Abstract
Squalene synthase (SQS) is a bifunctional enzyme that catalyzes the condensation of two molecules of farnesyl diphosphate (FPP) to give presqualene diphosphate (PSPP) and the subsequent rearrangement of PSPP to squalene. These reactions constitute the first pathway-specific steps in hopane biosynthesis in Bacteria and sterol biosynthesis in Eukarya. The genes encoding SQS were isolated from the hopane-producing bacteria Thermosynechococcus elongatus BP-1, Bradyrhizobium japonicum, and Zymomonas mobilis and cloned into an Escherichia coli expression system. The expressed proteins with a His6 tag were found exclusively in inclusion bodies when no additives were used in the buffer. After extensive optimization, soluble recombinant T. elongatus BP-1 SQS was obtained when cells were disrupted and purified in buffers containing glycerol. The recombinant B. japonicum and Z. mobilis SQSs could not be solubilized under any of the expression and purification conditions used. Purified T. elongatus His6-SQS gave a single band at 42 kDa by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and molecular ion at m/z 41886 by electrospray mass spectrometry. Incubation with FPP and NADPH gave squalene as the sole product. Incubation of the enzyme with [14C]FPP in the absence of NADPH gave PSPP. The enzyme requires Mg2+ for activity, has an optimum pH of 7.6, and is strongly stimulated by detergent. Under optimal conditions, the Km of FPP is 0.97 ± 0.10 μM and the kcat is 1.74 ± 0.04 s−1. Zaragozic acid A, a potent inhibitor of mammalian, fungal, and Saccharomyces cerevisiae SQSs, also inhibited recombinant T. elongatus BP-1 SQS, with a 50% inhibitory concentration of 95.5 ± 13.6 nM.
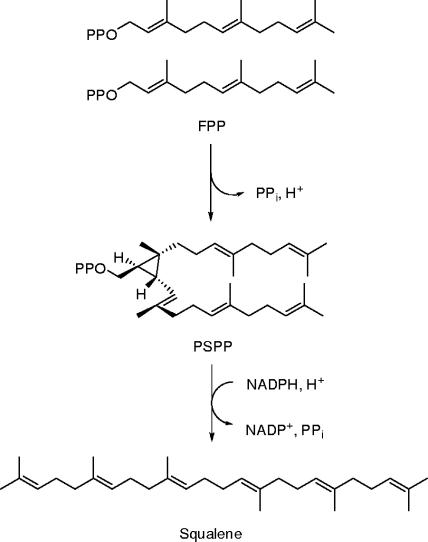
Squalene synthase ([SQS] farnesyl diphosphate:farnesyl diphosphate transferase; EC 2.5.1.21) catalyzes the condensation of two molecules of farnesyl diphosphate (FPP) to form a c1′-2-3-linked triterpene intermediate, presqualene diphosphate (PSPP), and the subsequent NADPH-dependent rearrangement and reduction of PSPP to produce squalene (SQ), as outlined in Fig. 1 (36, 37). These transformations are the first pathway-specific reactions in the hopanoid biosynthetic pathway in Bacteria (7, 33, 42, 48) and the sterol pathway in Eukarya (37). Studies of yeast SQS show that substrate addition is ordered with two molecules of FPP added first, followed by NADPH (24, 39). When NADPH is present in the buffer, PSPP is converted directly to SQ without dissociating from the active site. Formation of PSPP or a prior conformational change in SQS is the rate-limiting step in the overall conversion of FPP to SQ. PSPP is formed and released when NADPH is absent. Under these conditions, SQS catalyzes a slow “solvolysis” of PSPP to give triterpene alcohols and hydrocarbons with irregular isoprenoid skeletons (14).
FIG. 1.
Conversion of FPP to SQ.
SQS has been cloned from a variety of eukaryotes, including fungal (15, 21, 57), protozoan (45), rat (22), mouse (12), human (41, 50), and plant sources (27). The eukaryotic enzyme is associated with membranes, where it appears to be anchored by a hydrophobic membrane-spanning α-helix at the C terminus of the protein (47, 49, 57). The poor solubility of SQS has hampered attempts to purify native soluble protein. Small quantities of soluble SQS from yeast microsomes were purified with deoxycholate and detergents (1, 18). A purified sample of membrane-free, C-terminally truncated, rat hepatic microsomal SQS was obtained by trypsin digestion (47). A modestly soluble recombinant version of SQS was obtained from Saccharomyces cerevisiae (21, 57), Trypanosoma cruzi (45), rats (22), and humans (52) by truncation of the coding region for the C-terminal α-helix.
Hopanoids are pentacyclic triterpene lipids found in many bacteria (42), including a number of nitrogen-fixing organisms, and several species of cyanobacteria (7, 33, 48). Hopanoids are localized in bacterial membranes, where they exert many of the same stabilizing effects as membrane sterols in eukaryotes (16, 31). In contrast to eukaryotic SQS, little is known about the bacterial enzyme. Predicted secondary structures from amino acid sequences for the bacterial enzyme do not show the C-terminal α-helix seen in the eukaryotic proteins. There are no reports of solubilization or purification of a native or recombinant bacterial SQS. We now describe construction of an Escherichia coli clone with the full-length gene for SQS (tll1096) from the thermophilic cyanobacterium Thermosynechococcus elongatus BP-1 and the solubilization, purification, and characterization of the recombinant enzyme.
MATERIALS AND METHODS
Materials.
Genomic DNA for T. elongatus BP-1 was provided by Satoshi Tabata (Kazusa DNA Research Institute, Japan) (25). Bradyrhizobium japonicum USDA 110 (NRRL B-4361) and Zymomonas mobilis ZM4 (ATCC 31821) strains were obtained from Agricultural Research Service Culture Collection at the U.S. Department of Agriculture and American Type Culture Collection, respectively (17, 46). Magnesium chloride, Tween 80, dithiothreitol (DTT), bovine serum albumin (BSA), SQ, NADPH, and zaragozic acid A were purchased from Sigma. EDTA-free protease inhibitor cocktail tablets were purchased from Roche Diagnostics. Ni-nitrilotriacetic acid (NTA) agarose resin was purchased from Qiagen. All restriction endonucleases and T4 DNA ligase were purchased from New England Biolabs. pGEM-T Easy vector and Taq DNA polymerase were purchased from Promega. Deoxynucleoside triphosphates, a BenchMark protein ladder, and an Easy-DNA Genomic DNA Isolation Kit were purchased from Invitrogen. Easy-A PCR cloning enzyme, Pfu Ultra DNA polymerase, and XL1-Blue competent cells were purchased from Stratagene. pET28b(+) vector, BL21(DE3), and Rosetta (DE3) competent cells were purchased from Novagen. Microcon and Centriprep centrifugal filter devices were purchased from Millipore. Attempts to refold SQS after solubilization of inclusion bodies were conducted with a Spin-column Membrane Protein Folding Screen Kit from ProFoldin Protein Folding Service. Recombinant E. coli FPP synthase was provided by Mo Chen (unpublished data). Geranyl diphosphate (GPP) and FPP were synthesized by the procedure of Davisson et al. (8). [1-14C]isopentenyl diphosphate (IPP) and [1-3H]FPP were purchased from GE Healthcare.
General methods.
Mini-preparations of plasmid DNA for restriction analysis were obtained by using a Qiagen plasmid mini-prep kit. Genomic DNA was prepared from B. japonicum and Z. mobilis cells using an Easy-DNA Genomic DNA Isolation Kit. DNA fragments were purified by agarose gel electrophoresis using a GFX PCR DNA and gel purification kit from GE Healthcare. Restriction digestion, ligation, and transformation of competent cells were conducted as described by Sambrook et al. (44). PCR was performed using a PTC-200 Petier Thermal Cycler from MJ Research. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed using the discontinuous buffer system of Laemmli (19). Gels were stained with Gelcode blue stain reagent from Pierce. Protein concentrations were determined with a Coomassie plus protein assay reagent (Pierce) using a BSA standard. Oligonucleotide primers were synthesized by Integrated DNA Technologies. DNA was sequenced at the Health Sciences Center Sequencing Facility, Eccles Institute of Human Genetics, University of Utah. The molecular masses of proteins were determined by positive-ion electrospray mass spectrometry in the University of Utah Chemistry Department mass spectrometry laboratory. Homology searches were performed at the National Center for Biotechnology Information website. Pairwise sequence alignments were performed with the Vector NTI (Informax) and EMBL-EBI (European Bioinformatics Institute) servers (www.ebi.ac.uk/Tools/clustalw/index.html) using the ClustalW algorithm.
Cloning of bacterial SQS.
Nested PCR primers were used to amplify DNA fragments containing the SQS-coding gene tll1096 (EMBL nucleotide sequence accession no. BAC08649) with Easy-A PCR cloning enzyme from T. elongatus BP-1 genomic DNA as follows: forward primer, 5′-TTGTGGAAGGCATCCCCTGCTGGGTTGCGATC-3′; reverse primer, 5′-TAGCGATGACTGGCGGCAAACTCAGCACGG-3′. PCR conditions (25 cycles) were as follows: initial denaturation at 95°C for 120 s, denaturation at 95°C for 45 s, annealing at 66.4°C for 45 s, extension at 72°C for 90 s, and a final extension at 72°C for 7 min. Additional PCR primers were designed incorporating a 5′ NdeI site (underlined) and a 3′ HindIII site (underlined) complementary to tll1096 from nested PCR products as follows: NdeI 5′-CATATGCGTGTAGGAGTGAACC-3′ and HindIII 5′-AAGCTTATTAAAGTCCGCCAAGGATGAAAGG-3′. The stop codon was mutated from TAG to TAA. The start and stop codons are shown in bold. PCR conditions were identical to those described for nested PCR, except that the annealing steps were modified (52.7°C for 45 s). The gel-purified 1.1-kb PCR product was tailed with an A residue and ligated into the subcloning vector pGEM-T Easy using T4 DNA ligase to give pgBP1Q. pgBP1Q was transformed to XL1-Blue by electroporation for blue-white screening. Individual colonies from Luria-Bertani (LB)-ampicillin (100 μg/ml)-tetracycline (12 μg/ml) agar plates were picked, and the plasmid DNA was isolated. pgBP1Q was sequenced to verify that the 1.1-kb insert was identical to the deposited sequence for tll1096 from T. elongatus BP-1 (25).
pgBP1Q was digested with NdeI and HindIII and ligated using T4 DNA ligase into the doubly digested expression vector pET28b(+) with the same restriction enzymes to give the expression plasmid ptBP1QX. After tll1096 was subcloned to pET28b(+), the resulting plasmid (ptBP1QX) was transformed to XL1-Blue by electroporation. Individual colonies from LB-kanamycin (LB-Kan; 30 μg/ml) agar plates were picked, and the plasmid DNA was isolated and sequenced. This construct encoded a version of T. elongatus BP-1 SQS with an N-terminal His6 tag and a thrombin proteolytic site.
B. japonicum strain USDA 110 was cultivated in a yeast-mannitol medium to prepare genomic DNA as previously described (23). Nested PCR was used to amplify DNA fragments containing the long-version (EMBL nucleotide sequence accession no. BAC48266) and short-version (34) SQS genes from B. japonicum genomic DNA. Additional PCR primers were used to incorporate a 5′ NdeI site and a 3′ HindIII site complementary to the B. japonicum SQS gene from nested PCR products as follows for the long version of SQS (restriction sites are underlined, and start and stop codons are in boldface): for NdeI, 5′-[CATATGGGTGCCGCGGCGGCATCGCCG-3′; for HindIII, 5′-AAGCTTATTAAGCGTCATGTGCAGTCCCCGGTCTGG-3′. GGG next to the start codon in the forward primer was changed to GGT to introduce a silent mutation. For the short version of SQS, the primers were the following: for NdeI, 5′-CATATGACCTCTGCGAGCGAATTGCGATCCGGC-3′; for HindIII, 5′-AAGCTTATTAAGCGTCATGTGCAGTCCCCGGTCTGG-3′, were used. The stop codon was mutated from TAG to TAA. PCR conditions were identical to those used for nested PCR except for modification of the annealing steps (62.3°C for 45 s). Z. mobilis strain ZM4 was cultivated in RM medium (glucose [20 g/liter], yeast extract [10 g/liter], KH2PO4 [2 g/liter] [pH 6.0]) to prepare genomic DNA as previously described (4). The PCR primers used to incorporate a 5′ NdeI site and a 3′ HindIII site complementary to the Z. mobilis SQS gene (EMBL nucleotide sequence accession no. AAV89493) from Z. mobilis genomic DNA were as follows: for NdeI, 5′-CATATGGAAGGGGCGTGCGCAAGCACGTATAG-3′; for HindIII, 5′-AAGCTTATTAAGAAAATAATCTCCCTGCGGCGGCGAC-3′. The start and stop codons were mutated from GTG to ATG and TAG to TAA, respectively. PCR conditions were identical to those described for T. elongatus BP-1 SQS except for modifications to the annealing steps (58.3°C for 45 s). Procedures for cloning B. japonicum and Z. mobilis SQS after PCR were performed as described for T. elongatus BP-1 SQS cloning using pGEM-T Easy and pET28b(+) vectors. The final constructs encoded versions of B. japonicum and Z. mobilis SQSs with N-terminal His6 tags and thrombin proteolytic sites. The generated plasmids and E. coli strains are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| XL1-blue | Host for cloning vector; Tetr | Stratagene |
| XL-pgBP1Q | XL1-Blue containing pgBP1Q | This work |
| XL-ptBP1QX | XL1-Blue containing ptBP1QX | This work |
| XL-pgBJQlg | XL1-Blue containing pgBJQlg | This work |
| XL-ptBJQlgX | XL1-Blue containing ptBJQlgX | This work |
| XL-pgBJQst | XL1-Blue containing pgBJQst | This work |
| XL-ptBJQstX | XL1-Blue containing ptBJQstX | This work |
| XL-pgZMQ | XL1-Blue containing pgZMQ | This work |
| XL-ptZMQX | XL1-Blue containing ptZMQX | This work |
| Rosetta(DE3) | Host for protein expression; provides rare codon tRNAs; Camr | Novagen |
| ptBP1QXRO | Rosetta (DE3) containing ptBP1QX; Kanr Camr | This work |
| Thermosynechococcus elongatus BP-1 | Genomic DNA having SQS gene (tll1096) | S. Tabata; 25 |
| Bradyrhizobium japonicum USDA 110 | Proteobacteria strain having SQS gene | USDA |
| Zymomonas mobilis ZM4 | Proteobacteria strain having SQS gene (zmo0869) | ATCC |
| Plasmids | ||
| pGEM-T Easy | Cloning vector for PCR products; Ampr | Promega |
| pgBP1Q | pGEM-T Easy with PCR product containing ORF tll1096 | This work |
| pgBJQlg | pGEM-T Easy with PCR product containing ORF blr3001 | This work |
| pgBJQst | pGEM-T Easy with PCR product containing short version of SQS; bp 3312158-3313043 in B. japonicum genomea | This work |
| pgZMQ | pGEM-T Easy with PCR product containing ORF zmo0869 | This work |
| pET28b(+) | Expression vector with an N-terminal His6 tag; Kanr | Novagen |
| ptBP1QX | pET28b(+) with a 1.1-kb NdeI-HindIII insert corresponding to ORF tll1096 | This work |
| ptBJQlgX | pET28b(+) with a 1.0-kb NdeI-HindIII insert corresponding to ORF blr3001 | This work |
| ptBJQstX | pET28b(+) with a 0.9-kb NdeI-HindIII insert corresponding to short version of SQS | This work |
| ptZMQX | pET28b(+) with a 0.9-kb NdeI-HindIII insert corresponding to ORF zmo0869 | This work |
See reference 34.
Expression and purification of recombinant T. elongatus BP-1 SQS.
ptBP1QX was transformed into Rosetta (DE3) to create strain ptBP1QXRO. LB medium was used for all growth conditions (44). The bacterial strains used in this study are listed in Table 1 and were grown at 30°C in LB medium supplemented with chloramphenicol (Cam; 34 μg/ml) and Kan (30 μg/ml) as necessary. Cultures were grown by a three-stage fermentation protocol. LB-Cam-Kan cultures (5 ml) inoculated with a single colony of ptBP1QXRO were incubated by shaking at 250 rpm at 37°C for 10 h. These starter cultures were used to inoculate 100 ml of LB-Cam-Kan and grown overnight at 37°C. Inocula (10 ml) from the stage II culture were used to inoculate 1 liter of LB-Cam-Kan cultures. The cultures were grown to an optical density at 600 nm of approximately 0.6, induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside), and incubated for an additional 6 h at 30°C. Transformation and expression of ptBJQlgX, ptBJQstX, and ptZMQX were identical with the previous procedures for ptBP1QX.
For purification of recombinant T. elongatus BP-1 SQS, the cells from strain ptBP1QXRO were harvested by centrifugation for 20 min at 4°C at 5,000 × g. All steps in the purification were performed at 4°C. Cell paste (11 g) was suspended in 20 ml of lysis buffer consisting of 50 mM sodium phosphate (pH 8), 300 mM NaCl, 10 mM imidazole, 50% (vol/vol) glycerol, and a protease inhibitor cocktail tablet. The cells were disrupted by sonication (5 sets of 30-s pulses), and the resulting homogenate was centrifuged at 12,000 × g to remove cellular debris. Glycerol-free lysis buffer was added to the resulting supernatant to lower the glycerol concentration to 20%, Ni-NTA (7 ml in a 50% slurry) was added to the resulting mixture, and the suspension was swirled at 4°C at 100 rpm for 1 h on a rotary shaker. The slurry was poured into a 100-ml fritted glass column, and the flowthrough was collected. The Ni-NTA resin was washed with 30 ml of a 20% (vol/vol) glycerol, 50 mM imidazole, and 50 mM sodium phosphate buffer, pH 8.0, containing 300 mM NaCl; samples were further washed with 30 ml of 20% (vol/vol) glycerol, 70 mM imidazole, and 50 mM sodium phosphate buffer, pH 8.0, containing 300 mM NaCl. The recombinant protein was eluted with 20 ml of 20% (vol/vol) glycerol, 250 mM imidazole, and 50 mM sodium phosphate buffer, pH 8.0, containing 300 mM NaCl and analyzed by SDS-PAGE. The protein was concentrated via ultrafiltration (YM-10; Centriprep) and dialyzed against 20% (vol/vol) glycerol-50 mM HEPES, pH 7.5, containing 0.1 mM DTT to yield 10.3 mg of enzyme that gave a single band (>97% purity) by SDS-PAGE. The protein was flash-frozen and stored at −80°C until needed.
SQS assays.
SQS was assayed by modification of the procedure of Zhang and Poulter (58). In a typical assay, 10 μM [1-3H]FPP (50 μCi 3H/μmol) was incubated with 30 ng of SQS in 200 μl of 50 mM morpholinepropanesulfonic acid (MOPS) buffer, pH 7.2, containing 10 mM MgCl2, 1 mM DTT, 1 mg/ml BSA, 1 mM NADPH, and 1% (vol/vol) Tween 80 (buffer A). The reaction was initiated by adding 10 μl of a solution of SQS in the same buffer to 190 μl of the assay cocktail. The reaction mixture was incubated at 37°C for 5 min and quenched with 300 μl of methanol-40% (wt/vol) aqueous KOH (1:1, vol/vol). After NaCl (∼0.4 g) was added to saturate the mixture, ligroin (2 ml) containing 0.5% (vol/vol) SQ was added, and the resulting mixture was vortexed vigorously for 30 s. A 1-ml portion of the ligroin layer was loaded onto a 0.5- by 6-cm alumina column (80 to 200 mesh; Fisher), packed in a Pasteur pipette, and preequilibrated with 2 ml of ligroin containing 0.5% (vol/vol) SQ. The column was eluted with five 1-ml portions of toluene containing 0.5% (vol/vol) SQ. The radioactivity of the eluent was measured in 10 ml of Ultima Gold liquid scintillation cocktail (PerkinElmer) by liquid scintillation spectrometry (Packard).
Electrospray mass spectrometry of recombinant T. elongatus BP-1 SQS.
Concentrated protein (10 μl) was diluted into 500 μl of water-acetonitrile-formic acid (80:20:0.2) and concentrated via ultrafiltration (YM-10; Microcon). The protein was resuspended in the same solution, concentrated, and resuspended in 200 μl of water-acetonitrile-formic acid (50:50:0.2) to give a final protein concentration of 15 μM. Positive-ion electrospray mass spectrometry was performed on a Waters Micromass Quattro II triple-quadrupole mass spectrometer.
Product studies of recombinant T. elongatus BP-1 SQS with GC and GC-mass spectrometry.
FPP (50 μg; 0.12 μmol) and SQS (360 μg; 9.1 nmol) in 1 ml of 50 mM MOPS buffer, pH 7.2, containing 10 mM MgCl2 and 1 mM NADPH were incubated at 37°C for 2 h. NaCl (∼1 g) was added, and the mixture was extracted with 5 ml of methyl tert-butyl ether. The extracts were concentrated to ∼100 μl with a gentle stream of N2, and a 1-μl portion of the extract was analyzed by gas chromatography (GC) on an Agilent 6890N gas chromatograph with flame ionization detection using a 30-m by 0.318-mm (bore size) by 0.25-μm (film thickness) HP-5 capillary column (J & W Scientific). Compounds were eluted with a temperature gradient from 60 to 260°C at 10°C increase/min; temperature was then maintained at 260°C for 20 min with He, with a flow rate of 1.3 ml/min. Similar conditions were used to analyze samples by GC-mass spectrometry on an HP 5971A mass spectrometer. The mass spectra of eluted peaks were compared with authentic SQ.
Formation of [14C]PSPP and [14C]SQ by recombinant T. elongatus BP-1 SQS.
Formation of PSPP in the absence of NADPH by recombinant T. elongatus BP-1 SQS was measured using [1-14C]FPP synthesized by incubation of [1-14C]IPP and GPP with FPP synthase. The incubation mixture consisted of 18 μg of recombinant E. coli FPP synthase in 38 μl of 50 mM MOPS buffer, pH 7.2, 50 μM [1-14C]IPP (50 μCi 14C/μmol), 135 μM GPP, 10 mM MgCl2, 1 mM DTT, and 1 mg/ml BSA. After 10 min at 37°C, 0.6 μg of recombinant T. elongatus BP-1 SQS in 2 μl of MOPS buffer was added. The incubation was continued at 37°C for 2 h with occasional mixing before the reaction was quenched by the addition of 10 μl of 250 mM EDTA. A 5-μl portion of the resulting reaction mixture was spotted on a silica gel 60 F254 thin-layer chromatography (TLC) plate (Merck), which was developed with 30:70:13:8 chloroform-formic acid-pyridine-water. Radioactivity on the TLC plate was measured with a Typhoon 8600 imaging analyzer (GE Healthcare). In a parallel experiment, SQ formation was measured under the same conditions by adding 1 mM NADPH to the reaction mixture. The TLC plate was developed with 4:1 hexane-toluene.
Dependence on metal ions, NADPH, pH, temperature, and detergents.
Assays were typically run in 50 mM MOPS buffer, pH 7.2, containing 10 μM [1-3H]FPP (50 μCi 3H/μmol), 1 mM DTT, and 1 mg/ml BSA (buffer B). The reactions were worked up as described for the standard assay. For studies of Mg2+ dependence, the assays contained 0.2 to 30 mM MgCl2, 1 mM NADPH, and 1% (vol/vol) Tween 80 in buffer B. For studies of the NADPH dependence of SQS, assays contained 0 to 3.0 mM NADPH, 10 mM MgCl2, and 1% (vol/vol) Tween 80 in buffer B. Activity at pHs ranging from 5.3 to 9.0 at 37°C was measured in buffer A using a 50 mM concentration of a different buffer system, depending on the pH (pH 5.3, acetate; pH 5.7 to 6.4, morpholinethanesulfonic acid; pH 6.7 to 7.6, MOPS; pH 7.9 to 8.2, HEPES; pH 8.6 to 9.5, CHES [2-(cyclohexylamino) ethanesulphonic acid]). Activity at different temperatures (25 to 70°C) was measured using buffer A. The dependence of activity on Tween 80 concentration (0 to 3%, vol/vol) was measured in buffer B containing 1 mM NADPH and 10 mM MgCl2.
Steady-state kinetic measurements.
Assays were performed in duplicate in 50 mM MOPS buffer, pH 7.2, containing 10 mM MgCl2, 1 mM DTT, 1 mg/ml BSA, and 1% (vol/vol) Tween 80 at 37°C. For kinetic measurements to determine the Km and kcat of FPP (KmFPP and kcatFPP, respectively), the assay buffer contained 0.25 to 20 μM [1-3H]FPP (37.5 to 750 μCi 3H/μmol) and 1 mM NADPH. For the KmNADPH and kcatNADPH, the assay buffer contained 0.05 to 3 mM NADPH and 10 μM [1-3H]FPP (50 μCi 3H/μmol). Conversion of FPP to SQ was limited to 10% or less. Values for Km and kcat were calculated by fitting initial velocities at different substrate concentrations to the appropriate form of the Michaelis-Menten equation (39).
Inhibition of recombinant T. elongatus BP-1 SQS by zaragozic acid A.
The 50% inhibitory concentration of zaragozic acid A for T. elongatus BP-1 SQS was determined by measuring the conversion of FPP to SQ in the presence of different concentrations of zaragozic acid A. [1-3H]FPP (1 μM; 50 μCi 3H/μmol) and 0 to 160 nM zaragozic acid A were incubated with 5 ng of SQS in 200 μl of buffer A. The assays were then performed as described above in “SQS assays”.
RESULTS AND DISCUSSION
Sequence analysis of genes encoding bacterial SQS.
Genes encoding SQS have been cloned from a variety of eukaryotic sources, including yeast (15, 21, 57), protozoa (45), rat (22), mouse (12), human (41, 50), and plant (27). In bacteria, hopanoid biosynthesis gene clusters containing the gene for SQS, hpnC, were cloned from Z. mobilis and B. japonicum, and synthesis of SQ was detected in cell extracts of the transformants (34). We conducted a database search for proteins homologous to biochemically characterized SQSs from fungi, animals, and plants and found hits in cyanobacteria, proteobacteria, and archaea, including T. elongatus BP-1, Gloeobacter violaceus PCC 7421 (EMBL nucleotide sequence accession no. BAC91999) (26), Methylococcus capsulatus (EMBL nucleotide sequence accession no. CAA71097) (53), Halobacterium salinarum NRC-1 (EMBL nucleotide sequence accession no. AAG19173) (29), and Haloarcula marismortui ATCC 43049 (EMBL nucleotide sequence accession no. AAV46402) (3). Additional hits for SQS were found, as expected, in the hopane-producing bacteria B. japonicum, Z. mobilis, and M. capsulatus (33, 40, 53). T. elongates BP-1 also has a putative SQ-hopane cyclase (EMBL nucleotide sequence accession no. BAC09861).
Figure 2 shows amino acid alignments of representative SQSs from bacteria and eukaryotes. T. elongatus BP-1 SQS, which is typical of the bacterial SQSs, has ∼30% overall similarity with the mouse, rat, human, S. cerevisiae, and Schizosaccharomyces pombe proteins (Fig. 2) (15, 22, 27, 41, 50, 51, 57). Eukaryotic SQSs have four conserved regions (Fig. 2, I to IV), which kinetic studies with site-directed mutants and a crystal structure indicate are important for catalysis (10, 32). Regions I, II, and III are involved in the first half-reaction, the condensation of two molecules of FPP to give PSPP. Mutation of the highly conserved Tyr171 in region II of rat SQS appears to be particularly important (10). Regions I and III are likely involved in binding of the diphosphate units in FPP via bridging Mg2+ (10, 32). Region IV is thought to be required for the rearrangement of PSPP to SQ. Although SQS does not have a “typical” NADPH binding motif, likely candidates are the FCAIPQVMAIATL sequence found in region IV and the VKIRK sequence located downstream from region IV in the case of rat and human SQSs (10, 32). VKIRK is the most flexible part of human SQS in its crystal structure and is thought to be stabilized by NADPH binding (32). A related enzyme, phytoene synthase (PS), converts geranylgeranyl diphosphate to phytoene (13, 28). SQS and PS both catalyze the cyclopropanation and rearrangement reactions shown in Fig. 1. However, the rearrangement step in phytoene synthesis is terminated by loss of a proton, and the enzyme does not utilize NADPH (13, 50). Eukaryotic and bacterial PSs do not have sequences that correspond to the putative NADPH binding motifs found in SQSs.
FIG. 2.
Amino acid sequence alignments for SQSs from Rattus norvegicus (rSQS), Homo sapiens (hSQS), S. cerevisiae (ySQS), Arabidopsis thaliana (pSQS), Z. mobilis (zSQS), B. japonicum (bSQS), and T. elongatus BP-1 (tSQS). The ClustalW algorithm was used to generate the alignment (www.ebi.ac.uk/Tools/clustalw/index.html). Dashes indicate gaps that were introduced to maximize the alignment. The conserved regions I, II, III, and IV are indicated above the sequences.
The degree of similarity between eukaryotic and bacterial SQSs is highest in regions I and II and drops considerably in regions III and IV. The FCAIPQVMAIATL putative NADPH binding motif in region IV of eukaryotic SQSs is not highly conserved in the bacterial enzymes, while the VKIRK motif is well conserved in both groups. Finally, bacterial SQS does not have a consensus sequence for the membrane-spanning C-terminal α-helix that anchors eukaryotic SQS to membrane (47, 49, 57).
Cloning, solubilization, and purification of SQS.
Eukaryotic SQSs are associated with microsomes and can be solubilized with detergents. However, when expressed in E. coli, SQS is found in inclusion bodies and cannot be reconstituted to give soluble active enzyme. Zhang et al. discovered that soluble recombinant yeast SQS could be obtained by deletion of a putative C-terminal membrane-spanning α-helix (57). This approach has been used to obtain soluble recombinant enzyme from other eukaryotes (1, 18, 45, 47, 52). However, bacterial SQSs do not have a C-terminal sequence predicted to give a membrane-spanning helix, and the basis for membrane affiliation by the bacterial enzymes is not apparent.
Three bacteria, T. elongatus BP-1, B. japonicum, and Z. mobilis, were selected as sources for the SQS gene. Previously, SQ synthesis was detected in E. coli transformants harboring the hopane gene cluster from B. japonicum and Z. mobilis (34). In addition, a SQ-hopane cyclase from B. japonicum and Z. mobilis has been expressed in E. coli (33, 40). The moderate thermophile, T. elongatus, has been the source of genes for (E)-4-hydroxy-3-methylbut-2-enyl diphosphate synthase, an enzyme in the 2-C-methyl-d-erythritol phosphate pathway for isoprenoid biosynthesis (30), the circadian clock proteins (KaiA, KaiB, and KaiC), and photoreceptor proteins (9, 55, 56).
The DNA sequence for hpnC, the putative SQS gene in the B. japonicum hopanoid biosynthesis gene cluster (34), did not have the 84 bp at the N terminus reported in the genomic DNA sequence for the same bacterium (blr3001; EMBL nucleotide sequence accession no. BAC48266) (17). Both versions of B. japonicum SQS were cloned into an E. coli expression system. The DNA sequence for the Z. mobilis SQS gene from the hopanoid biosynthesis gene cluster (34) was also slightly different from the one (zmo0869; EMBL nucleotide sequence accession no. AAV89493) from Z. mobilis genomic DNA (37). hpnC from Z. mobilis SQS uses ATG as a start codon instead of GTG for zmo0869 (34, 46). Overall, 10 bp (12 amino acids in the protein) in hpnC from Z. mobilis SQS are different from those of zmo0869. These differences mostly code for amino acids found at the N terminus of the protein. The sequence of our PCR clone of Z. mobilis SQS matched the sequence for Z. mobilis genomic DNA (zmo0869) (Fig. 2). A comparison of flanking regions showed that our clone was located in the hopanoid biosynthesis gene cluster, as described previously (34, 46).
tll1096 from T. elongatus BP-1 has 1,080 bp and encodes a 359-amino-acid protein (∼39.9 kDa). The open reading frame (ORF) was cloned by PCR using T. elongatus BP-1 genomic DNA as a template. Initial attempts with primers containing NdeI and HindIII restriction sites failed. A 1.3-kb DNA fragment containing tll1096 was amplified using primers without the restriction sites. The gel-purified 1.3-kb DNA fragment was used as the template for a second round of PCR using primers to introduce the flanking NdeI and HindIII restriction sites. The PCR product was cloned into pGEM-T Easy to give pgBP1Q. The sequence of tll1096 in pgBP1Q was identical to that reported for T. elongatus BP-1 genomic DNA (25). tll1096 from pgBP1Q was subcloned into the expression vector pET28b(+) to give ptBP1QX, which encoded SQS with an N-terminal His6 tag. A parallel set of experiments was carried out for the SQS genes from B. japonicum and Z. mobilis.
IPTG-induced expression in ptBP1QX gave high levels of SQS as judged by SDS-PAGE of cell extracts. Initially, T. elongatus BP-1 SQS was found exclusively in inclusion bodies as an inactive enzyme. Attempts to solubilize and refold the protein under a variety of conditions were unsuccessful. Different media (LB or M9 minimal media), temperatures (18°C, 30°C, and 37°C), and E. coli hosts [BL21(DE3) and Rosetta (DE3)] gave similar results. Eventually, we were able to obtain soluble active T. elongatus BP-1 SQS (∼3 to 6% of total cytosolic protein) by adding glycerol (20 to 50% vol/vol) to the lysis and purification buffers and by lowering the incubation temperature to 30°C. Different buffer additives, including glycerol, Tween 20, Tween 80, NaCl, and polyethylene glycol, were examined, but none except glycerol gave active soluble T. elongatus BP-1 SQS. Glycerol seems to be more important for solubility rather than stability for the T. elongatus BP-1 SQS. Once BP-1 SQS is solubilized, the protein remains active and soluble even after glycerol is reduced or removed from the buffer. Unfortunately, similar attempts to obtain soluble recombinant SQS from B. japonicum and Z. mobilis failed.
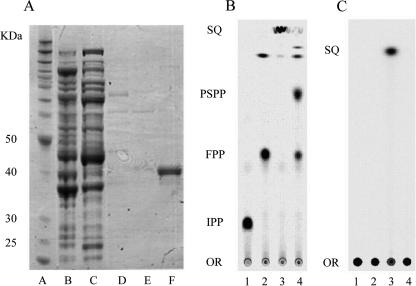
In a typical purification, cells were disrupted in buffer containing 50% (vol/vol) glycerol, and the glycerol concentration was lowered to 20% (vol/vol) to minimize problems associated with viscosity during subsequent steps. The cell extract was chromatographed on Ni-NTA. After a washing step with buffer containing 50 mM and 70 mM imidazole, SQS eluted as a sharp peak with buffer containing 250 mM imidazole. SDS gel electrophoresis of purified T. elongatus BP-1 SQS (2 to 3 mg/ml) gave a single band (>97% purity) at approximately 42 kDa, consistent with the predicted molecular mass for the His6-tagged enzyme (Fig. 3A). Purified protein was flash-frozen in liquid N2 and stored at −80°C until needed. The enzyme was stable for at least 6 months under these conditions.
FIG. 3.
Characterization of recombinant of T. elongatus BP-1 SQS. (A) SDS-PAGE of samples from the purification of recombinant T. elongatus BP-1 SQS. Lane A, molecular weight markers; lane B, cell extract from ptBP1QXRO; lane C, flowthrough; lane D, washed fraction with 50 mM imidazole-buffer; lane E, washed fraction with 70 mM imidazole-buffer; lane F, eluted SQS with 250 mM imidazole-buffer. (B and C) Autoradiograms of TLC plates of products from recombinant T. elongatus BP-1 SQS. In panel B, the TLC plate was developed with 30:70:13:8 chloroform-formic acid-pyridine-water. In panel C, development was with 4:1 hexane-toluene. Lane 1, [14C]IPP. Other lanes are as follows:[14C]IPP and GPP with recombinant E. coli FPP synthase (lane 2), with T. elongatus BP-1 SQS in the presence of NADPH (lane 3), and with T. elongatus BP-1 SQS in the absence of NADPH (lane 4). The positions of [14C]IPP, [14C]FPP, [14C]PSPP, [14C]SQ, and the chromatographic origin (OR) are indicated.
Product analysis.
Incubation of T. elongatus BP-1 SQS with FPP in the presence of NADPH gave a single product with a GC retention time of 24.5 min that comigrated with an authentic sample of SQ. A GC-mass spectrum of the product had a prominent peak at m/z 410 and a fragmentation pattern identical to that produced by an authentic sample of SQ.
Synthesis of PSPP by T. elongatus BP-1 SQS, when incubated with FPP in the absence of NADPH, was measured using [14C]FPP. [14C]FPP was synthesized from [14C]IPP and GPP using recombinant E. coli FPP synthase. Recombinant T. elongatus BP-1 SQS was then added to the same reaction mixture in the absence of NADPH to investigate PSPP formation. Product studies with TLC analysis required greater concentrations of substrate and T. elongatus BP-1 SQS than the kinetic assays because of the difference in the sensitivity of the two methods (2, 27, 38). Formation of PSPP was detected by TLC as a radioactive product (Fig. 3B) at an Rf of 0.73, which was identical to that of PSPP prepared with recombinant yeast SQS as described by Jarstfer et al. (14). SQ ran at the solvent front when the TLC plate was developed with 30:70:13:8 chloroform-formic acid-pyridine-water. Possible solvolysis products of FPP and PSPP were also detected at an Rf of ∼0.9 (Fig. 3B, lanes 2 and 4). Farnesol, neoridol, and hydrocarbons are known solvolysis products of FPP by FPP synthase (43). PSPP also undergoes SQS-catalyzed solvolysis to give a mixture of (Z)-dehydrosqualene, (R)-12-hydroxysqualene, and (10S,13S)-10-hydroxybotryococcene (14). In 4:1 hexane-toluene, SQ was detected at an Rf of 0.88, while IPP, FPP, and PSPP remained at the origin (Fig. 3C) (38). When NADPH was added to the incubation mixture, recombinant T. elongatus BP-1 SQS converted FPP directly to SQ without formation of detectable amounts of PSPP (Fig. 3C).
Characterization of recombinant T. elongatus BP-1 SQS.
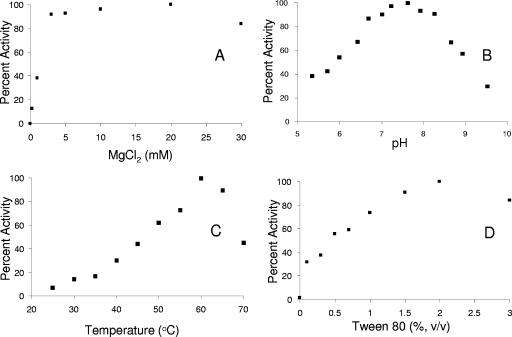
Positive-ion electrospray mass spectrometry of His6-SQS gave a peak corresponding to a mass of m/z 41886. This value is within experimental error of a predicted mass of 41,888 Da for the protein without the N-terminal Met, presumably removed by methionyl aminopeptidase in the E. coli expression system (11). The activity of T. elongatus BP-1 SQS increased in a hyperbolic manner with increasing concentrations of NADPH and Tween 80, with maximal activity at 2.0 mM NADPH and 2% (vol/vol) Tween 80 (Table 2 and Fig. 4D). Similar behavior is seen for the eukaryotic enzyme. Presumably the detergent facilitates turnover by providing a hydrophobic reservoir for the water-insoluble product of the reaction (57). Enzyme activity gradually increased from 25°C to a maximum at 60°C (Fig. 4C). A plot of activity versus pH gives a well-defined bell curve (Fig. 4B) with an optimum at pH 7.6, which is somewhat higher than the value of 7.2 reported for the yeast enzyme (57). T. elongatus BP-1 SQS has a strict requirement for Mg2+, with maximal activity at ∼20 mM (Fig. 4A). No activity was observed when the enzyme was assayed in Mg2+-free buffer. This behavior is typical for enzymes in the isoprenoid biosynthetic pathway that processes diphosphate substrates.
TABLE 2.
Kinetic constants for SQS
| SQS source | SQS type | kcat (s−1) | KmFPP (μM) | KmNADPH (μM) | kcat/KmFPP (μM−1 s−1) | Reference |
|---|---|---|---|---|---|---|
| Human | Microsomal | 2.3 | 52 | |||
| N- and C-terminally truncated | 1.44 | 2.8 | 0.51 | 52 | ||
| Rat | Microsomal | 1.8 | 52 | |||
| Yeast | C-terminally truncated | 0.53 | 2.5 | 530 | 0.21 | 21 |
| T. cruzi | Microsomal | 2.3 | 33 | 54 | ||
| N- and C-terminally truncated | 1.05 | 5.3 | 23 | 0.20 | 45 | |
| L. mexicana | Microsomal | 2.8 | 57 | 54 | ||
| T. elongatus BP-1 | 1.74 ± 0.04 | 0.97 ± 0.10 | 241 ± 13 | 1.80 ± 0.02 | This work |
FIG. 4.
Dependence of SQS activity from T. elongatus BP-1 on MgCl2 (A), pH (B), temperature (C), and Tween 80 (D).
The rate of SQ synthesis from FPP was linear at the limits of substrate consumption (1 to 15%) and reaction times (1 to 10 min) used for the assays. Michaelis constants for FPP and NADPH were determined at saturating concentrations of the other substrate, and the values are listed in Table 2. The catalytic efficiencies of the bacterial and eukaryotic enzymes are similar. KmFPP for T. elongatus BP-1 SQS is slightly lower than the values of the eukaryotic enzymes while kcatFPP is higher.
Zaragozic acid A, a fungal metabolite, is a potent inhibitor of mammalian and fungal SQSs, which are thought to mimic FPP and PSPP (5, 6, 35). Initially, zaragozic acid is a competitive inhibitor against FPP in rat SQS, which is followed by irreversible inactivation of the enzyme (20). When T. elongatus BP-1 SQS activity was measured in the presence of ∼Km concentration of FPP, zaragozic acid A showed dose-dependent inhibition. Zaragozic acid A is also a potent inhibitor of recombinant T. elongatus BP-1 SQS, with a 50% inhibitory concentration of 95.5 ± 13.6 nM.
Conclusions.
In summary, the tll1096 gene in T. elongatus BP-1 encodes SQS. The protein shows modest (∼30%) overall similarity to eukaryotic SQSs. The highest degree of similarity was seen in regions I and II, thought to be responsible for the conversion of FPP to PSPP. The similarity in regions III and IV is considerably lower. Bacterial SQSs, including the T. elongatus BP-1 enzyme, do not have the C-terminal membrane-spanning motif found in the eukaryotic proteins. When expressed in E. coli, the bacterial SQSs were located in inclusion bodies and could not be reconstituted to give soluble active enzymes. After considerable experimentation, we were able to solubilize and purify the recombinant His6-tagged enzyme from T. elongatus BP-1 (>97% homogeneity) by including high concentrations of glycerol in the disruption and purification buffers. We were not, however, able to find conditions that gave active soluble enzyme for the proteins from Z. mobilis and B. japonicum. The metal ion dependence, pH dependence, and kinetic properties of the T. elongatus enzyme were similar to those of its eukaryotic counterparts. The T. elongatus BP-1 SQS gene contains conserved regions similar to those associated with catalysis by the enzyme in Eukarya. Zaragozic acid A, a potent inhibitor of mammalian, fungal, and yeast SQS, is also a potent inhibitor of recombinant T. elongatus BP-1 SQS.
Acknowledgments
We thank Satoshi Tabata of Kazusa DNA Research Institute, Japan, for providing a sample of T. elongatus BP-1 genomic DNA and Mo Chen for providing the recombinant E. coli FPP synthase.
This work was supported by NIH grant GM 25521.
Footnotes
Published ahead of print on 28 March 2008.
REFERENCES
- 1.Agnew, W. S., and G. Popjak. 1978. Squalene synthetase. Solubilization from yeast microsomes of a phospholipid-requiring enzyme. J. Biol. Chem. 2534574-4583. [PubMed] [Google Scholar]
- 2.Akamine, S., K. Nakamori, S. A. Chechetka, M. Banba, Y. Umehara, H. Kouchi, K. Izui, and S. Hata. 2003. cDNA cloning, mRNA expression, and mutational analysis of the squalene synthase gene of Lotus japonicus. Biochim. Biophys. Acta 162697-101. [DOI] [PubMed] [Google Scholar]
- 3.Baliga, N. S., R. Bonneau, M. T. Facciotti, M. Pan, G. Glusman, E. W. Deutsch, P. Shannon, Y. Chiu, R. S. Weng, R. R. Gan, P. Hung, S. V. Date, E. Marcotte, L. Hood, and W. V. Ng. 2004. Genome sequence of Haloarcula marismortui: a halophilic archaeon from the Dead Sea. Genome Res. 142221-2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barrow, K. D., J. G. Collins, R. S. Norton, P. L. Rogers, and G. M. Smith. 1984. 31P nuclear magnetic resonance studies of the fermentation of glucose to ethanol by Zymomonas mobilis. J. Biol. Chem. 2595711-5716. [PubMed] [Google Scholar]
- 5.Bergstrom, J. D., C. Dufresne, G. F. Bills, M. Nallin-Omstead, and K. Byrne. 1995. Discovery, biosynthesis, and mechanism of action of the zaragozic acids: potent inhibitors of squalene synthase. Annu. Rev. Microbiol. 49607-639. [DOI] [PubMed] [Google Scholar]
- 6.Bergstrom, J. D., M. M. Kurtz, D. J. Rew, A. M. Amend, J. D. Karkas, R. G. Bostedor, V. S. Bansal, C. Dufresne, F. L. Van Middlesworth, O. D. Hensens, J. M. Liesch, D. L. Zink, K. E. Wilson, J. Onishi, J. A. Milligan, G. Bills, L. Kaplan, M. Nallin Omstead, R. G. Jenkins, L. Huang, M. S. Meinz, L. Quinn, R. W. Burg, Y. L. Kong, S. Mochales, M. Mojena, I. Martin, F. Pelaez, M. T. Diez, and A. W. Alberts. 1993. Zaragozic acids: a family of fungal metabolites that are picomolar competitive inhibitors of squalene synthase. Proc. Natl. Acad. Sci. USA 9080-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berry, A. M., O. T. Harriott, R. A. Moreau, S. F. Osman, D. R. Benson, and A. D. Jones. 1993. Hopanoid lipids compose the Frankia vesicle envelope, presumptive barrier of oxygen diffusion to nitrogenase. Proc. Natl. Acad. Sci. USA 906091-6094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davisson, V. J., A. B. Woodside, and C. D. Poulter. 1985. Synthesis of allylic and homoallylic isoprenoid pyrophosphates. Methods Enzymol. 110130-144. [DOI] [PubMed] [Google Scholar]
- 9.Fukushima, Y., K. Okajima, Y. Shibata, M. Ikeuchi, and S. Itoh. 2005. Primary intermediate in the photocycle of a blue-light sensory BLUF FAD-protein, Tll0078, of Thermosynechococcus elongatus BP-1. Biochemistry 445149-5158. [DOI] [PubMed] [Google Scholar]
- 10.Gu, P., Y. Ishii, T. A. Spencer, and I. Shechter. 1998. Function-structure studies and identification of three enzyme domains involved in the catalytic activity in rat hepatic squalene synthase. J. Biol. Chem. 27312515-12525. [DOI] [PubMed] [Google Scholar]
- 11.Hirel, P. H., M. J. Schmitter, P. Dessen, G. Fayat, and S. Blanquet. 1989. Extent of N-terminal methionine excision from Escherichia coli proteins is governed by the side-chain length of the penultimate amino acid. Proc. Natl. Acad. Sci. USA 868247-8251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Inoue, T., T. Osumi, and S. Hata. 1995. Molecular cloning and functional expression of a cDNA for mouse squalene synthase. Biochim. Biophys. Acta 126049-54. [DOI] [PubMed] [Google Scholar]
- 13.Iwata-Reuyl, D., S. K. Math, S. B. Desai, and C. D. Poulter. 2003. Bacterial phytoene synthase: molecular cloning, expression, and characterization of Erwinia herbicola phytoene synthase. Biochemistry 423359-3365. [DOI] [PubMed] [Google Scholar]
- 14.Jarstfer, M. B., D. L. Zhang, and C. D. Poulter. 2002. Recombinant squalene synthase. Synthesis of non-head-to-tail isoprenoids in the absence of NADPH. J. Am. Chem. Soc. 1248834-8845. [DOI] [PubMed] [Google Scholar]
- 15.Jennings, S. M., Y. H. Tsay, T. M. Fisch, and G. W. Robinson. 1991. Molecular cloning and characterization of the yeast gene for squalene synthetase. Proc. Natl. Acad. Sci. USA 886038-6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jurgens, U. J., P. Simonin, and M. Rohmer. 1992. Localization and distribution of hopanoids in membrane systems of the cyanobacterium Synechocystis PCC 6714. FEMS Microbiol. Lett. 71285-288. [DOI] [PubMed] [Google Scholar]
- 17.Kaneko, T., Y. Nakamura, S. Sato, K. Minamisawa, T. Uchiumi, S. Sasamoto, A. Watanabe, K. Idesawa, M. Iriguchi, K. Kawashima, M. Kohara, M. Matsumoto, S. Shimpo, H. Tsuruoka, T. Wada, M. Yamada, and S. Tabata. 2002. Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Res. 9189-197. [DOI] [PubMed] [Google Scholar]
- 18.Kuswik-Rabiega, G., and H. C. Rilling. 1987. Squalene synthetase. Solubilization and partial purification of squalene synthetase, copurification of presqualene pyrophosphate and squalene synthetase activities. J. Biol. Chem. 2621505-1509. [PubMed] [Google Scholar]
- 19.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227680-685. [DOI] [PubMed] [Google Scholar]
- 20.Lindsey, S., and H. J. Harwood. 1995. Inhibition of mammalian squalene synthetase activity by zaragozic acid A is a result of competitive inhibition followed by mechanism-based irreversible inactivation. J. Biol. Chem. 2709083-9096. [DOI] [PubMed] [Google Scholar]
- 21.LoGrasso, P. V., D. A. Soltis, and B. R. Boettcher. 1993. Overexpression, purification, and kinetic characterization of a carboxyl-terminal-truncated yeast squalene synthetase. Arch. Biochem. Biophys. 307193-199. [DOI] [PubMed] [Google Scholar]
- 22.McKenzie, T. L., G. Jiang, J. R. Straubhaar, D. G. Conrad, and I. Shechter. 1992. Molecular cloning, expression, and characterization of the cDNA for the rat hepatic squalene synthase. J. Biol. Chem. 26721368-21374. [PubMed] [Google Scholar]
- 23.Miller, K. J., R. S. Gore, R. Johnson, A. J. Benesi, and V. N. Reinhold. 1990. Cell-associated oligosaccharides of Bradyrhizobium spp. J. Bacteriol. 172136-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mookhtiar, K. A., S. S. Kalinowski, D. Zhang, and C. D. Poulter. 1994. Yeast squalene synthase. A mechanism for addition of substrates and activation by NADPH. J. Biol. Chem. 26911201-11207. [PubMed] [Google Scholar]
- 25.Nakamura, Y., T. Kaneko, S. Sato, M. Ikeuchi, H. Katoh, S. Sasamoto, A. Watanabe, M. Iriguchi, K. Kawashima, T. Kimura, Y. Kishida, C. Kiyokawa, M. Kohara, M. Matsumoto, A. Matsuno, N. Nakazaki, S. Shimpo, M. Sugimoto, C. Takeuchi, M. Yamada, and S. Tabata. 2002. Complete genome structure of the thermophilic cyanobacterium Thermosynechococcus elongatus BP-1. DNA Res. 9123-130. [DOI] [PubMed] [Google Scholar]
- 26.Nakamura, Y., T. Kaneko, S. Sato, M. Mimuro, H. Miyashita, T. Tsuchiya, S. Sasamoto, A. Watanabe, K. Kawashima, Y. Kishida, C. Kiyokawa, M. Kohara, M. Matsumoto, A. Matsuno, N. Nakazaki, S. Shimpo, C. Takeuchi, M. Yamada, and S. Tabata. 2003. Complete genome structure of Gloeobacter violaceus PCC 7421, a cyanobacterium that lacks thylakoids. DNA Res. 10137-145. [DOI] [PubMed] [Google Scholar]
- 27.Nakashima, T., T. Inoue, A. Oka, T. Nishino, T. Osumi, and S. Hata. 1995. Cloning, expression, and characterization of cDNAs encoding Arabidopsis thaliana squalene synthase. Proc. Natl. Acad. Sci. USA 922328-2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neudert, U., I. M. Martínez-Férez, P. D. Fraser, and G. Sandmann. 1998. Expression of an active phytoene synthase from Erwinia uredovora and biochemical properties of the enzyme. Biochim. Biophys. Acta 139251-58. [DOI] [PubMed] [Google Scholar]
- 29.Ng, W. V., S. P. Kennedy, G. G. Mahairas, B. Berquist, M. Pan, H. D. Shukla, S. R. Lasky, N. S. Baliga, V. Thorsson, J. Sbrogna, S. Swartzell, D. Weir, J. Hall, T. A. Dahl, R. Welti, Y. A. Goo, B. Leithauser, K. Keller, R. Cruz, M. J. Danson, D. W. Hough, D. G. Maddocks, P. E. Jablonski, M. P. Krebs, C. M. Angevine, H. Dale, T. A. Isenbarger, R. F. Peck, M. Pohlschroder, J. L. Spudich, K. W. Jung, M. Alam, T. Freitas, S. Hou, C. J. Daniels, P. P. Dennis, A. D. Omer, H. Ebhardt, T. M. Lowe, P. Liang, M. Riley, L. Hood, and S. DasSarma. 2000. Genome sequence of Halobacterium species NRC-1. Proc. Natl. Acad. Sci. USA 9712176-12181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okada, K., and T. Hase. 2005. Cyanobacterial non-mevalonate pathway: (E)-4-hydroxy-3-methylbut-2-enyl diphosphate synthase interacts with ferredoxin in Thermosynechococcus elongatus BP-1. J. Biol. Chem. 28020672-20679. [DOI] [PubMed] [Google Scholar]
- 31.Ourisson, G., M. Rohmer, and K. Poralla. 1987. Prokaryotic hopanoids and other polyterpenoid sterol surrogates. Annu. Rev. Microbiol. 41301-333. [DOI] [PubMed] [Google Scholar]
- 32.Pandit, J., D. E. Danley, G. K. Schulte, S. Mazzalupo, T. A. Pauly, C. M. Hayward, E. S. Hamanaka, J. F. Thompson, and H. J. Harwood, Jr. 2000. Crystal structure of human squalene synthase. A key enzyme in cholesterol biosynthesis. J. Biol. Chem. 27530610-30617. [DOI] [PubMed] [Google Scholar]
- 33.Perzl, M., P. Muller, K. Poralla, and E. L. Kannenberg. 1997. Squalene-hopene cyclase from Bradyrhizobium japonicum: cloning, expression, sequence analysis, and comparison to other triterpenoid cyclases. Microbiology 1431235-1242. [DOI] [PubMed] [Google Scholar]
- 34.Perzl, M., I. G. Reipen, S. Schmitz, K. Poralla, H. Sahm, G. A. Sprenger, and E. L. Kannenberg. 1998. Cloning of conserved genes from Zymomonas mobilis and Bradyrhizobium japonicum that function in the biosynthesis of hopanoid lipids. Biochim. Biophys. Acta 1393108-118. [DOI] [PubMed] [Google Scholar]
- 35.Petras, S. F., S. Lindsey, and H. J. Harwood. 1999. HMG-CoA reductase regulation: use of structurally diverse first half-reaction squalene synthetase inhibitors to characterize the site of mevalonate-derived nonsterol regulator production in cultured IM-9 cells. J. Lipid Res. 4024-38. [PubMed] [Google Scholar]
- 36.Poulter, C. D. 1990. Biosynthesis of non-head-to-tail terpenes. Formation of 1′-1 and 1′-3 linkages. Acc. Chem. Res. 2370-77. [Google Scholar]
- 37.Poulter, C. D., and H. C. Rilling. 1981. Conversion of farnesyl pyrophosphate to squalene, p. 413-441. In J. W. Porter and S. L. Spurgeon (ed.), Biosynthesis of isoprenoid compounds, vol. 1. Wiley, New York, NY. [Google Scholar]
- 38.Radisky, E. S. 1999. Elucidation of a kinetic mechanism for squalene synthase. Ph.D. thesis. University of Utah, Salt Lake City, UT.
- 39.Radisky, E. S., and C. D. Poulter. 2000. Squalene synthase: steady-state, pre-steady-state, and isotope-trapping studies. Biochemistry 391748-1760. [DOI] [PubMed] [Google Scholar]
- 40.Reipen, I. G., K. Poralla, H. Sahm, and G. A. Sprenger. 1995. Zymomonas mobilis squalene-hopene cyclase gene (shc): cloning, DNA sequence analysis, and expression in Escherichia coli. Microbiology 141155-161. [DOI] [PubMed] [Google Scholar]
- 41.Robinson, G. W., Y. H. Tsay, B. K. Kienzle, C. A. Smith-Monroy, and R. W. Bishop. 1993. Conservation between human and fungal squalene synthetases: similarities in structure, function, and regulation. Mol. Cell. Biol. 132706-2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rohmer, M., P. Bouvier-Nave, and G. Ourisson. 1984. Distribution of hopanoid triterpenes in prokaryotes. J. Gen. Microbiol. 1301137-1150. [Google Scholar]
- 43.Saito, A., and H. C. Rilling. 1981. The formation of cyclic sesquiterpenes from farnesyl pyrophosphate by prenyltransferase. Arch. Biochem. Biophys. 208508-511. [DOI] [PubMed] [Google Scholar]
- 44.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 45.Sealey-Cardona, M., S. Cammerer, S. Jones, L. M. Ruiz-Pérez, R. Brun, I. H. Gilbert, J. A. Urbina, and D. González-Pacanowska. 2007. Kinetic characterization of squalene synthase from Trypanosoma cruzi: selective inhibition by quinuclidine derivatives. Antimicrob. Agents Chemother. 512123-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seo, J. S., H. Chong, H. S. Park, K. O. Yoon, C. Jung, J. J. Kim, J. H. Hong, H. Kim, J. H. Kim, J. I. Kil, C. J. Park, H. M. Oh, J. S. Lee, S. J. Jin, H. W. Um, H. J. Lee, S. J. Oh, J. Y. Kim, H. L. Kang, S. Y. Lee, K. J. Lee, and H. S. Kang. 2005. The genome sequence of the ethanologenic bacterium Zymomonas mobilis ZM4. Nat. Biotechnol. 2363-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shechter, I., E. Klinger, M. L. Rucker, R. G. Engstrom, J. A. Spirito, M. A. Islam, B. R. Boettcher, and D. B. Weinstein. 1992. Solubilization, purification, and characterization of a truncated form of rat hepatic squalene synthetase. J. Biol. Chem. 2678628-8635. [PubMed] [Google Scholar]
- 48.Simonin, P., U. J. Jurgens, and M. Rohmer. 1996. Bacterial triterpenoids of the hopane series from the prochlorophyte Prochlorothrix hollandica and their intracellular localization. Eur. J. Biochem. 241865-871. [DOI] [PubMed] [Google Scholar]
- 49.Stamellos, K. D., J. E. Shackelford, I. Shechter, G. Jiang, D. Conrad, G. A. Keller, and S. K. Krisans. 1993. Subcellular localization of squalene synthase in rat hepatic cells. Biochemical and immunochemical evidence. J. Biol. Chem. 26812825-12836. [PubMed] [Google Scholar]
- 50.Summers, C., F. Karst, and A. D. Charles. 1993. Cloning, expression and characterisation of the cDNA encoding human hepatic squalene synthase, and its relationship to phytoene synthase. Gene. 136185-192. [DOI] [PubMed] [Google Scholar]
- 51.Tansey, T. R., and I. Shechter. 2001. Squalene synthase: structure and regulation. Prog. Nucleic Acid Res. Mol. Biol. 65157-195. [DOI] [PubMed] [Google Scholar]
- 52.Thompson, J. F., D. E. Danley, S. Mazzalupo, P. M. Milos, M. E. Lira, and H. J. Harwood, Jr. 1998. Truncation of human squalene synthase yields active, crystallizable protein. Arch. Biochem. Biophys. 350283-290. [DOI] [PubMed] [Google Scholar]
- 53.Tippelt, A., L. Jahnke, and K. Poralla. 1998. Squalene-hopene cyclase from Methylococcus capsulatus (Bath): a bacterium producing hopanoids and steroids. Biochim. Biophys. Acta 1391223-232. [DOI] [PubMed] [Google Scholar]
- 54.Urbina, J. A., J. L. Concepcion, S. Rangel, G. Visbal, and R. Lira. 2002. Squalene synthase as a chemotherapeutic target in Trypanosoma cruzi and Leishmania mexicana. Mol. Biochem. Parasitol. 12535-45. [DOI] [PubMed] [Google Scholar]
- 55.Uzumaki, T., M. Fujita, T. Nakatsu, F. Hayashi, H. Shibata, N. Itoh, H. Kato, and M. Ishiura. 2004. Crystal structure of the C-terminal clock-oscillator domain of the cyanobacterial KaiA protein. Nat. Struct. Mol. Biol. 11623-631. [DOI] [PubMed] [Google Scholar]
- 56.Vakonakis, I., J. Sun, T. Wu, A. Holzenburg, S. S. Golden, and A. C. LiWang. 2004. NMR structure of the KaiC-interacting C-terminal domain of KaiA, a circadian clock protein: implications for KaiA-KaiC interaction. Proc. Natl. Acad. Sci. USA 1011479-1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang, D., S. M. Jennings, G. W. Robinson, and C. D. Poulter. 1993. Yeast squalene synthase: expression, purification, and characterization of soluble recombinant enzyme. Arch. Biochem. Biophys. 304133-143. [DOI] [PubMed] [Google Scholar]
- 58.Zhang, D., and C. D. Poulter. 1995. Biosynthesis of non-head-to-tail isoprenoids. Synthesis of 1′-1 and 1′-3 structures by recombinant yeast squalene synthase J. Am. Chem. Soc. 1171641-1642. [Google Scholar]