°C, 56°C for 1 min, and 72°C for 30 s, and a dissociation curve consisting of incubation at 95°C for 1 min and 56°C for 30 s, with an incremental temperature increase to 95°C. Sybr green fluorescence data were collected following the 56°C primer annealing step in each of the 40 amplification cycles and throughout the dissociation curve. The presence of a single sharp peak in the dissociation curve indicated that one specific product had been amplified. The sizes of products from two representative reactions with each pair of primers were estimated by agarose gel electrophoresis. To determine the PCR amplification efficiency of each primer pair, 10-fold dilutions from 10 ng to 10−5 ng of a representative sample (cDNA derived from S. gordonii DL1 monoculture incubated for 3 h in CDM) were used as a template for Q-RT-PCR in three independent reactions. Only primer pairs that amplified a specific product of the predicted size with a reaction efficiency of >80% were included in this study.
The relative quantity of S. gordonii-derived cDNA in each sample was estimated by Q-RT-PCR using primers 16SSgF1 and 16SSgR1, which are specific for the S. gordonii 16S rRNA coding sequence. To compensate for variations in the efficiencies of RNA extraction and cDNA synthesis between samples, the expression of all other genes was normalized to the measured expression level of S. gordonii 16S rRNA genes. Control cDNA synthesis reactions without reverse transcriptase were performed for three independent preparations from S. gordonii monoculture cells incubated in CDM for 3 h. None of these gave significant levels of amplification in Q-RT-PCRs with any primer pair, indicating that RNA preparations were not contaminated with DNA. Negative controls without template were included in each Q-RT-PCR and did not amplify DNA.
Nucleotide sequence accession number.
Data from these experiments have been deposited in the GEO database under accession number GSE9478.
RESULTS
Coaggregation model for analysis of intergeneric communication between S. gordonii and A. naeslundii.
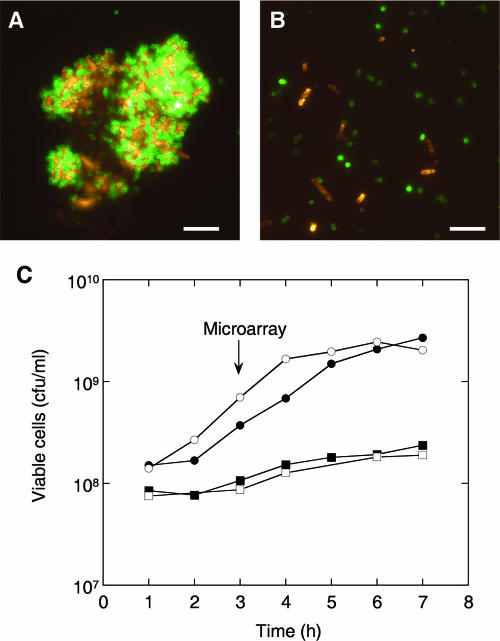
S. gordonii DL1 (Challis) coaggregates with many strains of A. naeslundii when cells are suspended in a standardized coaggregation buffer (4, 26) or in saliva (25). Preliminary experiments indicated that S. gordonii DL1 also formed stable coaggregation partnerships with A. naeslundii strains when cells were suspended in a variety of growth media. A. naeslundii MG1 (coaggregation group A [4]) and S. gordonii DL1 (coaggregation group 1) were selected for studies of intergeneric communication, since their complete genome sequences have been determined (http://cmr.tigr.org/tigr-scripts/CMR/GenomePage.cgi?org=gan and NCBI accession NC_009785, respectively). A microarray covering the genome of S. gordonii DL1 has recently been fabricated (55), and it is anticipated that microarrays for A. naeslundii MG1 will become available in the near future. Vortex mixing dense cell suspensions of S. gordonii DL1 and A. naeslundii MG1 (each strain at a concentration of ∼5 × 109 CFU/ml) in CDM resulted in the formation of coaggregates with almost-complete clarification of the medium. No visible change in the coaggregates occurred upon dilution of the medium (1:25). Using immunofluorescence microscopy, we observed that S. gordonii cells and A. naeslundii cells were intermingled throughout the coaggregates (Fig. 1A) and therefore had strong potential for intergeneric communication.
FIG. 1.
Growth of S. gordonii and A. naeslundii monocultures and coaggregates in CDM. (A and B) Fluorescence micrographs of mixed-species cultures after 3 h of growth. Cells were stained with antibodies against S. gordonii (green) or A. naeslundii (orange). The structure of an intact coaggregate (A) and the culture after gentle sonication to facilitate the enumeration of cells (B) are shown. Bars, 10 μm. (C) Growth of S. gordonii in monoculture (•) or coaggregates (○) and A. naeslundii in monoculture (▪) or coaggregates (□) was quantified using selective agar for A. naeslundii or nonselective agar and a limited incubation time (24 h) for S. gordonii. Samples for microarray analysis were taken at 3 h.
The impact of coaggregation with A. naeslundii on S. gordonii gene expression was assessed in batch monocultures or coaggregate cultures in CDM. For the quantification of each species in coaggregate cultures, samples were gently sonicated to separate single cells without cell lysis (Fig. 1B). Numbers of Actinomyces CFU were determined on Actinomyces selective agar; numbers of Streptococcus CFU were enumerated on nonselective agar with incubation for 24 h, at which time only S. gordonii colonies were visible (see Materials and Methods). The growth rate of S. gordonii was slightly higher in coaggregates than in monocultures, and the lag time was reduced (Fig. 1C). However, the final yields were similar in the presence and absence of A. naeslundii. A. naeslundii grew relatively slowly in CDM, and the growth rate was not affected by coaggregation with S. gordonii. For microarray analysis of S. gordonii gene expression, samples were taken 3 h after inoculation of the culture (Fig. 1C).
Microarray analysis of S. gordonii gene expression in monocultures and coaggregates.
Total RNA was purified from S. gordonii monocultures or from S. gordonii-A. naeslundii coaggregate cultures sampled at 3 h postinoculation. RNA was reverse transcribed, and cDNAs from the two populations of cells were competitively hybridized to a microarray containing 70-mer oligonucleotide probes for 2,195 predicted S. gordonii genes (see Materials and Methods). The proportion of cDNA derived from A. naeslundii in samples from coaggregates was estimated by Q-RT-PCR, using primers specific for genes encoding 16S rRNA from S. gordonii and A. naeslundii. These analyses indicated that cDNA from A. naeslundii comprised approximately 10 to 15% of the total cDNA (data not shown), which was in accordance with the proportion of A. naeslundii cells in coaggregate populations at 3 h (Fig. 1C). No significant hybridization of cDNA derived from A. naeslundii monocultures to the S. gordonii microarray was observed, and therefore, the presence of A. naeslundii RNA in samples from mixed cultures should not affect microarray analyses.
Initially, five genes that differed in expression from 2- to 3-fold between monocultures and coaggregates and five genes with >3-fold up- or down-regulation in coaggregates were selected for further analysis by Q-RT-PCR. Four of the five genes tested that were found by microarray analysis to be regulated between 2- and 3-fold appeared to be changed <1.5-fold by Q-RT-PCR and were not considered by this technique to be different between the two populations (data not shown). By contrast, all five tested genes that were regulated >3-fold by microarray analysis were also found to be regulated >3-fold by Q-RT-PCR. Therefore, genes with <3-fold regulation were excluded from further analysis. ORFs with >3-fold differences in expression between monocultures and coaggregate cultures are listed in Table 2. To test the statistical significance of the changes in gene expression, significance analysis of microarrays (53) was performed, using a Δ value of 0.50 and a resultant median false discovery rate of 0.36%. Of the 23 genes that were regulated >3-fold by coaggregation, 22 genes were considered significant by this test (Table 2).
TABLE 2.
Genes found to be strongly (>3-fold) regulated by coaggregation in a microarray screen
| ORF | Gene | Predicted function | Relative fold expression in coaggregates vs monoculturesa
|
|
|---|---|---|---|---|
| Microarray | Q-RT-PCRb | |||
| Up-regulated | ||||
| SGO_0292 | spxB | Pyruvate oxidase | 3.1 ± 0.7 | 4.7 ± 0.4 |
| SGO_1582 | bfbF | 6-Phospho-β-glucosidase | 4.3 ± 2.0 | 15.5 ± 3.9 |
| SGO_1581 | bfbG | Conserved hypothetical protein | 3.1 ± 0.4 | |
| SGO_1580 | bfbB | PTS, component EIIB | 4.2 ± 2.6c | |
| SGO_1579 | bfbR | Antiterminator, BglG family | 5.3 ± 2.2 | |
| SGO_1578 | bfbA | PTS, component EIIA | 4.3 ± 1.6 | |
| SGO_1577 | bfbD | Conserved hypothetical protein | 9.0 ± 2.4 | |
| SGO_1576 | bfbC | PTS, component EIIC | 7.8 ± 1.8 | 17.9 ± 1.9 |
| SGO_1308 | Conserved hypothetical protein | 3.2 ± 0.6 | ||
| Down-regulated | ||||
| SGO_0178 | rnpA | RNase P | −4.4 ± 0.2 | |
| SGO_0177 | Hypothetical protein | −5.0 ± 0.4 | ||
| SGO_0176 | argH | Arginosuccinate lyase | −13.9 ± 0.2 | |
| SGO_0175 | argG | Arginosuccinate synthase | −10.9 ± 0.3 | −20.9 ± 0.6 |
| SGO_1716 | Hpp family oligopeptide binding lipoprotein | −3.1 ± 0.1 | ||
| SGO_1590 | arcD | Arginine-ornithine antiporter | −4.2 ± 0.3 | −2.1 ± 0.4 |
| SGO_1569 | argC | N-Acetyl-γ-glutamyl-phosphate reductase | −15.5 ± 0.3 | −20.2 ± 0.8 |
| SGO_1568 | argJ | Bifunctional arginine biosynthesis protein | −11.8 ± 0.3 | |
| SGO_1567 | argB | N-Acetylglutamate kinase | −12.3 ± 0.2 | |
| SGO_1566 | argD | Acetylornithine aminotransferase | −7.7 ± 0.2 | −20.8 ± 0.6 |
| SGO_1109 | pyrB | Aspartate carbamoyltransferase | −3.5 ± 0.1 | |
| SGO_1108 | pyrP | Uracil permease | −4.5 ± 0.2 | |
| SGO_1104 | pyrAb | Carbamoyl phosphate synthase, large subunit | −4.6 ± 0.4 | −2.5 ± 0.1 |
| SGO_1103 | pyrAa | Carbamoyl phosphate synthase, small subunit | −4.3 ± 0.4 | |
Gene expression values represent geometric means ± standard deviations for three independent experiments.
The expression of representative genes was further analyzed by Q-RT-PCR. The absence of data indicates that expression levels were not determined.
Not considered significant by significance analysis of microarrays.
Nine genes were up-regulated in coaggregates; seven of these were consecutive ORFs on the chromosome (SGO_1576 to SGO_1582) and may represent a single operon (Table 2). This locus is involved in β-glucoside utilization and has been identified in screens for genes up-regulated in monoculture biofilms and for genes with increased expression in cells adhered to saliva-coated hydroxyapatite (20). The pyruvate oxidase gene spxB was up-regulated 3.1-fold in coaggregates. Pyruvate oxidase has been shown to have multiple roles in S. gordonii and related lactic acid-forming bacteria, including the production of H2O2 from pyruvate (45), H2O2 tolerance (40), virulence and attachment to host cells (47), binding to P. gingivalis (27), and stationary-phase survival (13). SGO_1308 was 3.2-fold increased in coaggregates and encodes a predicted protein of unknown function.
The majority (9/14) of genes down-regulated in coaggregates encode products involved in arginine and/or pyrimidine biosynthesis (Table 2). ORFs SGO_0175-SGO_0176 and SGO_1566 to SGO_1569 encode arginine biosynthesis enzymes, and SGO_1109 encodes a key enzyme in pyrimidine biosynthesis. ORFs SGO_1103 and SGO_1104, pyrAa and pyrAb (carbamoyl phosphate synthase subunits), are predicted to be involved in both pathways, since carbamoyl phosphate is required for the synthesis of arginine and pyrimidines. In some bacteria, pyrAa and pyrAb are specifically associated with pyrimidine biosynthesis, and a second carbamoyl phosphate synthase, encoded by carA and carB, is involved in arginine biosynthesis. However, genome sequence analysis indicated that carA and carB genes are not present in S. gordonii. Thus, in silico, the arginine and pyrimidine biosynthesis pathways appear to be linked in this organism.
SGO_1716 was down-regulated 3.1-fold in coaggregates. This ORF is located immediately upstream of and in the same direction as the hppH gene, encoding hexa/heptapeptide permease lipoprotein HppH (18). The expression of hppH was down-regulated 2.6-fold ± 0.2-fold in coaggregates, and it is possible that hppH and SGO_1716 form an operon. The polypeptide encoded by SGO_1716 has a secretion signal sequence containing a consensus motif for lipid modification, LAAC (15), and shares 61.2% amino acid identity with HppH. It is therefore likely that SGO_1716 is also involved in the uptake of short peptides. Two other genes encoding membrane transporters were down-regulated >3-fold in coaggregates: those for uptake of arginine (SGO_1590, arcD) and pyrimidines (SGO_1108, pyrP) (Table 2). SGO_0177 and SGO_0178, encoding a hypothetical protein and RNase P, respectively, were down-regulated four- to fivefold in coaggregates. These ORFs may be cotranscribed with argGH since they are immediately downstream of argH and are oriented in the same direction.
Genes and pathways in S. gordonii arginine metabolism.
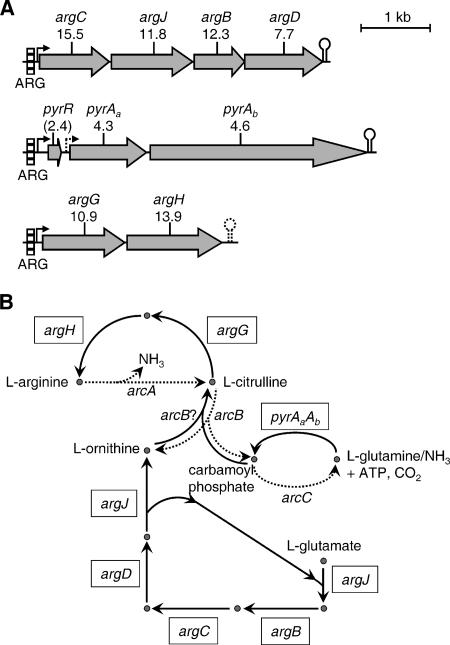
The greatest changes in gene expression resulting from coaggregation were in components of arginine biosynthesis, which were down-regulated up to 15-fold (Table 2). Furthermore, the regulation of arginine biosynthesis gene expression occurred at three distinct loci (ORFs SGO_0175-SGO_0176, SGO_1566 to SGO_1569, and SGO_1103-SGO_1104), indicating that this was a coordinated response to coaggregation. Analysis of these three loci, using MolQuest software for the prediction of promoter and Rho-independent terminator sequences, indicated that argCJBD, pyrRAaAb, and argGH are likely to form complete or partial operons (Fig. 2A). A putative promoter immediately upstream of pyrAa (5′-TTGAAA-N15-GTCAATAAT-3′) was predicted with low confidence, and it is possible that a stronger promoter upstream of pyrR directs the transcription of pyrRAaAb. A Rho-independent terminator was identified downstream of argH (5′-GAAAACATCGTGAAAAAGAGCGATGTTTTTGCTTTAT AAAAA-3′), which would form only a very weak stem-loop structure and may not be functional. Consensus ARG box elements that may be recognized by ArgR/AhrC-type transcriptional regulators were identified within 60 bp of promoters at each of the arginine biosynthesis gene loci (Fig. 2A). Transcriptional regulators of the ArgR/AhrC family are ubiquitous in eubacteria (32), and genomes of lactic acid bacteria, including Streptococcus spp., often encode multiple ArgR/AhrC homologues (35). A BLAST search of all predicted S. gordonii protein sequences with the amino acid sequence of B. subtilis AhrC returned three distinct matches, encoded by SGO_0697, SGO_1598, and SGO_2057, with scores of >70 and probabilities of hits occurring by chance (E values) of <1 × 10−14. Therefore, it is likely that S. gordonii has the capacity for transcriptional regulation at ARG box elements.
FIG. 2.
In silico analysis of arginine biosynthesis gene loci and arginine metabolism in S. gordonii. (A) Predicted operon structure of arginine biosynthesis genes. Promoters (forward arrows) were identified with the BProm module of MolQuest bioinformatics software, and terminators (double verticle line and loop) were found using the FindTerm module. Promoters and terminators identified with a high degree of confidence are indicated by solid lines, and weak elements are shown with a dashed line. ARG box consensus elements for recognition by ArgR-type transcriptional regulators were identified using Prodoric software and are marked by a striped rectangle. Genes and levels of down-regulation in coaggregated S. gordonii cells compared with those in monocultures are indicated (Table 2). The pyrR gene, encoding a putative regulator of pyrimidine metabolism, was <3-fold decreased in coaggregates (indicated by parentheses). (B) Arginine biosynthesis and catabolism pathways in S. gordonii and major intermediary compounds. Biosynthetic steps are indicated by solid lines, and dashed lines represent catabolic reactions. Arginine is produced from l-glutamate using carbamoyl phosphate. This in turn is synthesized by the glutamine-dependent carbamoyl phosphate synthase complex encoded by pyrAa and pyrAb. Note that the transfer of an N-acetyl group from N-acetyl-l-ornithine to l-glutamate, catalyzed by the argJ gene product, represents both the first and fifth steps in arginine biosynthesis. Arginine is degraded by the arginine dihydrolase system that yields two molecules of ammonia per arginine molecule. Genes that were strongly down-regulated in coaggregates are indicated with a box, and the functions encoded by these genes are listed in Table 2. The arc genes encode components of arginine catabolism as follows: arcA, arginine deiminase; arcB, ornithine carbamoyltransferase; and arcC, carbamate kinase. It is possible that the arcB gene product also catalyzes the anabolic synthesis of l-citrulline from l-ornithine and carbamoyl phosphate (see the text).
Genes encoding all enzymes for the synthesis of arginine from glutamate were down-regulated in coaggregated cultures compared with those in monocultures, with the exception of the gene encoding an ornithine carbamoyltransferase (EC 2.1.3.3) for the conversion of ornithine to citrulline (Fig. 2B). Ornithine carbamoyltransferase is evolutionarily related to aspartate carbamoyltransferase (28), involved in pyrimidine biosynthesis, and these enzymes can be distinguished by a characteristic signature sequence described at http://www.expasy.org/prosite (F-X-E/K-X-S-G/T-R-T; a glutamate residue at position 3 indicates that the enzyme recognizes aspartate as a substrate). A search of the S. gordonii genome in all six reading frames using the amino acid sequence of Lactococcus lactis anabolic ornithine carbamoyltransferase (ArgF) returned two sequences with BLAST scores of >100, ORFs SGO_1109 and SGO_1592. SGO_1109 is located within a cluster of pyrimidine biosynthesis genes and encodes an amino acid sequence containing the signature for aspartate carbamoyltransferase enzymes (PyrB). On the other hand, SGO_1592 encodes ArcB, an ornithine carbamoyltransferase enzyme associated with the catabolic arginine dihydrolase system (10). Therefore, there is no clear anabolic ornithine carbamoyltransferase encoded within the S. gordonii genome. Although no reports document that ArcB also has the capacity to catalyze the anabolic conversion of ornithine to citrulline for arginine biosynthesis in S. gordonii, we suggest that this might occur.
Extracellular arginine concentration is a stimulus for the coordinated regulation of arginine biosynthesis genes in S. gordonii.
Amino acid biosynthesis genes in bacteria are commonly regulated in response to the prevailing product concentration. The coordinated expression of arginine biosynthesis genes in response to exogenous arginine in E. coli was described over 40 years ago (31). Using Q-RT-PCR, the expression of S. gordonii arginine biosynthesis genes at three distinct loci (Fig. 2) was determined in cells cultured in arginine-replete (5 mM arginine) CDM, shifted to CDM without arginine, and incubated for a further 30 min. Under these conditions, the expression levels of argC, argG, and pyrAb were up-regulated 220-fold ± 1.8-fold, 130-fold ± 1.4-fold, and 9.8-fold ± 0.3-fold, respectively, in cells shifted to medium with no arginine compared with those of the genes in control cells resuspended in medium with high arginine concentrations. As a control, levels of amyB mRNA, encoding α-amylase, were monitored in each culture. The expression of amyB is increased in S. gordonii cells apposed to Veillonella atypica cells (12) but is not affected by coaggregation with A. naeslundii. Levels of amyB mRNA were not significantly regulated by arginine (1.5-fold ± 0.1-fold up-regulated in medium with no arginine).
The above data suggested that a major part of the response of S. gordonii to coaggregation might be caused by modulation of the arginine concentration in the external milieu by A. naeslundii cells. Therefore, the concentration of arginine in the bulk medium was determined following the growth of S. gordonii and A. naeslundii for 3 h in monoculture and coaggregate CDM (initial concentration, 0.5 mM arginine) cultures. In monoculture, S. gordonii consumed 57% ± 6.0% of the available extracellular arginine during this period, whereas A. naeslundii cells removed only 4.4% ± 3.0% of the arginine. In coaggregate cultures, arginine was depleted by 64% ± 3.6%. It should be noted that these measurements of global arginine concentrations will not have detected any local variation that may have occurred in arginine levels within or around S. gordonii cells in coaggregates.
Arginine biosynthesis in S. gordonii is inefficient.
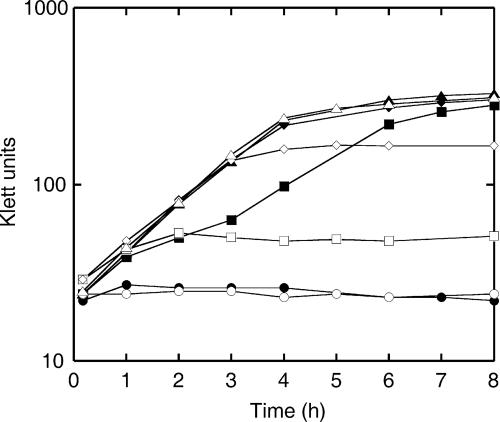
To determine whether S. gordonii has the capacity to synthesize arginine, a modified CDM without arginine was inoculated with S. gordonii and incubated at 37°C. No growth was observed in cultures incubated statically under atmospheric O2 for at least 72 h (data not shown). However, it has been shown that some streptococci that require arginine under aerobic conditions can grow anaerobically without exogenous arginine (51). Incubation of S. gordonii under strictly anaerobic conditions yielded high-cell-density cultures within 24 h, and growth was repeated over five subcultures (>40 generations). To analyze arginine biosynthesis in more detail, an isogenic argH::aphA3 mutant (strain PK3337) was generated by allelic exchange mutagenesis. Under aerobic atmosphere and at ≤5 mM arginine, the growth yield of S. gordonii PK3337 was proportional to the concentration of arginine in the medium, and maximum growth occurred with ≥1 mM arginine (Fig. 3). Wild-type S. gordonii DL1 grew to a high cell density within 8 h in a medium containing 0.1 mM arginine. However, the growth pattern was not a simple exponential increase in cell density. After an initial period of growth for 1 h, a lag phase of 2 h was observed before rapid growth resumed (Fig. 3). In standard CDM (0.5 mM arginine), S. gordonii DL1 grew exponentially for 4 h and reached a significantly higher cell density than strain PK3337, which entered stationary phase at 3 h. Thus, it appears that S. gordonii synthesizes arginine de novo during aerobic growth in CDM but is unable to initiate arginine biosynthesis in the absence of added arginine, indicating that the activation of the biosynthetic pathway for arginine is inefficient.
FIG. 3.
Effect of arginine availability on growth of S. gordonii DL1 or the PK3337 argH::aphA3 isogenic mutant. Symbols represent DL1 (•) or PK3337 (○) in 0 mM arginine, DL1 (▪) or PK3337 (□) in 0.1 mM arginine, DL1 (♦) or PK3337 (⋄) in 0.5 mM arginine, and DL1 (▴) or PK3337 (▵) in 1 mM arginine.
Coaggregation enables the growth of S. gordonii at low arginine concentrations.
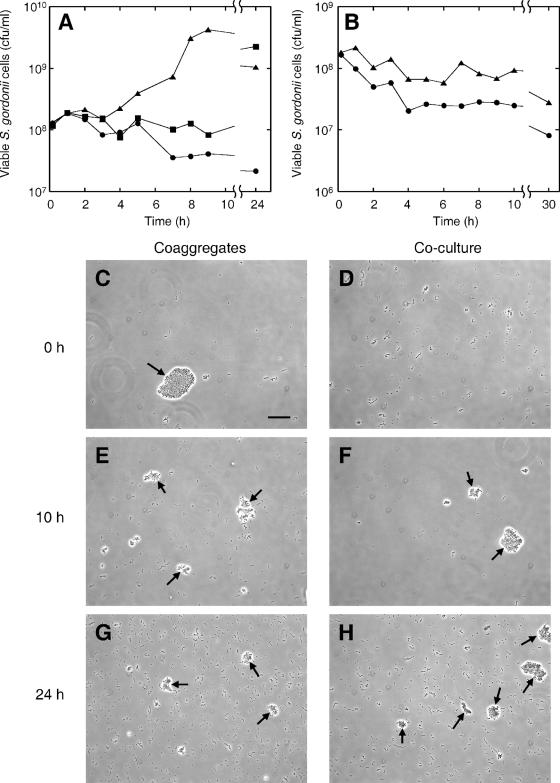
Since the expression of arginine biosynthesis genes was altered in S. gordonii cells in coaggregation with A. naeslundii, we hypothesized that coaggregation may affect the ability of S. gordonii to grow under aerobic conditions at low arginine concentrations that do not support efficient growth (i.e., ≤0.1 mM; Fig. 3). CDM containing reduced (0.025 mM) arginine was inoculated with ∼108 S. gordonii cells/ml or with a mixture of ∼108 S. gordonii cells/ml and ∼108 A. naeslundii cells/ml. Tubes were vortexed to induce coaggregate formation in the dual-species cultures and were incubated aerobically without shaking at 37°C. The growth of S. gordonii was monitored as described in Materials and Methods. S. gordonii DL1 monocultures did not grow in this medium, and viable cell numbers gradually decreased, reaching ∼2 × 107 cells/ml after 24 h (Fig. 4A). However, in coaggregates, S. gordonii cell numbers started to increase after ∼4 h and reached ∼4 × 109 cells/ml after 9 h. To test the role of coaggregation in enabling the growth of S. gordonii, a separate coculture containing S. gordonii cells and A. naeslundii cells that were not vortexed was set up and did not initially form coaggregates (compare Fig. 4C and D). Coaggregation occurred gradually during extended coculture incubation, and by 6 h, small clumps of ≤50 cells were observed (data not shown). After 10 h and 24 h, there was little difference between the number of coaggregates present in the coculture and that in the culture that was vortexed to induce coaggregation (Fig. 4E to H). No growth of S. gordonii was observed in cocultures during the early period of uninduced formation of small coaggregates, up to 9 h after inoculation. However, after prolonged incubation (24 h), high numbers of S. gordonii cells were detected in these cultures (>109 cells/ml), equivalent to the levels of S. gordonii cells in mixed-species cultures that were initially vortexed to induce coaggregates.
FIG. 4.
Effect of coaggregation or coculture on S. gordonii growth at low (0.025 mM) arginine concentrations. S. gordonii cells were incubated in monoculture or were mixed with A. naeslundii cells with or without vigorous vortexing to induce the formation of coaggregates. Growth of S. gordonii DL1 (A) in monoculture (•), coculture without vortexing (▪), or coaggregates (▴) or growth of S. gordonii PK3337 (B) in monoculture (•) or coaggregates (▴). (C to H) Phase-contrast micrographs of S. gordonii DL1 coaggregates (C, E, and G) and cocultures (D, F, and H) with A. naeslundii after 0 h (C and D), 10 h (E and F), or 24 h (G and H). Large coaggregates are indicated by arrows. Bar, 20 μM.
To investigate whether the S. gordonii arginine biosynthetic pathway was required for growth in coaggregates, the S. gordonii PK3337 argH::aphA3 strain was incubated in monoculture or in coaggregates with A. naeslundii as described above (Fig. 4B). The density of S. gordonii PK3337 cells decreased gradually in both monocultures and coaggregate cultures, indicating that these cells were not growing but were losing viability. Therefore, an intact arginine biosynthesis pathway is required for coaggregation-mediated growth of S. gordonii at low arginine concentrations.
Expression of S. gordonii arginine biosynthesis genes in coaggregates and cocultures.
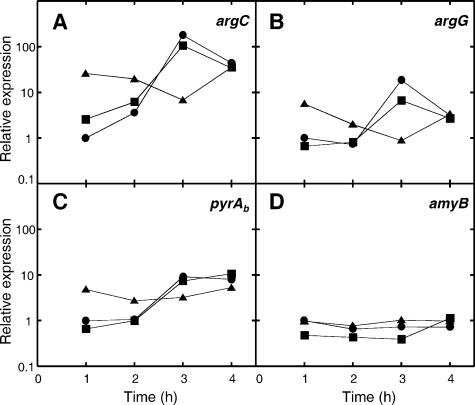
From the above-described experiments, it appeared that coaggregation with A. naeslundii, but not coculture in the absence of extensive coaggregation, facilitated arginine biosynthesis by S. gordonii. Q-RT-PCR was employed to assess the levels of expression of S. gordonii arginine biosynthesis genes in coaggregated and in cocultured streptococci and actinomyces compared with expression levels in S. gordonii monocultures (Fig. 5). Cells were cultured in standard CDM (0.5 mM arginine) and were sampled intermittently throughout exponential growth (Fig. 1). A marked increase in the expression levels of argC (50-fold increase), argG (26-fold), and pyrAb (9-fold) was observed in monocultures between 2 h and 3 h. Similar increases in argC, argG, and pyrAb expression levels were seen in cocultures that did not contain coaggregates (Fig. 5). By contrast, in coaggregated S. gordonii cells, argC, argG, and pyrAb mRNA levels were unchanged or slightly decreased (<3-fold) from 2 h to 3 h. Extracellular arginine had decreased from ∼0.5 mM initially to ∼0.2 mM by 3 h (see above). Differences in expression levels of S. gordonii arginine biosynthesis genes in monocultures or cocultures compared with those in coaggregates were apparent after just 1 h. At this time, the expression levels of argC, argG, and pyrAb were 26-, 6-, and 5-fold higher, respectively, in S. gordonii cells in coaggregation with A. naeslundii than in monocultures. The expression of argC, argG, and pyrAb in coaggregates remained relatively constant throughout growth, although argC and argG expression levels were decreased slightly at 3 h compared with those at 1 h (fourfold and sixfold, respectively). No significant differences in amyB expression were observed between any of the cultures at any time during growth (Fig. 5D).
FIG. 5.
Expression of arginine-related genes during growth of S. gordonii in CDM (0.5 mM arginine). The expression of arginine biosynthesis genes (argC, argG, and pyrAb) or a control gene (amyB) was monitored during growth in monoculture (•) or in coculture (▪) or coaggregation (▴) with A. naeslundii. The abundance of transcripts was quantified using Q-RT-PCR, and expression levels relative to those in the monoculture at 1 h are shown.
DISCUSSION
The most striking effect of coaggregation on the transcriptional profile of S. gordonii was the actinomyces-mediated stabilization of the expression of genes involved in arginine biosynthesis and uptake. Changes in the expression of pyrimidine biosynthesis/transport genes are likely to be linked to the regulation of arginine metabolism genes, since the biosynthetic pathways share a common intermediate, carbamoyl phosphate. SGO_1716 appears to encode an oligopeptide transporter component and thus may contribute to arginine metabolism. In addition, the regulation of ORFs SGO_0177 and SGO_0178 (immediately downstream of argGH) in coaggregates probably resulted from altered levels of argG promoter activity. Therefore, changes in arginine metabolism may underlie the majority of the observed gene regulation responses to coaggregation with A. naeslundii.
Coaggregation was essential for A. naeslundii-mediated changes in the expression of S. gordonii arginine biosynthesis genes; regulation was not observed in cocultures that did not contain coaggregates. Similarly, the initiation of S. gordonii growth in medium with little or no arginine was dependent on the formation of coaggregates. Therefore, the stimulus for changes in arginine metabolism cannot be a freely diffusible signaling molecule, and this system is distinct from the communication between V. atypica and S. gordonii (12) or from autoinducer 2-mediated mutualism between A. naeslundii and S. oralis (41). Measurements of arginine concentrations in the bulk medium provided clear evidence that S. gordonii imported arginine during growth and thus suggested that extracellular arginine might be a stimulus for gene regulation. This was confirmed in S. gordonii monocultures, since a shift from a high arginine concentration to no arginine resulted in a dramatic (10- to 200-fold, depending on the locus) up-regulation of arginine biosynthesis genes. It is therefore likely that arginine sensing played a major role in the communication between A. naeslundii and S. gordonii in coaggregates. A. naeslundii did not significantly modulate the global arginine concentration in the medium, and this is consistent with the absolute requirement for coaggregation to initiate changes in the S. gordonii arginine biosynthesis pathway.
It appears that coaggregation provides a local microenvironment in which arginine uptake and/or biosynthesis is facilitated in S. gordonii. Thus, S. gordonii cells may acquire arginine directly from A. naeslundii by a contact-dependent mechanism. For example, S. gordonii produces an extracellular arginine aminopeptidase (14) that could potentially scavenge arginine residues from proteins on the cell wall of A. naeslundii. However, A. naeslundii did not supply enough arginine to support the aerobic growth of an S. gordonii argH mutant in CDM containing 0.025 mM arginine (Fig. 4B). Alternatively, reduced O2 tension in the center of coaggregates might enhance the efficiency of arginine biosynthesis in S. gordonii. In this case, rapid up-regulation of arginine biosynthesis genes in cells protected from O2 would enable them to accumulate an intracellular pool of arginine that could be utilized once external arginine became scarce. This hypothesis would fit with the observation that S. gordonii arginine biosynthesis genes are initially up-regulated following coaggregation and then decline slightly as streptococci start to grow outside the coaggregates (Fig. 5). It would be interesting to determine whether enhanced S. gordonii arginine biosynthesis is dependent upon the metabolic activity of A. naeslundii cells in coaggregates or indeed whether oral bacteria other than A. naeslundii induce similar gene regulation in S. gordonii. Studies to resolve these issues are under way.
The regulation of arginine biosynthesis genes in bacteria is typically mediated by the binding of an ArgR/AhrC family transcriptional repressor to a consensus element, the ARG box, in the promoter/operator region of a gene or operon (32). Since ArgR/AhrC-type repressors are activated by binding to arginine, the transcription of arginine biosynthesis genes is repressed under arginine-replete conditions. It seemed likely that the coordinated regulation of S. gordonii arginine biosynthesis genes might be achieved by a similar mechanism. Therefore, we scanned the S. gordonii genome sequence for regions of similarity to a consensus position weight matrix based on confirmed Bacillus subtilis ARG boxes (32). Four of the five strongest matches that were within 60 bp of a putative σ70-dependent promoter element were sequences upstream of genes known to be involved in arginine metabolism and identified in this study as being coordinately regulated following coaggregation with A. naeslundii (Fig. 6). The S. gordonii genome sequence encodes three ArgR/AhrC-type regulators, designated ArgR, AhrC, and ArcR. We hypothesize that coaggregation with A. naeslundii affects the binding of one or more of these regulators to the ARG box sequences upstream of pyrR, argG, argC, and arcD and that this in turn modulates the expression of each operon. Experiments are currently under way to elucidate the functions of ArgR, AhrC, and ArcR in the regulation of arginine biosynthesis by generating and analyzing knockout mutants in each regulator individually and in combination.
Streptococci are nutritionally fastidious microorganisms that need multiple amino acids and vitamins for growth. Requirements for arginine vary among different species and strains and are influenced by environmental factors such as O2 tension and the presence of carbonate (6, 7, 51). In aerobic monocultures, we have found that >0.025 mM arginine is required for the initiation of arginine biosynthesis in S. gordonii. In medium containing low (0.1 mM) arginine concentrations, a lag phase was observed after the exogenous arginine had been consumed (Fig. 3), indicating that a period of adaptation occurred before arginine biosynthesis was initiated and rapid growth resumed. We suggest that this adaptation reflects a bottleneck in the arginine biosynthesis pathway caused by the poor expression of arcB at low arginine concentrations. Analysis of the S. gordonii genome indicated that a single ornithine carbamoyltransferase, encoded by arcB, might catalyze both the catabolic and the anabolic interconversion of ornithine and citrulline (Fig. 2B). The expression of arcB in S. gordonii is thought to be directed by the promoter upstream of arcA, which is subject to multiple regulatory controls: it is activated by ArcR in the presence of arginine and by Flp under anaerobic conditions and is repressed by CcpA in response to glucose or other phosphotransferase transport system (PTS) sugars (9, 58). Therefore, at low concentrations of arginine and in the presence of oxygen and glucose, the arcA promoter is inactive and arcB is poorly expressed. Once arginine biosynthesis is initiated, positive feedback may increase the expression of arcABC to a level where there is sufficient production of the ArcB enzyme for arginine biosynthesis to occur. Arginine catabolism might be avoided at this point by independent control of ArcA (arginine deiminase) activity or by rapid incorporation of newly synthesized arginine into proteins. This model suggests that a minimal level of arcB expression is essential for the maintenance of arginine homeostasis in S. gordonii and that if arcB transcript levels fall below this threshold, cells cannot synthesize enough arginine for growth. We are currently investigating the role of arcB in arginine biosynthesis by constructing a knockout mutant and a complemented strain in which arcB is under the control of a heterologous promoter.
A number of S. gordonii genes unrelated to arginine biosynthesis were up-regulated in response to coaggregation with A. naeslundii. For example, seven genes in the biofilm-related bfb locus were strongly (three- to ninefold) up-regulated. Using Q-RT-PCR to monitor gene expression, coaggregation was essential for this response; in cocultures of S. gordonii and A. naeslundii that were not vortexed to induce coggregation, the expression levels of bfb genes were equivalent to those in S. gordonii monocultures (data not shown). The bfb locus is one of several gene clusters encoding PTS components and associated β-glucosidases that have been shown to be inducible using an S. gordonii in vivo expression technology screening system (20). Increased promoter activity upstream of bfb genes was reported by 25% of in vivo expression technology clones retrieved from monoculture biofilms (20). The disruption of any one of three genes in this locus, bfbF (β-glucosidase), bfbB (PTS EIIB), and/or bfbR (antiterminator), resulted in strains deficient in monospecies biofilm formation on plastic surfaces in microtiter wells. Furthermore, several distinct clones with reporter gene insertions in the bfb locus were recovered from a model of adhesion to saliva-coated hydroxyapatite, indicating that bfb genes are up-regulated by adherence to a surface (20). Our results confirm that adhesion is a trigger for increased expression of bfb genes and demonstrate that coaggregation is an excellent model for investigating biofilms and surface contact. Other genes up-regulated in response to coaggregation were SGO_1308, encoding a hypothetical 154-amino-acid protein, and spxB, encoding pyruvate oxidase. Recently, it has been shown that SpxB is required for the binding of S. gordonii to P. gingivalis (27), and this protein may have a similar function in adherence to A. naeslundii. Alternatively, the up-regulation of spxB expression may enhance the ability of S. gordonii to compete with neighboring species by increasing H2O2 production.
In summary, we have shown that coaggregation with A. naeslundii stabilizes arginine metabolism in S. gordonii and reduces dependence on extracellular arginine. This is likely to be important in early dental plaque since the concentration of free arginine in saliva is extremely low, in the order of 0.004 mM to 0.03 mM (2, 49, 54). Even within a mature 48-h dental plaque biofilm, free arginine is present only at around 0.2 mM (16). S. gordonii can obtain arginine by degrading host salivary polypeptides such as proline-rich proteins (11). However, it is not clear that this can provide sufficient arginine for metabolism and growth, particularly in dense plaque biofilms where diffusion of proteins is limited. These data indicate that coaggregation may be fundamental for the successful colonization of oral surfaces by streptococci and may act as a switch to stimulate growth in multispecies biofilm environments.
Acknowledgments
This research was supported in part by the Intramural Research Program of the National Institute of Dental and Craniofacial Research, National Institutes of Health. Microarray fabrication and analysis were supported by Public Health Service grant DE11090 from the National Institute of Dental and Craniofacial Research awarded to M.M.V.
Footnotes
Published ahead of print on 21 March 2008.
REFERENCES
- 1.Aas, J. A., B. J. Paster, L. N. Stokes, I. Olsen, and F. E. Dewhirst. 2005. Defining the normal bacterial flora of the oral cavity. J. Clin. Microbiol. 435721-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brand, H. S., G. G. Jörning, R. A. Chamuleau, and L. Abraham-Inpijn. 1997. Effect of a protein-rich meal on urinary and salivary free amino acid concentrations in human subjects. Clin. Chim. Acta 26437-47. [DOI] [PubMed] [Google Scholar]
- 3.Cisar, J. O., S. H. Curl, P. E. Kolenbrander, and A. E. Vatter. 1983. Specific absence of type 2 fimbriae on a coaggregation-defective mutant of Actinomyces viscosus T14V. Infect. Immun. 40759-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cisar, J. O., P. E. Kolenbrander, and F. C. McIntire. 1979. Specificity of coaggregation reactions between human oral streptococci and strains of Actinomyces viscosus or Actinomyces naeslundii. Infect. Immun. 24742-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cisar, J. O., A. E. Vatter, and F. C. Mcintire. 1978. Identification of virulence-associated antigen on surface fibrils of Actinomyces viscosus T14. Infect. Immun. 19312-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cowman, R. A., M. M. Perrella, B. O. Adams, and R. J. Fitzgerald. 1975. Amino acid requirements and proteolytic activity of Streptococcus sanguis. Appl. Microbiol. 30374-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cowman, R. A., M. M. Perrella, and R. J. Fitzgerald. 1974. Influence of incubation atmosphere on growth and amino acid requirements of Streptococcus mutans. Appl. Microbiol. 2786-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diaz, P. I., N. I. Chalmers, A. H. Rickard, C. Kong, C. L. Milburn, R. J. Palmer, Jr., and P. E. Kolenbrander. 2006. Molecular characterization of subject-specific oral microflora during initial colonization of enamel. Appl. Environ. Microbiol. 722837-2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dong, Y., Y. Y. Chen, and R. A. Burne. 2004. Control of expression of the arginine deiminase operon of Streptococcus gordonii by CcpA and Flp. J. Bacteriol. 1862511-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong, Y., Y. Y. Chen, J. A. Snyder, and R. A. Burne. 2002. Isolation and molecular analysis of the gene cluster for the arginine deiminase system from Streptococcus gordonii DL1. Appl. Environ. Microbiol. 685549-5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drobni, M., T. Li, C. Krüger, V. Loimaranta, M. Kilian, L. Hammarström, H. Jörnvall, T. Bergman, and N. Strömberg. 2006. Host-derived pentapeptide affecting adhesion, proliferation, and local pH in biofilm communities composed of Streptococcus and Actinomyces species. Infect. Immun. 746293-6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Egland, P. G., R. J. Palmer, Jr., and P. E. Kolenbrander. 2004. Interspecies communication in Streptococcus gordonii-Veillonella atypica biofilms: signaling in flow conditions requires juxtaposition. Proc. Natl. Acad. Sci. USA 10116917-16922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goffin, P., L. Muscariello, F. Lorquet, A. Stukkens, D. Prozzi, M. Sacco, M. Kleerebezem, and P. Hols. 2006. Involvement of pyruvate oxidase activity and acetate production in the survival of Lactobacillus plantarum during the stationary phase of aerobic growth. Appl. Environ. Microbiol. 727933-7940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldstein, J. M., D. Nelson, T. Kordula, J. A. Mayo, and J. Travis. 2002. Extracellular arginine aminopeptidase from Streptococcus gordonii FSS2. Infect. Immun. 70836-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hayashi, S., and H. C. Wu. 1990. Lipoproteins in bacteria. J. Bioenerg. Biomembr. 22451-471. [DOI] [PubMed] [Google Scholar]
- 16.Higham, S. M., and W. M. Edgar. 1989. Human dental plaque pH, and the organic acid and free amino acid profiles in plaque fluid, after sucrose rinsing. Arch. Oral Biol. 34329-334. [DOI] [PubMed] [Google Scholar]
- 17.Hsu, S. D., J. O. Cisar, A. L. Sandberg, and M. Kilian. 1994. Adhesive properties of viridans streptoccocal species. Microb. Ecol. Health Dis. 7125-137. [Google Scholar]
- 18.Jenkinson, H. F., R. A. Baker, and G. W. Tannock. 1996. A binding-lipoprotein-dependent oligopeptide transport system in Streptococcus gordonii essential for uptake of hexa- and heptapeptides. J. Bacteriol. 17868-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jenkinson, H. F., and R. J. Lamont. 1997. Streptococcal adhesion and colonization. Crit. Rev. Oral Biol. Med. 8175-200. [DOI] [PubMed] [Google Scholar]
- 20.Kiliç, A. O., L. Tao, Y. Zhang, Y. Lei, A. Khammanivong, and M. C. Herzberg. 2004. Involvement of Streptococcus gordonii β-glucoside metabolism systems in adhesion, biofilm formation, and in vivo gene expression. J. Bacteriol. 1864246-4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kolenbrander, P. E. 1993. Coaggregation of human oral bacteria: potential role in the accretion of dental plaque. J. Appl. Bacteriol. 74(Suppl.)79S-86S. [DOI] [PubMed] [Google Scholar]
- 22.Kolenbrander, P. E. 2000. Oral microbial communities: biofilms, interactions, and genetic systems. Annu. Rev. Microbiol. 54413-437. [DOI] [PubMed] [Google Scholar]
- 23.Kolenbrander, P. E. 1989. Surface recognition among oral bacteria: multigeneric coaggregations and their mediators. Crit. Rev. Microbiol. 17137-159. [DOI] [PubMed] [Google Scholar]
- 24.Kolenbrander, P. E., R. J. Palmer, Jr., A. H. Rickard, N. S. Jakubovics, N. I. Chalmers, and P. I. Diaz. 2006. Bacterial interactions and successions during plaque development. Periodontol. 2000 4247-79. [DOI] [PubMed] [Google Scholar]
- 25.Kolenbrander, P. E., and C. S. Phucas. 1984. Effect of saliva on coaggregation of oral Actinomyces and Streptococcus species. Infect. Immun. 44228-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kolenbrander, P. E., and B. L. Williams. 1981. Lactose-reversible coaggregation between oral actinomycetes and Streptococcus sanguis. Infect. Immun. 3395-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuboniwa, M., G. D. Tribble, C. E. James, A. O. Kiliç, L. Tao, M. C. Herzberg, S. Shizukuishi, and R. J. Lamont. 2006. Streptococcus gordonii utilizes several distinct gene functions to recruit Porphyromonas gingivalis into a mixed community. Mol. Microbiol. 60121-139. [DOI] [PubMed] [Google Scholar]
- 28.Labedan, B., A. Boyen, M. Baetens, D. Charlier, P. Chen, R. Cunin, V. Durbeco, N. Glansdorff, G. Herve, C. Legrain, Z. Liang, C. Purcarea, M. Roovers, R. Sanchez, T. L. Toong, M. Van de Casteele, F. van Vliet, Y. Xu, and Y. F. Zhang. 1999. The evolutionary history of carbamoyltransferases: a complex set of paralogous genes was already present in the last universal common ancestor. J. Mol. Evol. 49461-473. [DOI] [PubMed] [Google Scholar]
- 29.Lewis, R., D. McKenzie, J. Bagg, and A. Dickie. 1995. Experience with a novel selective medium for isolation of Actinomyces spp. from medical and dental specimens. J. Clin. Microbiol. 331613-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li, J., E. J. Helmerhorst, C. W. Leone, R. F. Troxler, T. Yaskell, A. D. Haffajee, S. S. Socransky, and F. G. Oppenheim. 2004. Identification of early microbial colonizers in human dental biofilm. J. Appl. Microbiol. 971311-1318. [DOI] [PubMed] [Google Scholar]
- 31.Maas, W. K. 1994. The arginine repressor of Escherichia coli. Microbiol. Rev. 58631-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Makarova, K. S., A. A. Mironov, and M. S. Gelfand. 2001. Conservation of the binding site for the arginine repressor in all bacterial lineages. Genome Biol. 2research0013.1-research0013.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mashburn, L. M., A. M. Jett, D. R. Akins, and M. Whiteley. 2005. Staphylococcus aureus serves as an iron source for Pseudomonas aeruginosa during in vivo coculture. J. Bacteriol. 187554-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McIntire, F. C., L. K. Crosby, A. E. Vatter, J. O. Cisar, M. R. McNeil, C. A. Bush, S. S. Tjoa, and P. V. Fennessey. 1988. A polysaccharide from Streptococcus sanguis 34 that inhibits coaggregation of S. sanguis 34 with Actinomyces viscosus T14V. J. Bacteriol. 1702229-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nicoloff, H., F. Arsène-Ploetze, C. Malandain, M. Kleerebezem, and F. Bringel. 2004. Two arginine repressors regulate arginine biosynthesis in Lactobacillus plantarum. J. Bacteriol. 1866059-6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nyvad, B., and M. Kilian. 1990. Comparison of the initial streptococcal microflora on dental enamel in caries-active and in caries-inactive individuals. Caries Res. 24267-272. [DOI] [PubMed] [Google Scholar]
- 37.Nyvad, B., and M. Kilian. 1987. Microbiology of the early colonization of human enamel and root surfaces in vivo. Scand. J. Dent. Res. 95369-380. [DOI] [PubMed] [Google Scholar]
- 38.Palmer, R. J., Jr., S. M. Gordon, J. O. Cisar, and P. E. Kolenbrander. 2003. Coaggregation-mediated interactions of streptococci and actinomyces detected in initial human dental plaque. J. Bacteriol. 1853400-3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Palmer, R. J., Jr., K. Kazmerzak, M. C. Hansen, and P. E. Kolenbrander. 2001. Mutualism versus independence: strategies of mixed-species oral biofilms in vitro using saliva as the sole nutrient source. Infect. Immun. 695794-5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pericone, C. D., S. Park, J. A. Imlay, and J. N. Weiser. 2003. Factors contributing to hydrogen peroxide resistance in Streptococcus pneumoniae include pyruvate oxidase (SpxB) and avoidance of the toxic effects of the Fenton reaction. J. Bacteriol. 1856815-6825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rickard, A. H., R. J. Palmer, Jr., D. S. Blehert, S. R. Campagna, M. F. Semmelhack, P. G. Egland, B. L. Bassler, and P. E. Kolenbrander. 2006. Autoinducer 2: a concentration-dependent signal for mutualistic bacterial biofilm growth. Mol. Microbiol. 601446-1456. [DOI] [PubMed] [Google Scholar]
- 42.Rozen, S., and H. Skaletsky. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol 132365-386. [DOI] [PubMed] [Google Scholar]
- 43.Saeed, A. I., N. K. Bhagabati, J. C. Braisted, W. Liang, V. Sharov, E. A. Howe, J. Li, M. Thiagarajan, J. A. White, and J. Quackenbush. 2006. TM4 microarray software suite. Methods Enzymol. 411134-193. [DOI] [PubMed] [Google Scholar]
- 44.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 45.Sedewitz, B., K. H. Schleifer, and F. Götz. 1984. Purification and biochemical characterization of pyruvate oxidase from Lactobacillus plantarum. J. Bacteriol. 160273-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Simionato, M. R., C. M. Tucker, M. Kuboniwa, G. Lamont, D. R. Demuth, G. D. Tribble, and R. J. Lamont. 2006. Porphyromonas gingivalis genes involved in community development with Streptococcus gordonii. Infect. Immun. 746419-6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spellerberg, B., D. R. Cundell, J. Sandros, B. J. Pearce, I. Idanpaan-Heikkila, C. Rosenow, and H. R. Masure. 1996. Pyruvate oxidase, as a determinant of virulence in Streptococcus pneumoniae. Mol. Microbiol. 19803-813. [DOI] [PubMed] [Google Scholar]
- 48.Stoodley, P., K. Sauer, D. G. Davies, and J. W. Costerton. 2002. Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 56187-209. [DOI] [PubMed] [Google Scholar]
- 49.Syrjanen, S. M., L. Alakuijala, P. Alakuijala, S. O. Markkanen, and H. Markkanen. 1990. Free amino acid levels in oral fluids of normal subjects and patients with periodontal disease. Arch. Oral Biol. 35189-193. [DOI] [PubMed] [Google Scholar]
- 50.Tao, L., D. J. LeBlanc, and J. J. Ferretti. 1992. Novel streptococcal-integration shuttle vectors for gene cloning and inactivation. Gene 120105-110. [DOI] [PubMed] [Google Scholar]
- 51.Terleckyj, B., and G. D. Shockman. 1975. Amino acid requirements of Streptococcus mutans and other oral streptococci. Infect. Immun. 11656-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Terleckyj, B., N. P. Willett, and G. D. Shockman. 1975. Growth of several cariogenic strains of oral streptococci in a chemically defined medium. Infect. Immun. 11649-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 985116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Van Wuyckhuyse, B. C., H. E. Perinpanayagam, D. Bevacqua, R. F. Raubertas, R. J. Billings, W. H. Bowen, and L. A. Tabak. 1995. Association of free arginine and lysine concentrations in human parotid saliva with caries experience. J. Dent. Res. 74686-690. [DOI] [PubMed] [Google Scholar]
- 55.Vickerman, M. M., S. Iobst, A. M. Jesionowski, and S. R. Gill. 2007. Genome-wide transcriptional changes in Streptococcus gordonii in response to competence signaling peptide. J. Bacteriol. 1897799-7807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xie, H., G. S. Cook, J. W. Costerton, G. Bruce, T. M. Rose, and R. J. Lamont. 2000. Intergeneric communication in dental plaque biofilms. J. Bacteriol. 1827067-7069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xie, H., X. Lin, B. Y. Wang, J. Wu, and R. J. Lamont. 2007. Identification of a signalling molecule involved in bacterial intergeneric communication. Microbiology 1533228-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zeng, L., Y. Dong, and R. A. Burne. 2006. Characterization of cis-acting sites controlling arginine deiminase gene expression in Streptococcus gordonii. J. Bacteriol. 188941-949. [DOI] [PMC free article] [PubMed] [Google Scholar]