Abstract
Adeno-associated virus (AAV) serotypes differ broadly in transduction efficacies and tissue tropisms and thus hold enormous potential as vectors for human gene therapy. In reality, however, their use in patients is restricted by prevalent anti-AAV immunity or by their inadequate performance in specific targets, exemplified by the AAV type 2 (AAV-2) prototype in the liver. Here, we attempted to merge desirable qualities of multiple natural AAV isolates by an adapted DNA family shuffling technology to create a complex library of hybrid capsids from eight different wild-type viruses. Selection on primary or transformed human hepatocytes yielded pools of hybrids from five of the starting serotypes: 2, 4, 5, 8, and 9. More stringent selection with pooled human antisera (intravenous immunoglobulin [IVIG]) then led to the selection of a single type 2/type 8/type 9 chimera, AAV-DJ, distinguished from its closest natural relative (AAV-2) by 60 capsid amino acids. Recombinant AAV-DJ vectors outperformed eight standard AAV serotypes in culture and greatly surpassed AAV-2 in livers of naïve and IVIG-immunized mice. A heparin binding domain in AAV-DJ was found to limit biodistribution to the liver (and a few other tissues) and to affect vector dose response and antibody neutralization. Moreover, we report the first successful in vivo biopanning of AAV capsids by using a new AAV-DJ-derived viral peptide display library. Two peptides enriched after serial passaging in mouse lungs mediated the retargeting of AAV-DJ vectors to distinct alveolar cells. Our study validates DNA family shuffling and viral peptide display as two powerful and compatible approaches to the molecular evolution of novel AAV vectors for human gene therapy applications.
A large number of inherited or acquired diseases remain promising targets for human gene therapy. One vector that has shown outstanding potential thus far in numerous preclinical and clinical evaluations is based on nonpathogenic adeno-associated virus (AAV). A unique asset among various properties that make AAV especially attractive over its competitors, such as adenoviral or lentiviral vectors, is the availability of a vast number of natural isolates which differ significantly in their properties (24). We and others have shown previously that the function of an AAV vector particle is determined mainly by the capsid protein and that viral Rep proteins and genomic packaging elements are largely interchangeable (24, 27, 85). Paradoxically, the ever-increasing repertoire of naturally occurring and synthetically generated AAV capsid sequences (>300 to date) is currently creating a dilemma for the rational selection of the optimal serotype for a given application. The importance of finding the ideal capsid for efficient and safe gene transfer has been exemplified in many preclinical studies, as well as in a clinical trial using the AAV type 2 (AAV-2) prototype in human liver tissue (36, 47). In one previous study, the treatment of patients with severe hemophilia B with recombinant AAV-2 expressing human factor IX (hFIX) resulted in mildly elevated, yet therapeutic, levels of this blood coagulation factor. However, expression was short lived, and the hFIX decline was accompanied by a transient asymptomatic increase of liver transaminases, due to a T-cell immune response against the AAV-2 capsid (47). Also, preexisting neutralizing anti-AAV-2 antibodies (frequent in humans) in these individuals likely inhibited the linear vector dose response previously observed in animals.
We and others have suggested previously that the use of novel AAV serotypes, in particular, nonhuman isolates, will help to overcome some of these problems (19, 24, 63). Important examples are AAV-8 and AAV-9, which can transduce mouse liver far better than AAV-2, albeit the difference in dogs or primates is less clear (17, 52, 54, 75). The potential for the complete transduction of liver tissue and perhaps other tissues makes these two non-AAV-2 serotypes also particularly interesting for therapeutic RNA interference (RNAi) (28). We recently demonstrated the feasibility of efficiently and persistently suppressing hepatitis B virus with RNAi from a double-stranded AAV-8 vector (28). On the other hand, a potential drawback of AAV-8 and AAV-9 is their lack of specific tissue tropism (34, 52). The resulting frequent vector dissemination into all organs, including the brain, even from low peripheral doses in mice or monkeys (52, 54) is a particular concern for RNAi therapies in which control over vector biodistribution and the limitation of off-target effects will be imperative for the success of the approach (28).
In order to overcome the constraints of wild-type AAV serotypes, numerous groups have recently begun to develop novel strategies to engineer “designer” AAVs tailored for the therapeutic transduction of clinically relevant organs (reviewed in detail in references 9, 12, 35, 41, 51, and 85). Briefly, the variety of strategies can be grouped into indirect or chemical approaches and direct physical modification strategies. In the indirect approaches, specific molecules (e.g., bispecific antibodies [6] or avidin-coupled ligands [4]) are allowed to react with the viral surface (biotinylated in the case of avidin [4]), as well as a cellular receptor, forming a conjugate ideally able to retarget the capsid to a refractory cell type. Yet, numerous pharmacological problems, such as concerns about in vivo complex stability and difficulties in upscaling complex manufacturing, continue to prevent the broad adaptation of these approaches. Alternative, more powerful strategies rely on the direct physical modification of the AAV capsid protein and gene. Early examples include the generation of mosaic AAV capsids via the mixing of helper plasmids carrying capsid genes from distinct serotypes, such as AAV-1 and AAV-2 (30) and pairwise combinations of AAV-1 through AAV-5 (62). Similar mosaics were generated previously via a marker rescue approach, yielding AAV-2/AAV-3 recombinants with unique properties (8). A related strategy is the rational creation of chimeric virions via domain swapping among multiple parental serotypes, involving either entire capsid loops or parts thereof or individual residues. Notable examples include AAV-1/AAV-2 chimeras with improved tropism in muscle tissue (31), with one of these chimeras presently being studied in a phase I clinical trial for the treatment of Duchenne muscular dystrophy (85). Most recently, our own group described a battery of unique chimeras comprising elements from serotypes 2 and 8, which were exploited to identify capsid subdomains responsible for efficient AAV transduction in murine liver tissue in vivo (64).
A special type of chimeric capsids are those containing foreign proteins or peptides inserted into various positions of the virion shell. The methods and strategies used are widely diverse, and again, we refer to comprehensive reviews (12, 35, 41). Noteworthy here are approaches to fuse targeting ligands to the N termini of AAV capsid proteins (ideally, VP2 [45, 83]), or more powerful, to insert short peptides (up to 14 amino acids [21], typically 7) into exposed regions of the assembled virion. This strategy is referred to as viral display, in analogy to phage display, and has already been used extensively to retarget AAV-2 virions to a multitude of refractory or hard-to-infect cell types, such as vascular endothelial, smooth muscle, and pancreatic islet cells (43, 55, 77, 81, 82) and various tumor lines (22, 58, 65, 66). It has particularly benefited from comprehensive mutational analyses by various groups (e.g., references 21, 33, 56, and 83) that have resulted in the identification of prominent locations within the AAV-2 capsid tolerating peptide insertion. Most notable is the heparin binding domain (HBD), consisting of a total of four arginine (R) residues and one lysine residue, with R585 and R588 representing the most crucial components (37, 56). Numerous groups have now consistently shown that the insertion of 7-mer peptides into this region not only is frequently well tolerated and efficiently mediates virus retargeting, but also provides the extra benefit that the endogenous AAV-2 tropism can be abolished, thus enhancing target specificity (e.g., reference 21).
In addition to identifying sites for vector engineering, some of the mutational AAV studies directly yielded novel capsid variants with potential benefits for clinical use. A remarkable case was a recent study by Lochrie et al. (42) in which a set of 127 AAV-2 variants with point or insertion mutations were generated and screened for multiple properties. Several capsids were isolated which differed from the wild-type AAV-2 capsid in having better in vitro transduction efficiencies (albeit being equally efficient in vivo) or, clinically most relevant, higher-level resistance to individual or pooled human antisera. Nonetheless, the limitations of the approach also became clear, most notably, the extreme effort required to generate and manually screen a large number of mutants, which in fact prevented the interesting analyses of all possible combinations of beneficial point mutations in further capsids.
Indeed, the factors of time and labor are the main reasons why an increasing number of groups have recently begun to develop novel means for AAV vector evolution that no longer rely on the rational modification of the AAV-2 capsid. Instead, the new combinatorial methodologies allow for the far more efficient creation and selection of interesting candidates in a library-based high-throughput format. Thus far, two different strategies have been reported, both principally expanding on previously developed techniques. One is the use of viral display libraries, in which random 7-mer peptides are inserted into the AAV-2 HBD (at amino acid 587 or 588), yielding between 4 × 106 and 1.7 × 108 capsids potentially exposing new ligands on their surfaces (50, 58, 76). Subsequent iterative selection on diverse cell types refractory to the wild type, e.g., coronary artery endothelial cells, cardiomyoblasts, and carcinoma, leukemia, and megakaryocytic cell lines, led to enrichment with peptide mutants with increased target specificities and efficacies (48, 50, 58, 76). The second library type, independently described by two groups in 2006, relies on error-prone PCR amplification of the AAV-2 capsid gene (46, 59). Similar to the methods in earlier mutational studies, this approach resulted in the identification of AAV-2 point mutations (usually up to two per capsid) which yielded mutants that differed from the wild type in having mildly enhanced efficacies in vitro and/or improved transduction efficiencies in the presence of neutralizing anti-AAV-2 antibodies either generated in rabbits or preexisting in individual human sera.
Here, for the first time, we introduce the technology of DNA family shuffling into the realm of AAV vector evolution. The basic concept of this technology is the in vitro recombination of related parental genes with >50% homology, which are first fragmented and then reassembled based on partial homology, resulting in libraries of chimeric genes. Iterative amplification under pressure can then yield hybrids not only combining parental assets, but also ideally exhibiting novel and synergistic properties (70, 71). DNA family shuffling has been used extensively in recent years to evolve and improve all types of proteins, from markers and enzymes to vaccines (e.g., references 10, 13-15, and 39). Importantly, a set of reports also suggested its power to enhance viral gene therapy vectors by creating retro- or lentiviruses with improved stability or efficacy compared to that of the parental wild types (57, 61, 69). Here, we describe the novel use of DNA family shuffling for the highly efficient molecular interbreeding of eight multispecies AAVs to create chimeric capsids and, moreover, document its compatibility and synergism with existing AAV vector evolution technology.
MATERIALS AND METHODS
Plasmids for generation of shuffled AAV capsid library.
Plasmids containing full-length capsid genes (cap) of seven different AAV serotypes were present in the lab (for AAV-2, AAV-4, and AAV-5) or kindly provided by James Wilson (for AAV-8 and AAV-9) or Jay Chiorini and Rob Kotin (for avian and bovine AAV). Goat AAV was not available as a molecular clone early in our study and was therefore partly synthesized (GeneArt, Regensburg, Germany) as an 888-nucleotide (nt) fragment (nt 1023 to 1910). This subclone corresponds to the entire right half of the goat AAV capsid protein, which comprises all 42 reported (3) differences between goat AAV and AAV-5. The other seven cap genes were initially amplified via PCR and subcloned into pBluescript II SK (Stratagene). The purpose was to flank all cap genes with sites for the unique restriction enzyme PacI (5′) or AscI (3′) to facilitate the later cloning of shuffled cap genes into a wild-type AAV plasmid (see below). All primers also contained either a HindIII (5′) or a SpeI (3′) site to allow directed cloning into pBluescript (none of the four restriction enzymes cut in any parental cap gene). A 20-nt signature region was inserted between the two restriction sites in each primer to provide conserved primer binding sites for the later PCR amplification of shuffled genes. The resulting sequence of the forward primer was 5′ GGACTC AAGCTT GTCTGAGTGACTAGCATTCG TTAATTAA CAGGT ATG N22 3′ (the HindIII site is in bold, the PacI site is in bold italics, the signature region is underlined, and N22 denotes the first 22 nt of each cap gene following the ATG start codon). Likewise, the reverse primer was 5′ CGTGAG ACTAGT GCTTACTGAAGCTCACTGAG GGCGCGCC TTA N22 3′ (the SpeI site is in bold, the AcsI site is in bold italics, the signature region is underlined, and N22 denotes the last 22 nt of each cap gene up to the TAA stop codon).
In parallel, a wild-type cap recipient plasmid was engineered to contain the AAV-2 packaging elements (inverted terminal repeats [ITRs]) flanking the AAV-2 rep gene (encoding AAV replication proteins), together with PacI and AscI sites for cap cloning and the AAV-2 polyadenylation site. Therefore, AAV-2 rep (nt 191 to 2189) was PCR amplified using primers containing BglII sites and then subcloned into pTRUF3 (carrying AAV-2 ITRs with adjacent BglII sites) (88). The forward primer used was 5′ CGAACC AGATCT GTCCTGTATTAGAGGTCACGTGAG 3′ (the BglII site is in bold, and AAV-2 nt 191 is underlined), and the reverse primer was 5′ GGTAGC AGATCT GTTCGACCGCAGCCTTTCGAATGTCCGG TTTATT GATTA GGCGCGCC CTGGACTC TTAATTAA CATTTATTGTTCAAAGATGC 3′ (the BglII site is in bold, the polyadenylation signal is underlined, the AscI site is in bold italics, the PacI site is underlined and shown in bold italics, and the AAV-2 rep stop codon is underlined and shown in italics). Note that this procedure changed the AAV-2 SwaI site (downstream of the rep stop codon) into a PacI site.
DNA family shuffling of AAV capsid genes.
For DNA family shuffling, we basically utilized a two-step protocol in which the parental genes were first fragmented using DNase I enzyme and then reassembled into a full-length gene via primerless PCR (69, 71). This PCR was followed by a second PCR including primers binding outside of the cap genes, allowing the subcloning of the products into the wild-type recipient ITR-rep plasmid (see above and Fig. 1B). Initially, we isolated all cap genes from our subclones via HindIII/SpeI digestion (EcoRI digestion for goat AAV) and then optimized the reaction conditions as follows. We tested various DNase I concentrations and incubation times, aiming to obtain a pool of fragments between 0.2 and 1.0 kb in size (which gave the best results in later steps). The optimal conditions were found to be 1 μg of each cap gene, 1 μl of 1:200-prediluted DNase I (10 U/μl; Roche), 50 mM Tris-Cl (pH 7.4), and 1 mM MgCl2 in a total volume of 50 μl. The reaction mixture was incubated for 2 min at room temperature, and then the reaction was stopped by heat inactivation at 75°C for 10 min. Fragments of the desired sizes were isolated by running the entire reaction mixture on a 1% agarose gel (total final mixture volume, ∼60 μl). We next optimized the reassembly PCR by testing various DNA polymerases (Platinum Pfx [Invitrogen], DeepVent [NEB], and Taq [Amersham]) and respective conditions. Best results were obtained using PuReTaq ready-to-go PCR beads (Amersham) and the following conditions: 25 μl of purified cap fragments and a program of 4 min at 95°C; 40 cycles of 1 min at 95°C, 1 min at 50°C, and 3 min at 72°C; 10 min at 72°C; and 10 min at 4°C. Agarose gel (1%) analysis of 1 μl from this reaction mixture typically showed a smear in the area up to the 5-kb marker and no distinct bands. The same three polymerases listed above were then evaluated for the primer-containing second PCR mixture, and the following conditions were found to be optimal: 1 μl of Platinum Pfx, 2 μl of the product from the first PCR, 1 mM MgSO4, 1 μg of each primer (see below), and 0.3 mM (each) deoxynucleoside triphosphates in a total volume of 50 μl and a program of 5 min at 94°C; 40 cycles of 30 s at 94°C, 1 min at 55°C, and 3 min at 68°C; 10 min at 68°C; and 10 min at 4°C. The primers used bound to the 20-nt signature regions described above. This reaction gave a distinct ∼2.2-kb full-length cap band (on 1% agarose gel), which was purified (60 μl total) and cloned (4 μl) by using the Zero Blunt TOPO PCR cloning kit (with electrocompetent Escherichia coli TOP10 cells; Invitrogen, Carlsbad, CA). This intermediate cloning step significantly enhanced the yield of shuffled cap genes compared to that of efforts to clone the PCR product directly via conventional means (data not shown). The shuffled cap genes were then released from the TOPO plasmid via PacI and AscI double digestion and cloned into the appropriately digested ITR-rep recipient plasmid.
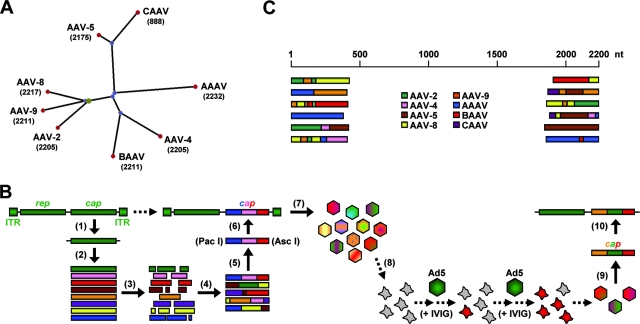
FIG. 1.
Generation of an AAV capsid library via DNA family shuffling. (A) Phylogram tree (created using PhyloDraw [http://pearl.cs.pusan.ac.kr/phylodraw/#test]) showing the eight AAV serotypes used as parents for DNA family shuffling (numbers denote lengths of capsid genes, in nucleotides). Branch lengths are proportional to the amounts of evolutionary change, calculated in ClustalW (http://www.ebi.ac.uk/clustalw/#). CAAV, AAAV, and BAAV, caprine, avian, and bovine AAVs, respectively. (B) Individual steps for generation of the library (scheme). Full-length cap genes were PCR amplified and subcloned for further amplification (1) and then isolated (2) and DNase I digested (3). Two consecutive PCRs without (4) or with (5) conserved primers were performed to reassemble shuffled full-length cap genes. These genes were inserted (6) into a plasmid carrying AAV-2 ITRs and a rep gene. The transfection of 293 cells with the resulting plasmid library (7) together with an adenoviral helper resulted in a viral library. One possible selection scheme used in this study was the coinfection of cultured liver cells (8) with the library and helper adenovirus under stringent conditions, resulting in the amplification of specific AAV capsids. Viral DNA can then be isolated (9) and cloned into an AAV helper plasmid carrying the AAV-2 rep gene without ITRs (10) for subsequent vector production. Ad5, helper adenovirus type 5. (C) Examples of shuffled cap genes in an initial small-scale library. DNA was extracted from 24 randomly chosen clones, and 5′ and 3′ ends of the individual cap genes were sequenced (using T3/T7 primers binding in the plasmid backbone). Shown, per end, are six representative alignments with the eight parents.
Performing all these reactions under minimal conditions (with respect to volumes and amounts), we obtained a library of approximately 3 × 104 bacterial colonies. The upscaling of each step (including final plating onto 100 15-cm plates) resulted in a final library of ∼7 × 105 plasmid clones. The integrity, genetic diversity, and functionality of the library were confirmed by DNA sequencing and small-scale expression studies (see Fig. 1C and Fig. S1 and S2 in the supplemental material).
Selective in vitro amplification of the shuffled capsid library.
We prepared a viral library by transfecting 293 cells in 50 T225 flasks (∼109 cells) with 50 μg of plasmid from the bacterial library per flask, together with 25 μg of an adenoviral helper plasmid (23). The resulting hybrid viruses were concentrated, purified, and titrated as described previously for recombinant AAV (23, 53). The final library used in this study had a particle titer (viral genome concentration) of 8.2 × 1011/ml (total volume, 3 ml). Various amounts of purified shuffled AAV were then incubated with the different cell lines (in 6-cm dishes), together with various amounts of helper adenovirus type 5. Ideally, the adenovirus would lyse the cells within 3 days, giving the AAV sufficient time to replicate. The AAV amounts were adjusted empirically so that we obtained minimal signals in Western blot analyses of cell extracts. This strategy helped to optimize the stringency of our library in each amplification round by ensuring that (ideally) a single viral genome was delivered to each cell and subsequently packaged into the capsid expressed from this particular viral genome. In one set of experiments, the library was additionally subjected to intravenous immunoglobulin (IVIG) pressure during amplification. Therefore, various volumes of the library and IVIG (GamimuneN [10%]; Bayer, Elkhart, IN) were mixed (see Fig. S3 in the supplemental material for examples), and the mixtures were incubated for 1 h at 37°C and then added to the cells. After overnight incubation, the cells were washed and superinfected with adenovirus. The wash step was included to avoid helper virus inactivation by the IVIG. As before, AAV amplification was controlled by Western blotting after each round, and only supernatants giving minimal expression were used for subsequent infections. The increasing IVIG resistance of the library during consecutive passages allowed us to continuously escalate the IVIG doses (see Fig. S3 in the supplemental material). All amplification experiments comprised five infection cycles (adenovirus was heat inactivated between each and then added fresh, to avoid uncontrolled amplification). Finally, viral DNA was purified from the supernatant by using a DNA extractor kit (Wako, Japan), and AAV cap genes were PCR amplified by using DeepVent polymerase and primers 5′ GATCTGGTCAATGTGGATTTGGATG 3′ (binding in AAV-2 rep upstream of the PacI site used for cap cloning) and 5′ GACCGCAGCCTTTCGAATGTCCG 3′ (binding downstream of the AscI site and the polyadenylation signal). The resulting blunt-ended cap genes were subcloned using the Zero Blunt TOPO PCR cloning kit for sequencing (Invitrogen), and DNA from individual clones (96 per cell line per amplification round) was prepared. To assemble full-length cap sequences, we first used T3 and T7 primers to obtain the 5′ and 3′ ends of each clone and then designed individual primers (data not shown) to acquire the remaining sequence. Alignments (DNA and protein) with the sequences of the eight parental viruses were performed using BLAST and VectorNTI 10/AlignX software (Invitrogen).
Helper plasmid cloning and vector particle production.
Helper plasmids expressing wild-type AAV-2, AAV-8, or AAV-9 cap together with AAV-2 rep genes, as well as AAV-2-based vector plasmids expressing the hFIX gene from a liver-specific promoter or the elongation factor 1α promoter, lacZ from a cytomegalovirus (CMV) promoter, or the luciferase gene from a simian virus 40 promoter, have all been described previously (18, 19, 28, 52). The generation of two self-complementary vector plasmids expressing either the gfp gene from a CMV promoter or the human alpha-1 antitrypsin (hAAT) gene from a Rous sarcoma virus promoter will be described in detail elsewhere (D. Grimm, L. Wang, J. S. Lee, T. A. Storm, and M. A. Kay, unpublished data). For the cloning of helper plasmids expressing shuffled cap genes, the entire AAV-8 cap gene was removed from the AAV-8 helper construct by cutting with SwaI and PmeI (both create blunt ends; SwaI cuts 9 nt upstream of the VP1 gene start codon, and PmeI cuts 53 nt downstream of the polyadenylation signal). The novel cap genes were amplified from the respective TOPO constructs (see above) via PCR using the forward primer 5′ AAAT CAGGT N25 3′ (the underlined nucleotide sequence restored the SwaI site to maintain correct reading frames, and N25 denotes the first 25 nt of each cap gene, ATGGCTGCCGATGGTTATCTTCCAG for AAV-DJ, AAV-2, AAV-8, and AAV-9). The reverse primer was 5′ AAAC GAATTCGCCCTTCGCAGAGACCAAAGTTCAACTGAAACGAATCAACCGG TTTATT GATTAACAGGCAA N23 3′ (the nucleotides restoring the PmeI site are underlined, the polyadenylation signal is shown in bold, and N23 denotes the last [3′] 23 nt of the shuffled capsid genes, TTACAGATTACGGGTGAGGTAAC for AAV-DJ [3′-to-5′ orientation]). PCRs were performed using DeepVent DNA polymerase (NEB), creating blunt ends allowing straightforward insertion into the linearized AAV-8 helper plasmid. Insert junctions and correct orientation were confirmed via DNA sequencing (Biotech Core). Vector production and particle titration (by dot blotting) were performed as described previously (53); yields for all vectors including AAV-DJ and the HBD mutants typically exceeded 6 × 1013 total physical particles per 50 T225 flasks (2 × 109 cells). HBD mutant plasmids (see Fig. 6A) were generated by A585X and B588Y site-directed mutagenesis using a QuikChange II kit (Stratagene), with A and B representing the native residues and X and Y representing the new residues. The gain or loss of heparin binding ability was confirmed via a standard heparin binding assay using type I heparin agarose (Sigma) (data not shown). Due to an unknown deficiency, the AAV-9/AAV-2 chimeric mutant (AAV-9/2) could not be produced in sufficient amounts for in vivo evaluation.
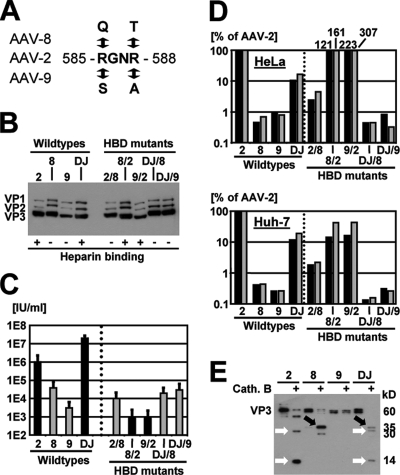
FIG. 6.
In vitro analyses of AAV-DJ and HBD mutants. (A) Two arginine residues (numbers refer to positions in AAV-2) in AAV-2, AAV-8, AAV-9, or AAV-DJ were mutagenized to eliminate or introduce an HBD (37). (B) Western blots (using B1 antibody) confirming correct VP protein expression from all HBD mutants. AAV-8 and AAV-DJ (wild types and mutants) expressed proteins more strongly than AAV-2 or AAV-9, for reasons unknown. +, with; −, without. (C) Titration of infectious particles on 293 cells confirmed the role of the HBD in infection in culture. The mutation of the HBD in AAV-2 or AAV-DJ reduced infectivity, measured the ratio of total to infectious AAV particles, by 2 to 3 logs. However, including an HBD in AAV-8 and AAV-9 did not further increase the infectivity of these vectors. (D) Results from cell binding assays confirming the role of the HBD in attachment to cultured cells (HeLa or Huh-7). The drop in binding with the AAV-2 and AAV-DJ mutants correlated well with the transduction data presented in panel C. Surprisingly, the HBD-positive AAV-8 and AAV-9 mutants bound severalfold more efficiently than AAV-2 on HeLa cells and, in all cases, far better than wild types 8 and 9 but transduced much less efficiently. Cell attachment and transduction thus do not necessarily correlate, suggesting that additional intracellular factors and steps contributed to the superior transduction efficiency of AAV-DJ. (E) AAV particle digestion with the endosomal proteinase cathepsin B (cath. B) (2) yielded distinct patterns for the individual serotypes in a Western blot analysis using polyclonal anti-AAV-2 VP serum. AAV-DJ showed a hybrid pattern with bands from AAV-2 and AAV-8 (white and black arrows, respectively), further supporting the idea that its properties resulted from synergistic or additive effects from its parents (cell binding from AAV-2 and rapid uncoating from AAV-8).
Helper plasmids for peptide display.
Prior to the generation of AAV peptide display libraries, we mutated the AAV-DJ helper plasmid to permit the insertion of oligonucleotides encoding seven amino acids after residue R588. In parallel, we created (using the cloning strategy for cap described above) and also mutated an AAV-2 helper. We basically adapted a multistep mutagenesis strategy first described for AAV-2 by Müller and colleagues (50), with the exception that we additionally mutated R585 into a glutamine (as in AAV-8). Accordingly, our mutagenesis primers were identical to those described above, except for the final (third) primer pair, which was 5′ CAACCTCCAGCAAGGCCAGAGAGGCCAAGGCCCAGGCGGCCACCGCAG 3′ (nucleotides changing R585 to Q585 are underlined and shown in bold) and 5′ CTGCGGTGGCCGCCTGGGCCTTGGCCTCTCTGGCCTTGCTGGAGGTTG 3′. The resulting AAV-2/AAV-DJ helper plasmids carried two unique SfiI sites permitting the straightforward insertion of 21-mer oligonucleotides encoding specific peptides. Notably, another SfiI site normally present in the AAV-2 rep gene was absent in the helper plasmid backbone used here (it had to be mutated in the original cloning strategy [50]; see also below). The oligonucleotides corresponding to the two lead peptides in this study were as follows: for the peptide NSSRDLG, 5′ AGGCAACTCAAGCCGAGACCTAGGAGCCCAGG 3′ and 5′ GGGCTCCTAGGTCTCGGCTTGAGTTGCCTCTC 3′, and for MVNNFEW, 5′ AGGCATGGTCAACAATTTTGAGTGGGCCCAGG 3′ and 5′ GGGCCCACTCAAAATTGTTGACCATGCCTCTC 3′ (nucleotides encoding the respective seven amino acids are underlined and shown in bold). Each oligonucleotide pair was annealed and then ligated into the SfiI-cut AAV-DJ helper plasmid. The presence of the correct inserts was confirmed upon sequencing with the HBD primer 5′ GTCATGATTACAGACGAAGAGGAAATC 3′ (binding upstream of the HBD-encoding sequences in the AAV-2 and AAV-DJ cap genes).
Creation of viral peptide display libraries based on AAV-2 and AAV-DJ.
The actual libraries of infectious AAV plasmids carrying random 21-mer oligonucleotides were again created in a multistep approach, similar to the generation of our shuffled library. First, we destroyed the SfiI site in wild-type AAV-2 rep in the context of the pΔTR18 plasmid (carrying AAV-2 rep and cap) (27). Primers used were 5′ GTGAGTAAGGCACCGGAGGCCC 3′ and 5′ GGGCCTCCGGTGCCTTACTCAC 3′ (the nucleotide change disrupting the SfiI site is underlined and shown in bold). The mutated rep gene was then PCR amplified using the same BglII site-containing forward primer described above and, as the reverse primer, 5′ GGTAGC AGATCT GTTTAAAC CATTTATTGTTCAAAGATGCAGTC 3′ (the BglII site is in bold, the PmeI site is underlined and shown in bold italics, and the AAV-2 rep stop codon is underlined and shown in italics). This fragment was inserted into BglII-cut pTRUF3 to become flanked by AAV-2 ITRs. The resulting construct was linearized by cutting with HindIII and PmeI, and the mutated AAV-2 and AAV-DJ cap genes (containing two adjacent SfiI sites but lacking any insert) were inserted as HindIII/PmeI fragments. In a final step, we then cloned a library of 21-mer oligonucleotides encoding random peptides into the two plasmids. The 21-mers were flanked by sequences comprising BglI sites, so that after cleavage they would be compatible with the SfiI-cut AAV rep-cap plasmids. The full sequence of the forward primer was 5′ CAGTCGGCCAGAGAGGC (NNB)7 GCCCAGGCGGCTGACGAG 3′ (BglI sites are underlined and shown in bold), with B representing the nucleotide T, C, or G (see Results). To create a double-stranded oligonucleotide for actual BglI cleavage and cloning, primer 5′ CTCGTCAGCCGCCTGG 3′ was used in a second-strand synthesis reaction as described previously (50). The presence of random 21-mer inserts or of 21-mer inserts selected after biopanning (see below) in individual clones was confirmed using the HBD sequencing primer.
AAV protein analyses.
Western blot and immunofluorescence analyses were carried out as reported previously (29) by using the monoclonal B1 antibody (useful because its 8-amino-acid epitope [see Fig. 2D and Fig. S4 in the supplemental material] is largely conserved across known AAV serotypes) for the detection of immobilized AAV capsid proteins. For immunofluorescence studies, polyclonal antisera were diluted 1:200 in 1× phosphate-buffered saline (PBS) while monoclonal antibodies (B1, A20, and 303.9) were used undiluted. Atomic structure models were created using DeepView Swiss-PdbViewer software version 3.7 (www.expasy.org/spdbv) and VIPER (viperdb.scripps.edu/oligomer_multi.php).
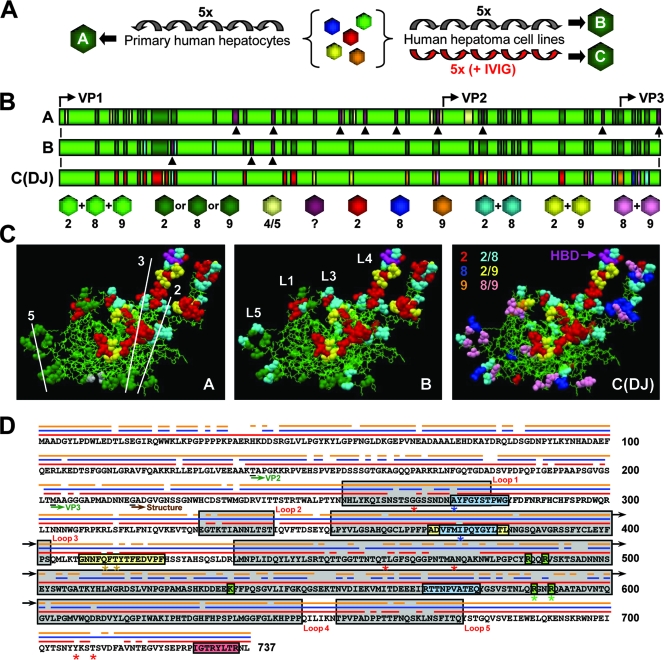
FIG. 2.
Molecular evolution of AAV vectors via DNA family shuffling. (A) The AAV capsid library was serially amplified on primary or transformed human liver cells. Purified human Igs (IVIG) were added to increase the selection pressure and to force vector evolution. Each scheme yielded a distinct pool of viral capsids (pools A to C). The alignment of ≥96 clones per pool with the sequences of the eight parental viruses confirmed the enrichment with specific sequences in association with increasing selection pressure. 5x, five times. (B) First 217 amino acids of the VP1 capsid protein for each pool. Colors represent the relationships to the parental strains (serotypes 2, 4, 5, 8, and 9), as also shown and detailed in Fig. S3 in the supplemental material. Arrowheads represent point mutations. Start codons for all three capsid proteins are shown. Pool C contained a single clone, designated AAV-DJ. (C) Putative atomic structure for each pool (the previously reported AAV-2 structure [Protein Data Bank file 1LP3 {http://www.rcsb.org}] was used as the basis for modeling; this structure lacks the residues represented in panel B). Thin green lines indicate sequence homology among the AAV-2, AAV-8, and AAV-9 parents. Residues shown as colored balls were derived from a subset of parental strains (see panel B for color codes; note that beige symbolizes AAV-5 in pool A here). AAV capsid symmetry axes (pool A) and four of the five loops (pool B) are shown. The location of two arginines as part of the conserved HBD (37) at the tip of loop IV is shown for AAV-DJ (pool C). (D) Capsid protein sequence of AAV-DJ. The three parental viruses are shown as thin lines above the sequence (AAV-2, red; AAV-8, blue; AAV-9, orange). Locations of the capsid loops, VP start codons, and the first residue of the atomic structure are indicated. A20 and B1 epitopes are boxed in blue and red, respectively (two mutations in the A20 epitope are shown by blue asterisks). Two recently identified immunogenic AAV-2 peptides (47) are boxed in yellow (AAV-DJ carries three point mutations, indicated by asterisks). Residues in green boxes form the conserved AAV-2 HBD (asterisks denote two arginines mutagenized in this study). Red asterisks denote residues previously discovered using other methods and believed to determine AAV-2 immunogenicity (see the text).
In vitro transduction, binding, and cleavage assays.
All transformed cell lines were maintained in Dulbecco's modified Eagle's medium (Gibco) containing 10% fetal calf serum, 2 mM l-glutamine, and 50 IU each of penicillin and streptomycin/ml at 37°C in 5% CO2. Fresh primary human hepatocytes (in 6-well plates without Matrigel) were obtained from Admet (Durham, NC) and maintained in hepatocyte basal medium (Cambrex, Walkersville, MD) with recommended supplements. The titration of gfp-expressing recombinant AAV particles was performed in 96-well plates (27), following the normalization of each virus stock to 2 × 109 particles/ml. For in vitro neutralization studies, 50-μl aliquots of each vector preparation were incubated with serial 10-fold dilutions of IVIG or mouse sera (following a 1-h heat inactivation step at 56°C) for 1 h at 37°C prior to titration on cells. Titers of neutralizing antibodies were calculated as reported previously (29). Details of the cell binding and cathepsin B digestion were reported recently (2).
In vivo biopanning.
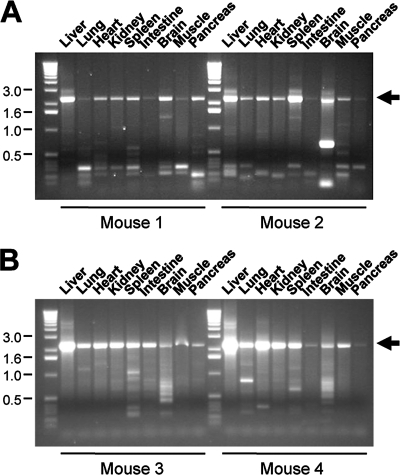
Wild-type FVB mice (6 to 8 weeks old; Jackson Laboratory, Bar Harbor, ME) were inoculated with the shuffled or peptide-displaying libraries at the doses indicated below. The mice also received different volumes (see Results) of wild-type adenovirus stocks purchased from the American Type Culture Collection (ATCC; Manassas, VA): human adenovirus C deposited as adenovirus 5 (catalog number VR-5) and mouse adenovirus (catalog number VR-550). Total inoculum volumes were 300 μl of 1× PBS for liver panning (injected via the tail vein) and 50 μl of 1× PBS for lung panning. For the latter, the mice were briefly anesthesized using an isoflurane vaporizer and placed on their backs. The virus suspension was then carefully pipetted directly onto both nostrils, resulting in the rapid aspiration of the suspension within a few seconds. Typically, 7 days postinfection, the mice were sacrificed and the organs were harvested, minced, and frozen in aliquots in liquid nitrogen. Total genomic DNA was prepared (as reported in reference 27) from one aliquot for subsequent PCR amplification of AAV DNA by using Platinum Pfx polymerase (Invitrogen) and specific primers flanking the entire capsid gene. For the shuffled cap genes, the primers were identical to those used before for in vitro biopanning. For the peptide-encoding cap genes, we used the same forward primer but a serotype-specific reverse primer: for AAV-2, 5′ TTACAGATTACGAGTCAGGTATC 3′, and for AAV-DJ, 5′ TTACAGATTACGGGTGAGGTAAC 3′. As before, the cap genes were then cloned using the Zero Blunt TOPO PCR cloning kit and sequenced using either T3 and T7 primers for the shuffled genes or the HBD sequencing primer for the peptide-expressing clones. Another aliquot was freeze-thawed three times in liquid nitrogen in 200 μl of 1× PBS and additionally homogenized to release intact AAV particles. Tissue debris was spun down (16,000 × g for 5 min), and the supernatant was heat inactivated (30 min at 65°C to kill amplified adenovirus) and then used for reinoculation into new mice (for liver tissue, up to 100 μl, and for lung tissue, 25 μl), together with freshly added helper adenovirus. In some cases (see below), the supernatant (50 μl) was depleted of murine immunoglobulin G (IgG) prior to reinfection by using a commercial kit (ProteoPrep immunoaffinity albumin and IgG depletion kit) according to the instructions provided by the manufacturer (Sigma, St. Louis, MO).
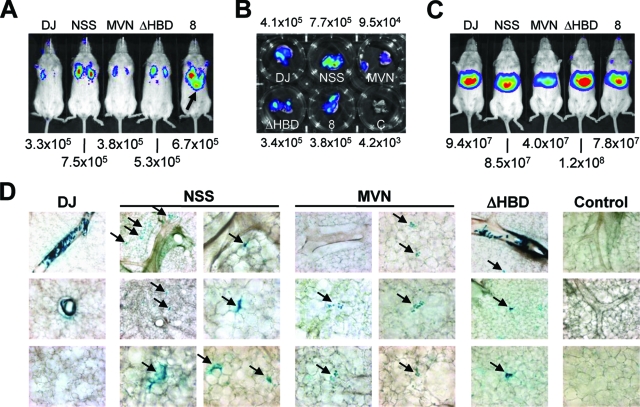
Expression studies with mice.
Wild-type female C57BL/6 mice (6 to 8 weeks old) were purchased from the Jackson Laboratory (Bar Harbor, ME). Recombinant AAVs expressing the hFIX or hAAT genes in 300 μl of 1× PBS were administered via tail vein infusion. Blood was collected at the indicated time points via retro-orbital bleeding, and plasma hFIX or hAAT levels were determined via an enzyme-linked immunosorbent assay as described previously (28, 52). Recombinant AAVs expressing the firefly luciferase gene in 300 μl of 1× PBS were infused via the tail vein for liver transduction or administered nasally in 50 μl of 1× PBS for lung transduction. Luciferase experiments were conducted with wild-type female FVB/NJ mice (6 to 8 weeks old) from the Jackson Laboratory (Bar Harbor, ME), and expression in live animals was monitored as described previously (28). Recombinant AAV-DJ variants expressing the lacZ gene were also given nasally. Two weeks later, animals were euthanized, the tracheas were cannulated, and lungs were inflated with 2% low-melting-point agarose and then sectioned with a Vibratome sectioning system into 100-μm slices, which were fixed overnight at 4°C in 4% paraformaldehyde. For β-galactosidase detection, lung slices were washed three times for 5 min each time in a solution of 1× PBS, 2 mM MgCl2, 0.02% NP-40, and 0.01% sodium deoxycholate and then incubated overnight at room temperature in the dark in a solution containing the β-galactosidase substrate X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside). Lungs were postfixed in 4% paraformaldehyde and stored at 4°C in 1× PBS. All procedures were approved by the Animal Care Committee at Stanford University. Genomic DNA was extracted from mouse tissues and analyzed via Southern blotting using hFIX gene-specific probes as reported previously (53).
Immunologic in vivo assays.
For passive immunization studies, mice were injected intravenously (via the tail vein) with 40 μl (low dose) or 200 μl (high dose) of IVIG (100 mg/ml) diluted in 1× PBS in a total volume of 300 μl and, 24 h later, infused (via the tail vein) with 2 × 1011 recombinant hFIX gene-expressing AAV particles. For cross-neutralization studies, mice were immunized against individual AAV serotypes by the peripheral infusion of 1011 recombinant hAAT gene-expressing particles. Three weeks later, mouse sera were collected for in vitro neutralization assays before the mice were reinfused with 1011 (or 5 × 1011 for AAV-2) (see below) recombinant hFIX gene-expressing AAV particles. In an experiment for which the data are not shown, the two AAV injections were carried out in the reverse order (first hFIX-expressing particles and then hAAT-expressing particles); the conclusions substantiated those presented below (see Fig. 9C).
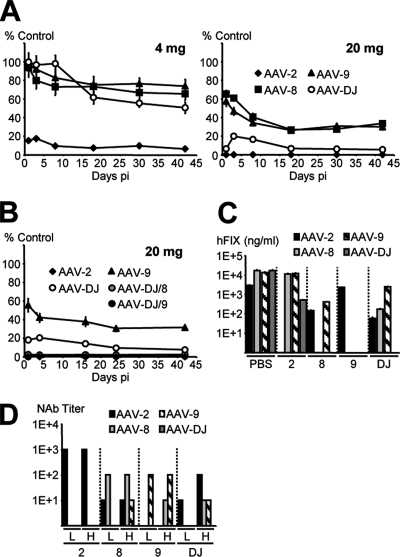
FIG. 9.
In vivo and in vitro neutralization of AAV-DJ and wild-type AAVs. (A) Mice (n = 4 per group) passively immunized with IVIG (4 or 20 mg) were injected with hFIX-expressing AAV. Plasma hFIX levels per virus and time point are shown as percentages of corresponding levels in control mice (those receiving PBS instead of IVIG). (B) Mice (n = 4 per group) immunized with the higher IVIG dose were also injected with the AAV-DJ HBD mutants. AAV-2, AAV-9, and AAV-DJ were included as controls. hFIX expression from the HBD mutants was marginal, comparable to that from AAV-2. (C) Mice (n = 4 per group) were injected with PBS or 1011 particles of hAAT-expressing AAV-2, AAV-8, AAV-9, or AAV-DJ (x axis), and 3 weeks later, they were reinjected with 1011 particles of hFIX-expressing viruses (5 × 1011 for the least efficient AAV-2, due to the enzyme-linked immunosorbent assay detection limit of ∼10 ng/ml). Shown are stable hFIX levels for each group as measured 6 weeks after the second injection. (D) Sera were taken from the mice described in the legend to panel C at the time of reinjection (bars H [higher dose]), as well as from a parallel group injected with a lower dose (bars L) of 2 × 1010 particles. Titers of neutralizing antibodies (NAb) against the wild-type AAVs or AAV-DJ were determined as detailed in Materials and Methods. pi, postinjection.
RESULTS
Generation of an AAV capsid library via DNA family shuffling.
A chimeric AAV capsid library was generated from eight parental wild-type viruses of human, primate, or nonprimate origin (Fig. 1A). In line with our primary goal, to evolve novel AAV capsids on liver cells in vitro, they were chosen based on performance in culture (AAV-2) or in liver tissue (AAV-8 and AAV-9), their substantial sequence divergence from AAV-2 (AAV-4, AAV-5, and avian, bovine, and caprine AAVs), or their low prevalence in the human population (all but AAV-2 and AAV-5). The conditions for AAV DNA family shuffling were established in small-scale studies (schematically depicted in Fig. 1B and C) in order to optimize parameters, including the choice of DNA polymerases for the PCRs and the lengths of the subgenomic capsid fragments (see Materials and Methods for details). The best conditions were then upscaled to yield a hybrid capsid-encoding plasmid library with a complexity of ∼7 × 105 distinct sequences. Packaging via the cotransfection of 293 cells with an adenoviral helper plasmid resulted in the final viral library, with a particle titer of ∼8.2 × 1011 genomes/ml (total volume, 4 ml from ∼109 cells, i.e., ∼3,300 particles per cell).
The isolation and sequencing of 48 randomly chosen clones confirmed extensive genetic diversity and validated the presence of all eight parental viruses in the final pool (see Fig. S1 and S2 in the supplemental material). Importantly, there was no apparent bias toward particular serotypes or combinations thereof in our library, nor did we obtain evidence for preferred hot spots for recombination. Instead, all eight parents were found in a random pattern, which is the ideal result. We were especially pleased to find recombinants with elements from very diverse serotypes, such as AAV-4 and AAV-5 (see data for clone S8 in Fig. S1 in the supplemental material), as it confirmed the potential of DNA family shuffling to create hybrids from parents differing by as much as ∼50%. As a result, the capsids in our library reflected and recapitulated the diversity of natural AAVs, exemplified by the fact that the levels of clonal homology to the AAV-2 prototype ranged anywhere from 46 to 93% (see Fig. S1 and S2 in the supplemental material). This wide sequence diversity positively distinguishes our methodology from previous AAV libraries, in which all resulting particles remained over 99% identical to the single parental virus, usually AAV-2 (46, 50, 58, 59, 76) (see also Discussion). Last but not least, we observed several point mutations in individual clones but found no evidence for lethal mutations or frameshifts. This outcome was in line with our expectations, as a hallmark of DNA family shuffling is the in-frame recombination of related functional sequences. This result further distinguishes our methodology from prior AAV evolution approaches, particularly those based on error-prone PCR, in which the offspring were frequently not viable (46, 59).
Stringent selection of AAV variants on human liver cells.
To screen for capsids with enhanced efficiency in liver cells, we serially (five times) amplified our library on human primary hepatocytes (Fig. 2A, pool A) or hepatoma cells (Huh-7 and HepG2) (Fig. 2A, pools B and C) as detailed in Materials and Methods. The extraction and sequencing of viral DNA from up to 192 clones per cell type (384 clones total) yielded 369 full-length capsid genes, whose compositions are shown in Fig. 2B and C. Strikingly, all clones showed predominant homology to five of the eight starting viruses, serotypes 2, 4, 5, 8, and 9, and had retained an HBD from the AAV-2 parent. Notably, this domain, whose function is binding to the primary AAV-2 receptor heparan sulfate proteoglycan (72), was clearly underrepresented in the unselected library, where it was found in only 3 of 48 clones (∼6%) (see, e.g., data for clone S8 in Fig. S2 in the supplemental material), in line with the random presence of AAV-2 sequences. The enrichment with and conservation of the HBD during selection on cultured cells suggested its crucial role for in vitro transduction. Indeed, vectors made from 10 individual capsids gave infectious titers similar to those of wild-type AAV-2 and exceeding those of HBD-deficient serotypes 8 and 9 (data not shown).
Despite the similarity of pools A (primary cells) and B (cell lines), we recovered only a single capsid sequence twice (Fig. 3, lane A), while all other 367 clones differed from each other by at least three amino acids (defined as our redundancy cutoff). In a direct comparison of the two pools, we noted the increased accumulation of serotype-specific residues, together with a reduction of random point mutations, in the capsids from the hepatocyte cell lines (Fig. 2B and C). This finding likely reflected the fact that HepG2 and Huh-7 cell lines are substantially more homogenous than primary human hepatocytes, which often vary among batches and donors. Nonetheless, the clonal heterogeneity even in the more evolved pool B prevented the reasonable selection and study of single sequences. To further force the evolution of individual capsids, we thus applied additional strong negative selection pressure to our library via incubation with pooled human antisera (IVIG) prior to reamplification (see Fig. S3 in the supplemental material). The high-level neutralizing activity of our particular IVIG batch (GamimuneN [10%]; Bayer) against multiple serotypes, especially AAV-2, implied its potential to eliminate capsids displaying prevalent epitopes from the library. Any surviving capsids were deemed to be useful in humans, with regard to the high frequency of neutralizing anti-AAV-2 antibodies in the population (63).
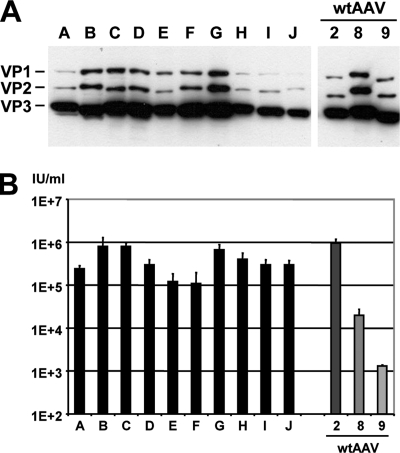
FIG. 3.
In vitro analysis of selected shuffled capsids. Following five consecutive amplifications of the AAV library on primary human hepatocytes, viral DNA was extracted from 10 randomly chosen clones (with the exception of the clone corresponding to lanes A, which was recovered twice from pool A), and the cap genes were subcloned into an AAV helper plasmid. (A) Western blot (using B1 antibody) showing differences in the expression levels and sizes of the individual VP proteins compared to those of wild-type AAVs (wtAAV). (B) Results from titration of infectious gfp-expressing particles. The helper plasmids described above were used to package a gfp-expressing AAV vector plasmid, and titers of recombinant particles in crude cell extracts were determined (n = 3) as detailed in Materials and Methods. All shuffled clones gave higher titers than the AAV-8 or AAV-9 helpers. No linear correlation between VP protein expression levels (A) and titers (B) could be made, suggesting that the various chimeras differed in their packaging efficacy, infectivity, and/or other parameters.
The sequencing of 96 clones after five passages under IVIG pressure revealed successful enrichment with a single chimera. This clone, termed AAV-DJ, displayed predominant sequence homology to serotypes 2, 8, and 9 (Fig. 2B and D) at levels (85 to 92%) similar to those of the homology of these wild types to one another (Table 1). Notably, AAV-DJ was distinguished from its closest natural relative, AAV-2, by a total of 60 amino acids (>8% of the VP1 capsid protein). It was thus substantially more divergent than, and compared highly favorably to, the bulk of previously evolved capsids, which typically differed from their single parent by only up to seven residues (depending on the library type, but in all cases corresponding to <1% of the capsid protein). AAV-DJ also showed ∼60% identity to the other five parental viruses, explained by the fact that all eight wild-type AAVs used in our study were at least ∼50% homologous to one another (Table 1). As a result, many individual residues in the AAV-DJ sequence could not be assigned to a particular parent.
TABLE 1.
Sequence homology of AAV-DJ and wild-type AAV capsid proteinsa
| Vector | % Homology of capsid protein of indicated vector to capsid protein of:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| DJ (737) | 2 (735) | 8 (738) | 9 (736) | 4 (734) | 5 (724) | A (743) | B (736) | C (296) | |
| DJ | 100 | ||||||||
| 2 | 92 | 100 | |||||||
| 8 | 88 | 82 | 100 | ||||||
| 9 | 85 | 81 | 85 | 100 | |||||
| 4 | 61 | 60 | 63 | 62 | 100 | ||||
| 5 | 57 | 57 | 58 | 57 | 53 | 100 | |||
| A | 57 | 57 | 57 | 58 | 54 | 54 | 100 | ||
| B | 58 | 59 | 58 | 59 | 76 | 55 | 54 | 100 | |
| C | 47 | 46 | 49 | 45 | 42 | 86 | 44 | 43 | 100 |
Shown are percentages of homology between the capsid protein of AAV-DJ and those of the eight parental wild-type AAVs. Numbers in parentheses indicate overall lengths of the various capsid proteins (in amino acids). AAV-DJ showed the highest levels of homology to wild types 2, 8, and 9 but also ∼50 to 60% homology to the other five parents. High degrees of homology of 81 to 85% were also evident for the three most efficient (in vitro and/or in murine liver) wild types, AAV-2, AAV-8, and AAV-9. Not surprisingly, these three serotypes were the predominant AAV-DJ parents (see the text). Notably, all five other wild types showed ∼40 to 60% homology to one another or to the AAV-2-AAV-8-AAV-9 group, exemplifying the overall close relationship of all naturally occurring AAVs. The levels of homology were higher for the even more closely related AAV-4 and bovine AAV, as well as for AAV-5 and goat AAV, as reported before. Note that only a subfragment (888 bp) of goat AAV which covered the diverse loops III and IV was used in this study, explaining the seemingly lower degrees of homology of other isolates to this serotype. A, avian AAV; B, bovine AAV; C, caprine (goat) AAV; 2, 8, 9, 4, and 5, wild-type AAV serotypes.
Clearly, AAV-DJ was more evolved than the clones obtained in the absence of IVIG, as was already evident from the data in Fig. 2B and C and as was further confirmed by sequence homology comparisons to pool A (from primary human hepatocytes). Indeed, the 10 clones in pool A displayed higher relative similarities to AAV-2, while AAV-DJ was more homologous to AAV-8 (Table 2). This finding validated our initial assumption that IVIG pressure would lead to an elimination of AAV-2 epitopes from our library. Concurrently, AAV-DJ was only ∼88 to 90% homologous to pool A, which further highlights its divergence from capsids evolved under less stringent conditions (the use of heterogeneous primary cells and the lack of IVIG pressure for pool A).
TABLE 2.
Homology of pool A and C and wild-type AAV capsid genes and proteinsa
| Vector | % Homology of capsid gene/protein of indicated vector to gene/protein of:
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| DJ (2,214/737) | A4 (2,211/736) | A11 (2,208/735) | D4 (2,208/735) | C8 (2,211/736) | G6 (2,208/735) | G5 (2,217/738) | B3 (2,211/736) | B4 (2,208/735) | G12 (2,208/735) | C3 (2,211/736) | |
| DJ | 100/100 | 89/90 | 89/91 | 89/91 | 87/88 | 89/91 | 88/91 | 88/90 | 91/93 | 88/88 | 88/89 |
| 2 | 90/92 | 92/94 | 90/93 | 94/96 | 88/90 | 96/96 | 93/94 | 90/93 | 91/94 | 92/93 | 94/95 |
| 8 | 87/88 | 82/84 | 81/84 | 82/83 | 82/85 | 81/84 | 85/87 | 83/87 | 84/85 | 81/83 | 81/84 |
| 9 | 81/85 | 81/83 | 87/88 | 83/85 | 85/85 | 81/84 | 80/84 | 84/86 | 82/86 | 82/84 | 80/83 |
Shown are the percentages of homology between pairs of full-length AAV capsid genes (first numbers) and proteins (second numbers). The designations in the column heads indicate 10 individual clones from pool A (selected on primary human hepatocytes in the absence of IVIG) (Fig. 2A). The numbers in parentheses show the total lengths of each capsid gene (first number, in nucleotides) and protein (second number, in amino acids). Clone A4 was independently recovered twice from pool A. Data for AAV-DJ are included for comparison. With the exception of clone C8, all clones from pool A showed higher levels of homology to wild-type AAV-2 (underlined values) than did AAV-DJ. In contrast, of all the clones, AAV-DJ showed the highest degree of homology to wild-type AAV-8 (underlined values). Both findings together suggest that stringent selection under IVIG pressure (i.e., that in the case of AAV-DJ) led to the elimination of AAV-2 sequences from the library and to a concurrent accumulation of AAV-8 residues. Also note that pool A (10 clones) and pool C (AAV-DJ) are on average only ∼88 to 90% homologous (first row), which is lower than the degree of homology of pool A clones to wild-type AAV-2 (∼92 to 94%) (second row). This result further highlights the divergence of AAV-DJ not only from wild-type AAVs but also from capsids evolved under less stringent conditions. 2, 8, and 9, wild-type AAV serotypes.
Molecular evolution affects mostly exposed capsid regions.
AAV capsids are complex three-dimensional protein structures, suggesting that the majority of amino acid changes resulting from our evolution process would occur on the exterior of the virion, at positions accessible to the selection pressure. Indeed, the bulk of the 60 non-AAV-2 residues in AAV-DJ were located in the loops extruding from the particle, especially in the major loop IV (Fig. 2C and D and also see Fig. S4 in the supplemental material). Consequently, the overall identity of AAV-DJ to its eight parents dropped from ∼31% (for the total VP1 protein) to only ∼18% in this exposed capsid region. Importantly, the AAV capsid loops contain most of the residues critical for natural receptor binding or for antibody recognition or escape. This arrangement explains the observed lack of AAV-DJ detection by the AAV-2-specific A20 antibody, as the conformational epitope of the A20 antibody (80) was dispersed over three of the five capsid loops and disrupted by two point mutations in AAV-DJ (Fig. 4). In contrast, the highly conserved residues constituting the capsid core remained mostly unchanged in AAV-DJ, as their inaccessible location on the inside of the assembled particle protected them from synthetic (or natural) AAV evolution. This arrangement also explains why all eight parental AAVs in our study (and perhaps all naturally occurring AAVs) showed ∼31% overall identity and at least 50% homology in pairwise comparisons (see Fig. S4 in the supplemental material and Table 1).
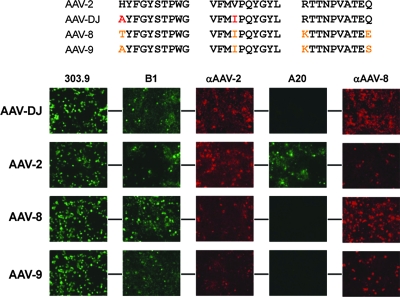
FIG. 4.
Presentation of epitopes on the AAV-DJ capsid. Shown at the top are the putative AAV-2 epitope for the monoclonal antibody A20 (a conformational epitope comprising three distinct peptides [see also Fig. 2D]) and the corresponding sequences in AAV-DJ, AAV-8, and AAV-9 (amino acid changes compared to the sequence of AAV-2 are highlighted in red [AAV-DJ] or orange [AAV-8 and AAV-9]). The bottom panels show results from immunofluorescence studies of cells cotransfected with the various helper constructs and an adenoviral helper plasmid (to boost gene expression from the AAV plasmids). The two amino acid changes in AAV-DJ were already sufficient to abolish the binding of the A20 antibody, validating and narrowing down the A20 epitope (80) and thus exemplifying the potential of DNA family shuffling as a reverse-genetics tool. AAV-DJ cross-reacted with both the polyclonal anti-AAV-2 and anti-AAV-8 sera, as expected from its chimeric structure. Similar cross-reactivity with these sera was also observed for wild types 2, 8, and 9. The fact that AAV-DJ, AAV-8, and AAV-9 were detected by the B1 antibody (initially raised against AAV-2 capsid proteins) was not surprising considering the high degree of conservation of its epitope in natural AAVs (see Fig. S4 in the supplemental material). All mono- and polyclonal anti-AAV antibodies were described previously (78, 79), except for the polyclonal rabbit anti-AAV-8 antiserum. 303.9, anti-Rep; B1, anti-VP; αAAV-2, anti-AAV-2 VP serum; A20, anti-AAV-2 capsids; αAAV-8, anti-AAV-8 VP serum.
The critical roles of the residues in the capsid loops were further apparent upon alignments of AAV-DJ sequences with those of the 10 clones from pool A (Fig. 5). As mentioned above (Table 2), most clones from pool A had preserved AAV-2 sequences, while AAV-DJ was more related to serotypes 8 and 9 than the pool A clones were. In analogy to the results of the previous wild-type comparison, we noted that the greatest sequence diversity occurred in the exposed regions, with many of the changes clustered within loops I, IV, and V. Intriguingly, our alignments confirmed 6 of the 12 previously reported hypervariable regions (HVRs) in the AAV capsid gene (11, 16) but, moreover, identified several further hot spots of sequence diversity (Fig. 5 and also see Fig. S4 in the supplemental material). Most of them were located in the N termini of VP1 and VP2, while others were dispersed among the areas corresponding to HVRs 2 to 5. Our observation of multiple differences between the N termini of AAV-DJ and pool A was not surprising, as the N-terminal region is temporarily exposed during the AAV life cycle (7, 40, 68) and thus potentially subject to evolution pressure.
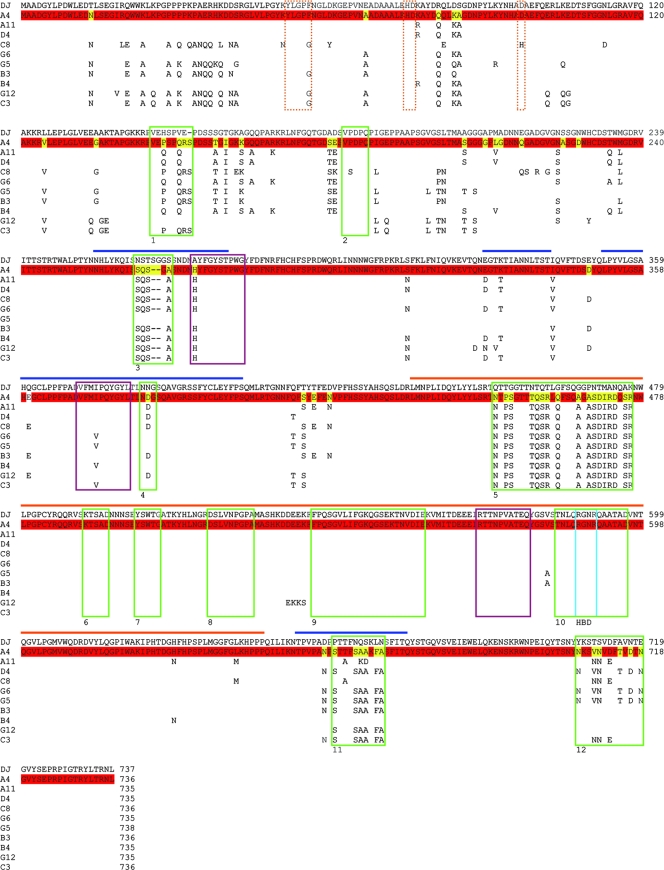
FIG. 5.
Protein sequence alignments for AAV-DJ and 10 clones from pool A. Shown are full protein alignments for the 10 clones described in the legend to Fig. 3. (The clones correspond to lanes A to J in Fig. 3 and are listed in order; e.g., clone A4 corresponds to lanes A, and clone A11 corresponds to lanes B, etc.). AAV-DJ served as the standard to highlight the different degrees of evolution between pools A and C (in which clones were selected with [pool C] or without [pool A] IVIG [Fig. 2A]). The clone A4 sequence is shown as a full sequence, as clone A4 was recovered twice from pool A. Residues identical in A4 and AAV-DJ are colored in red, while changes are shown in yellow. Only amino acids divergent from AAV-DJ are shown for the other nine clones (for the origin of these residues, see the wild-type sequences in Fig. S4 in the supplemental material and also see the text). The horizontal bars indicate the capsid loops (see Fig. S4 in the supplemental material). Note that many changes in the 10 clones from pool A were clustered in these loops, as could be expected. Moreover, the gene regions corresponding to six of these cluster regions were identical to 6 of the 12 previously reported HVRs (HVRs 1, 3, 4, 5, 11, and 12 among the previously described HVRs 1 to 12, identified by green boxes) in the AAV capsid gene. Intriguingly, our alignments also identified several further regions of sequence diversity not described before, especially in the VP1 and VP2 N termini. The dotted orange lines mark a highly conserved phospholipase 2A domain in the VP1 N terminus; note the unique amino acid change (D97H) in clone C8 (yet the capsid was infectious [see Fig. 3]). The three purple boxes show the A20 epitope; note that almost all clones had fully maintained the respective consensus sequence from AAV-2 (H in the first and V in the second part of the epitope) but that AAV-DJ had not (compare Fig. 4). The blue box shows the location of two of the five residues (two arginines) which constitute the HBD; they were fully conserved in all clones.
Notably, the phospholipase 2A domain in the VP1 N terminus, critical for particle infectivity (7, 86), remained highly conserved, with the single exception of clone C8 from pool A. In striking contrast to the N termini of the rest of the capsids, the AAV-DJ capsid N terminus was almost entirely derived from AAV-2, while the clones in pool A carried multiple dispersed residues from AAV-8 or AAV-9. These amino acids, especially the new clusters identified by our alignments, and their roles in the infection cycle should be very interesting targets for future AAV studies. These findings and considerations highlight the vast potential of DNA family shuffling, not only as a means to evolve viral vectors, but also as a functional genomics tool, useful to unravel basic virus biology.
In summary, we have cloned, sequenced (fully or partially), and compared a total of 513 candidates from our library before and after various selection schemes. Of the 465 clones from pools A to C, one was recovered twice (from pool A), while the 96 clones from pool C were completely identical (AAV-DJ). All other clones differed from one another by at least three amino acids and were not identified among the 48 clones from the unselected library. Likewise, AAV-DJ was found neither in pools A and B nor in the original library, confirming its specific evolution under stringent IVIG selection.
Recombinant AAV-DJ vectors mediate superior in vitro transduction.
We next generated gfp-expressing vectors from the AAV-DJ capsid gene and compared their in vitro infectivities to those of the eight most commonly used wild-type AAVs (serotypes 1 through 6, 8, and 9), including five of the AAV-DJ parents (serotypes 2, 4, 5, 8, and 9). Impressively, titration on 14 cell types from different species and tissues, including primary human hepatocytes, melanoma cells, and embryonic stem cells, showed that AAV-DJ vectors were not only superior to all HBD-negative wild-type viruses (up to 100,000-fold better than AAV-8 or AAV-9), but also substantially better than AAV-2 (Table 3 and data not shown). Ratios of total to infectious particles were frequently far below 500, highlighting the extreme efficiency of AAV-DJ in vitro and suggesting its particular usefulness for ex vivo gene transfer applications. The only exceptions on which AAV-DJ was not most efficient were human monocytes and dendritic cells (Table 3). On these cells, AAV-1 and AAV-6 outperformed the other vectors, albeit AAV-DJ was among the most efficient capsids. Our data for AAV-1 confirm and extend the findings of a recent study in which this serotype also surpassed AAV-2 to AAV-5 on murine hematopoietic stem cells (87). As expected, AAV-DJ transduction was largely unaffected by IVIG (similar to AAV-8 and AAV-9 transduction) (data not shown).
TABLE 3.
In vitro infectivities of AAV-DJ and wild-type vectorsa
| Cell line | Tissue or cell type | Infectivity of vector:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AAV-1 | AAV-2 | AAV-3 | AAV-4 | AAV-5 | AAV-6 | AAV-8 | AAV-9 | AAV-DJ | AAV-DJ/8 | ||
| Huh-7 | hu liver | 4e3 | 5e2 | 2e4 | 2e6 | 4e5 | 5e3 | 7e4 | 7e6 | 1e2 | 3e5 |
| 293 | hu kidney | 2e3 | 5e2 | 2e4 | 7e5 | 4e5 | 1e4 | 7e4 | 7e5 | 1e2 | 2e5 |
| HeLa | hu cervix | 7e4 | 2e3 | 1e5 | 2e6 | 3e4 | 2e5 | 1e6 | 2e6 | 3e2 | 1e6 |
| HepG2 | hu liver | 2e6 | 5e4 | 3e5 | 2e7 | 3e6 | 1e6 | 2e7 | ND | 4e3 | 1e7 |
| Hep1A | mu liver | 1e4 | 2e3 | 1e6 | 2e5 | 2e6 | 2e5 | 1e6 | 2e7 | 5e2 | 2e6 |
| 911 | hu retina | 6e3 | 1e3 | 9e3 | 5e5 | 7e5 | 6e3 | 1e6 | ND | 2e2 | 4e5 |
| CHO | ha ovary | 1e4 | 1e4 | 7e4 | 7e5 | 3e3 | 2e4 | 1e5 | 1e6 | 4e1 | 2e5 |
| COS | si kidney | 3e3 | 1e3 | 3e3 | 3e4 | 2e4 | 7e3 | 5e4 | 2e5 | 2e2 | 3e5 |
| MeWo | hu skin | 2e3 | 2e2 | 1e3 | 7e4 | 3e3 | 2e3 | 2e4 | 1e5 | 7e0 | 2e4 |
| NIH3T3 | mu fibroblasts | 2e5 | 2e4 | 7e5 | 7e5 | 7e6 | 2e5 | 7e6 | ND | 4e3 | 2e7 |
| A549 | hu lung | 7e4 | 1e4 | 5e4 | ND | 2e6 | 1e5 | 2e6 | 7e6 | 1e3 | 2e7 |
| HT1180 | hu fibroblasts | 5e4 | 1e4 | 1e5 | 7e6 | 3e6 | 3e4 | 2e6 | 1e7 | 3e3 | 5e6 |
| Monocytes | hu primary monocytes | 9e5 | 1e7 | ND | ND | 8e6 | 7e5 | ND | ND | 1e7 | ND |
| Immature DC | hu monocyte-derived DC | 8e5 | 2e7 | ND | ND | 9e6 | 7e5 | ND | ND | 1e7 | ND |
| Mature DC | hu monocyte-derived DC | 9e5 | 2e7 | ND | ND | 6e6 | 6e5 | ND | ND | 2e7 | ND |
Each cell line was infected with 10-fold serial dilutions of each serotype, AAV-DJ, or the mutant AAV-DJ/8 expressing a gfp reporter gene. Vector preparations were normalized to contain 2 × 109 total (vector DNA-containing) particles per ml prior to infection. Three days later, green fluorescent protein-expressing cells were counted and infectious titers were determined by taking into account the dilution factor. Numbers shown are average ratios (rounded) of total to infectious AAV particles from at least three independent titrations. Lower numbers indicate higher levels of infectivity. For each cell line, values corresponding to the most efficient AAV are underlined, while boldface indicates the lowest level of efficiency. AAV-DJ vectors showed the highest levels of infectivity on all tested cell lines. hu, human; mu, murine; ha, hamster; si, simian; DC, dendritic cells; ND, not detectable (>2 × 107).
To investigate the role of the AAV-DJ HBD, we mutated two crucial arginines (37, 56) into the respective residues in AAV-8 or AAV-9 (Fig. 6A and B). Green fluorescent protein expression from the resulting mutants was reduced by several orders of magnitude and was as low as that from serotypes 8 and 9 (Fig. 6C and Table 3) (results for mutants DJ/8 and DJ/9 were identical). The drop in infectivity correlated well with reduced binding to cells (Fig. 6D). However, cell attachment alone cannot explain the unusual infectivity of AAV-DJ, as AAV-2 actually bound 10-fold more efficiently. We rather assume a synergistic or additive effect from sharing beneficial properties from all AAV-DJ parents, resulting in the enhancement of multiple steps in AAV-DJ transduction. One likely outcome was the combination of efficient primary receptor binding (from AAV-2, compared to AAV-8 and AAV-9) with rapid virus processing and uncoating (from AAV-8) (73) (Fig. 6E). A further explanation may be that the juxtaposition of subunits from different parents created a unique property, such as the use of an unidentified coreceptor for faster capsid internalization (see also Discussion and Fig. 12 below). An important role in infectivity was likely also played by the unique AAV-DJ N terminus, which differed from those of all other recovered clones (see above).
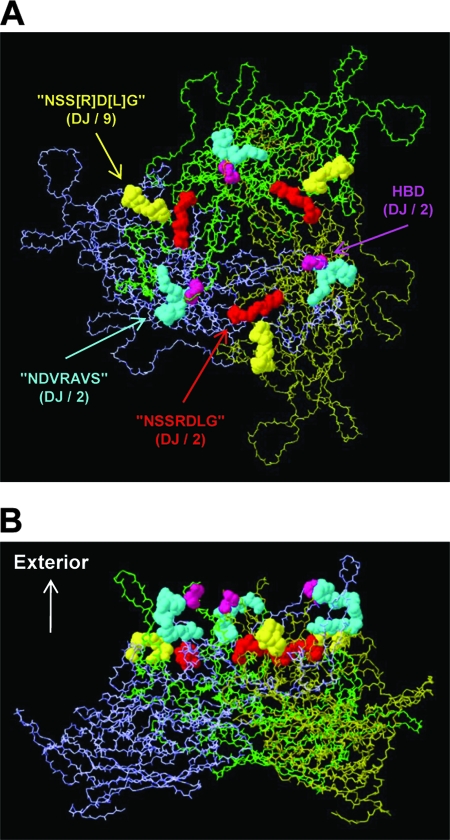
FIG. 12.
Model of an AAV VP3 trimer (panel A, top view down the threefold symmetry axis; panel B, side view) created using Swiss-PdbViewer (www.expasy.org/spdbv/text/getpc.htm) and the VIPER oligomer generator (viperdb.scripps.edu/oligomer_multi.php), with the following parameters: T=1 capsid structure; selected matrices A5, A6, and A17; and Protein Data Bank file no. 1LP3 (Fig. 2C) for the AAV-2 sequence. Sequence motifs that may contribute to AAV receptor binding are colored as follows (serotypes with the highest degree of conservation are shown in parentheses): purple, HPD; red, motif with similarity to the NSSRDLG peptide (in AAV-DJ and AAV-2, 534-NGRDSL-539; numbers refer to the AAV-DJ sequence) (compare Fig. 2D); blue, motif with similarity to the NDVRAVS peptide (in AAV-DJ and AAV-2, 505-RVS[KT]SADNNNS-516); and yellow, another motif with (partial) similarity to the NSSRDLG peptide (in AAV-DJ and AAV-9, 277-SGGSSNDN-284). Note how all four motifs are exposed on the capsid surface (B) and how pairs of motifs are located in close proximity to each other (yellow and red motifs or blue and purple motifs), as well as near the HBD (purple), suggesting that they may act cooperatively in receptor binding. Intriguingly, AAV-DJ has the only capsid combining all four motifs in one sequence, perhaps contributing to its high level of efficacy (see the text).
AAV-DJ yields robust hFIX expression in mouse liver tissue.
Based on the high level of efficacy of the AAV-DJ capsid in vitro, we became interested in evaluating vectors based on this novel chimera in mouse liver tissue in vivo. For this purpose, we produced recombinant AAV-DJ particles expressing the hFIX gene from a robust liver-specific promoter (53). Controls were wild-type capsids of serotypes 2, 8, and 9 and mutants DJ/8, DJ/9, and 2/8 (HBD negative) and 8/2 (HBD positive) (see above and Fig. 6A and D). Immunocompetent C57BL/6 mice were infused via the tail vein with particle doses of each virus ranging over four orders of magnitude (5 × 109 to 1 × 1012 particles), and plasma hFIX levels were monitored for up to 4 months.
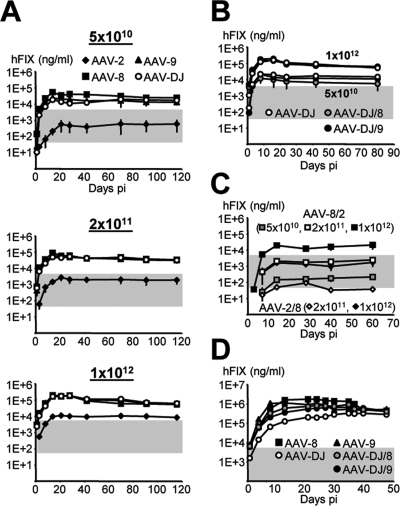
We observed dose-dependent expression from the AAV-DJ capsid at levels equivalent to those from AAV-8 and AAV-9, the best two naturally identified AAVs for liver tissue reported thus far (18, 19, 52) (Fig. 7A and data not shown for the 5 × 109 dose). All three viruses readily outperformed the AAV-2 prototype at any dose and expressed over 100% of normal hFIX levels already after the intravenous injection of 5 × 1010 particles (AAV-2 matched these levels only at a dose of 1012 particles, i.e., a 20-fold-higher dose). Quantification and analyses of persisting vector DNA confirmed the similarity of AAV-DJ, AAV-8, and AAV-9 and their comparable degrees of superiority over AAV-2 (see Fig. S5 in the supplemental material). These results were verified in analogous experiments using two alternative expression cassettes (data not shown).
FIG. 7.
hFIX expression from AAV-DJ in mice. (A) Dose-dependent and liver-specific hFIX expression. C57BL/6 mice (n = 3 to 8) were infused with all four hFIX-expressing vectors via peripheral tail vein injection. Gray shading indicates the range from 1 to 100% of normal hFIX levels in humans (0.05 to 5 μg/ml). Levels over 1% are considered to be therapeutic in hemophiliacs. Note that AAV-8, AAV-9, and AAV-DJ vectors exceeded the 100% level already at the lowest dose, whereas AAV-2 required a 20-fold-higher dose. (B) hFIX expression from the AAV-DJ HBD mutants (n = 3 per group). Shown are results from two representative doses; there was no significant difference from the results for AAV-DJ. (C) In contrast, the AAV-2 or AAV-8 HBD mutants expressed less hFIX than the corresponding wild types (n = 3 per group). (D) AAV-DJ showed unique transduction kinetics at a maximum dose of 7 × 1012 particles. The onset of gene expression was delayed compared to that from AAV-8 or AAV-9, yet hFIX levels became similar after ∼40 days (n = 3 per group). The AAV-DJ HBD mutants showed intermediate kinetics; stable hFIX levels were eventually also similar to those from AAV-8 and AAV-9 (and AAV-DJ). pi, postinjection.
Curiously, the two DJ HBD mutants were indistinguishable from AAV-DJ (and serotypes 8 and 9) at these doses, while the corresponding AAV-2 mutant (2/8) was inferior to wild-type AAV-2 (Fig. 7B and C). Different expression levels for AAV-8 and the HBD-positive 8/2 mutant were also noted, albeit here, the wild type performed better (Fig. 7C). This finding suggested an essential function of heparin binding for liver gene expression from AAV-2, corroborating previous findings with this serotype (37), but a redundancy for more efficient natural or synthetic capsids (those of AAV-DJ and serotypes 8 and 9). Moreover, together with our data from the in vitro assays (see above), these results exemplified the differential effects of the HBD on AAV transduction in culture and in organisms.
The AAV HBD plays a multifaceted role in vivo.
Interestingly, additional studies indicated an even more complex role for the HBD in vivo. At a maximal dose of 7 × 1012 particles, the transduction profiles of the most efficient viruses became unique. All HBD-negative variants showed faster transduction kinetics than AAV-DJ, although all viruses eventually (after ∼1.5 months) gave similar expression levels (Fig. 7D). A similarly slow response at extreme particle doses with a resulting lag phase had previously been reported for AAV-2 (53, 67) and was confirmed here (data not shown). The fact that AAV-DJ and AAV-2 share the HBD suggests a common molecular mechanism involving this domain, likely at the level of post-vector entry (e.g., particle trafficking or uncoating). However, this idea warrants further investigation in view of studies reporting blunted dose responses also for AAV-1 and AAV-5, which both lack a consensus HBD (52).
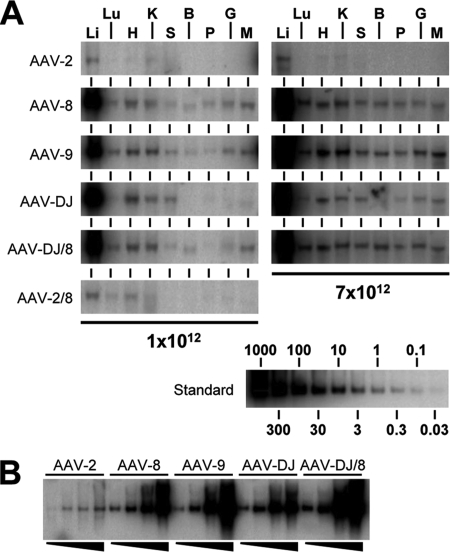
A second function of the HBD became apparent upon analyses of vector DNA biodistribution (Fig. 8A and Table 4). We corroborated previous reports of unrestricted tropism of AAV-8 and AAV-9 (HBD negative) (17, 18, 34, 52, 60), which readily transduced all tested tissues at a dose of 1012 particles per mouse. In striking contrast, AAV-2 and likewise AAV-DJ (both HBD positive) were restricted to liver tissue and, to a lesser extent, heart, kidney, and spleen tissues and were near or below the detection limit in all other tissues. In fact, the quantification of double-stranded vector DNA (using liver tissue as an internal standard for each group) showed that AAV-DJ transduced lung, brain, pancreas, and gut tissue about two- to fourfold less efficiently than wild types 8 and 9 (Table 4). The effect of the HBD on viral tropism was best exemplified by comparing AAV-DJ to the DJ/8 mutant: HBD deletion alleviated the liver restriction and expanded transduction to all nonhepatic tissues, including the brain, identical to the transduction patterns of AAV-8 and AAV-9. These findings not only corroborate but also may help explain a series of reports on the wide tissue dissemination of vectors based on HBD-negative natural serotypes (AAV-1 and AAV-4 to AAV-9) in mice, dogs, and monkeys (17, 29, 37, 52, 60), in contrast to that of the HBD-positive AAV-2. Notably, AAV-DJ also transduced nonhepatic tissues at the maximum dose of 7 × 1012 particles but still to a lesser extent than the HBD-negative viruses, in particular AAV-9 (Fig. 8A and Table 4). Importantly, even at this dose, brain and also lung transduction remained marginal.
FIG. 8.
Vector DNA biodistribution and dose response. (A) Genomic DNA extracted from nine tissue types (li, liver; lu, lung; h, heart; k, kidney; s, spleen; b, brain; p, pancreas; g, gut; and m, muscle) was analyzed for the presence of hFIX-expressing vector DNA. The results and the reference standard shown are representative of data for the two highest doses used here. The AAV-DJ transduction pattern was more restricted to liver, heart, kindey, and spleen tissues than those of AAV-8, AAV-9, and the HBD mutants. At the highest dose (7 × 1012 particles), AAV-DJ spillover into nonhepatic tissues was also less obvious than that of the other vectors. The HBD-negative AAV-2/8 mutant gave increased heart transduction compared to wild-type AAV-2, confirming previous data (37) (an unknown production deficiency prevented evaluation at the highest dose). (B) Comparison of vector DNA levels in liver following transduction with increasing particle doses (from left to right, 5 × 1010, 2 × 1011, 1 × 1012, and 7 × 1012 particles). AAV-DJ showed a blunted response at the highest dose, likely correlating with its slower onset of gene expression (Fig. 7D).
TABLE 4.
Relative levels of transduction of nonhepatic tissues with AAV vectorsa
| Vector | Dose (no. of particles) | Level of AAV DNA in:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Lung | Heart | Kidney | Spleen | Brain | Pancreas | Gut | Muscle | ||
| AAV-2 | 1e12 | ND | 0.7 ± 0.1 | 0.8 ± 0.1 | 0.2 ± 0.0 | ND | ND | ND | ND |
| 7e12 | ND | 1.5 ± 0.03 | 2.0 ± 0.3 | 1.0 ± 0.2 | ND | ND | ND | ND | |
| AAV-8 | 1e12 | 0.5 ± 0.0 | 1.2 ± 0.2 | 0.9 ± 0.2 | 0.3 ± 0.0 | 0.2 ± 0.0 | 0.2 ± 0.0 | 0.3 ± 0.0 | 0.7 ± 0.1 |
| 7e12 | 2.5 ± 0.3 | 2.5 ± 0.2 | 2.6 ± 0.3 | 1.5 ± 0.2 | 1.5 ± 0.2 | 1.2 ± 0.2 | 1.2 ± 0.2 | 1.9 ± 0.2 | |
| AAV-9 | 1e12 | 0.7 ± 0.1 | 1.3 ± 0.2 | 1.1 ± 0.2 | 0.4 ± 0.0 | 0.2 ± 0.0 | 0.2 ± 0.0 | 0.3 ± 0.0 | 0.8 ± 0.1 |
| 7e12 | 2.6 ± 0.3 | 3.6 ± 0.4 | 3.8 ± 0.4 | 1.5 ± 0.2 | 1.8 ± 0.2 | 1.3 ± 0.2 | 1.9 ± 0.2 | 3.0 ± 0.3 | |
| AAV-DJ | 1e12 | 0.2 ± 0.0 | 1.3 ± 0.2 | 0.8 ± 0.2 | 0.5 ± 0.1 | ND | 0.1 ± 0.0 | 0.1 ± 0.0 | 0.2 ± 0.0 |
| 7e12 | 0.6 ± 0.1 | 2.3 ± 0.2 | 2.1 ± 0.2 | 1.5 ± 0.2 | 0.4 ± 0.0 | 0.5 ± 0.0 | 0.5 ± 0.0 | 0.8 ± 0.1 | |
| AAV-DJ/8 | 1e12 | 0.6 ± 0.0 | 1.3 ± 0.2 | 0.8 ± 0.2 | 0.2 ± 0.0 | 0.2 ± 0.0 | 0.1 ± 0.0 | 0.2 ± 0.0 | 0.7 ± 0.1 |
| 7e12 | 2.6 ± 0.3 | 2.5 ± 0.3 | 2.3 ± 0.3 | 1.6 ± 0.3 | 1.8 ± 0.2 | 1.2 ± 0.2 | 1.3 ± 0.2 | 2.0 ± 0.2 | |
Vector copy numbers (per diploid genomic equivalent) were determined via phosphorimager scan analyses of Southern blots as shown in Fig. 8A. At least three independent mice per applied dose were analyzed. Copy numbers are shown as average percentages (rounded to one decimal place) ± standard deviations relative to copy numbers in liver tissue within each group, allowing comparison between vectors and doses. For AAV-2, most signals were below the detection limit of the Southern blot analyses (∼0.03 copies of double-stranded AAV DNA per cell), preventing the calculation of relative transduction in these cases (ND, not determined). Underlining highlights values for doses or tissues for which relative AAV-DJ transduction levels differed by at least twofold from those for serotypes 8 and 9, as well as for the AAV-DJ HBD mutant.
Additional side-by-side comparison of all liver vector DNA levels showed similar dose responses for the non-AAV-2 viruses at doses between 5 × 1010 and 1 × 1012 particles (Fig. 8B), in agreement with our expression data (see above). However, at 7 × 1012 particles, the HBD-negative viruses persisted at slightly higher copy numbers than AAV-DJ. The data in Fig. 7D, as well as our previous findings with AAV-8 (52), imply that this outcome resulted from faster transduction with the HBD-negative viruses than with the HBD-positive viruses. This effect in turn likely increased the steady-state levels of viral DNA. Because the levels of expression from all viruses eventually became similar (Fig. 7D), we further hypothesize that the majority of additional genomes were silenced over time. While the latter conclusion is also supported by the results of our earlier work (52), more detailed studies are needed to prove the ensuing opposite idea, that a higher proportion of vector DNA copies from AAV-DJ transduction than from transduction with HBD-negative viruses remains transcriptionally active in the liver.
AAV-DJ partly escapes humoral neutralization in mice.
We next quantified liver transduction in the presence of human serum to assess AAV-DJ's ability to evade neutralization in vivo. Therefore, we passively immunized mice with IVIG prior to the infusion of hFIX-expressing AAV-2, AAV-8, AAV-9, or AAV-DJ. As predicted, AAV-2 expression was nearly completely abolished. In contrast, transduction with AAV-DJ, AAV-8, or AAV-9 was inhibited in a dose-dependent manner, with AAV-DJ showing intermediate resistance at the high IVIG dose and efficient evasion (similar to that by serotypes 8 and 9) at the low IVIG dose (Fig. 9A). These results were essentially confirmed with a second independent IVIG batch from another vendor (Carimune [12%]; Behring AG) (data not shown). Interestingly, the DJ HBD mutants were fully neutralized, comparable to AAV-2 (Fig. 9B). This observation suggested that the HBD affected the display of epitopes on the capsid, thus implying yet another role of this domain for the AAV particle. Alternatively, published data for wild-type AAVs (17), as well as our own preliminary findings (data not shown), indicate that this domain may determine in vivo vector pharmacokinetics by enhancing the clearance of HBD-positive capsids from the blood and, thus, reducing their exposure to neutralizing antibodies.
Lastly, we assessed the feasibility of repeatedly administering the different viruses to mice to evaluate capsid cross-neutralization (Fig. 9C). As expected, we saw no gene expression upon the reinfusion of any of the capsids into animals already treated with the same serotype. However, we noted that AAV-8 and AAV-9 also efficiently blocked each other, substantiating prior data (18). This result may argue against the use of vectors based on these wild types in readministration protocols, although the vectors could be combined with AAV-2. In contrast, primary infusion with AAV-DJ allowed subsequent expression (at up to 18% of the respective PBS control) from AAV-2, AAV-8, or AAV-9, likely due to the fact that AAV-DJ shares only a limited number of epitopes with each wild-type virus. Surprisingly, in the reverse experiment, AAV-DJ vectors were inhibited in animals immunized with AAV-8 or AAV-9 while giving detectable expression in AAV-2-treated mice. This result implied a stronger or broader immune response from primary infusion with serotype 8 or 9 than from that with serotype 2. Intriguingly, AAV-DJ was more resistant to the corresponding mouse sera in culture than in vivo (Fig. 9D), and we also noted less cross-reactivity between AAV-8 and AAV-9 in culture than in vivo. These findings are perhaps due to the display of distinct epitopes in vitro and in vivo. This scenario may likewise explain the different degrees of neutralization of all three viruses with IVIG in cells and in mice.
Creation of a viral peptide display library based on AAV-DJ.
An intriguing property of the AAV-DJ capsid revealed by our in vitro assays is the inverse correlation of cell binding and transduction efficacies (Fig. 6). Together with the unique endosomal cathepsin B cleavage pattern, this finding suggests that the superior efficacy of the AAV-DJ capsid is due at least partially to faster uptake or enhanced intracellular processing. Conversely, attachment receptor binding seems less limiting for AAV-DJ transduction (in the presence of a functional HBD) than for transduction with the other vectors, as implied by the consistently high potency of AAV-DJ in multiple distinct cell types. This finding suggested AAV-DJ as a promising candidate for tissue retargeting approaches via viral peptide display, in analogy to prior work with the AAV-2 prototype. Importantly, by using AAV-DJ as a parental virus, we hoped to overcome the two main drawbacks of AAV-2: the high prevalence of neutralizing antibodies and the typically slow intracellular processing limiting transduction efficacy.
In addition to the use of an optimized capsid, we further threefold improved our strategy over those described in the prior reports by Michelfelder et al., Müller et al., Perabo et al., and Waterkamp et al. (48, 50, 58, 76). Firstly, we wanted to ensure that the HBD in AAV-DJ was nonfunctional to increase the likelihood that any new cell tropism was truly mediated by the displayed peptides. Previously, the loss of the heparin binding capability of the AAV-2 capsid had been achieved indirectly and incompletely, through steric hindrance and conformational effects resulting from the peptide insertion. Here, we additionally mutated one of the two critical arginines in this domain (R585) to fully abolish binding to heparin or the heparan sulfate proteoglycan receptor. Secondly, we assembled our random peptide library based on an (NNB)7 oligonucleotide, where B symbolizes the nucleotide C, G, or T. Compared to the (NNK)7 scheme (where K represents G or T) used in three of the four previous studies (48, 50, 76), our design further decreases the statistical risk of the unwanted incorporation of a stop codon (only one, TAG, remains possible among 48 triplets), from 3.1% (NNK) to 2.1% (NNB). Notably, our library still closely mimicks the natural amino acid distribution (see Fig. S6 in the supplemental material). Thirdly and most importantly, we reckoned that using AAV-DJ as a highly efficient parental capsid would permit the biopanning of our library under the physiologically most relevant conditions, i.e., in mice in vivo. This was a critical improvement over all previously reported selection schemes, in which AAV libraries were panned exclusively on cultured cells in vitro. For this goal, we chose the lung as a second target because it offers the extra benefit that it is a natural site of adenovirus infection, implying the feasibility of productively coamplifying AAV and helper virus particles in this organ in vivo.
The AAV-DJ peptide display library was constructed by modifying the basic three-step cloning scheme devised for AAV-2 by Müller et al. (50), with the two mentioned improvements of an additional R585Q mutation and the use of (NNB)7 21-mers for peptide coding. However, we maintained the concept of displaying the targeting peptide ligands after residue R588, a locale successfully used by many groups. The resulting plasmid library had a diversity of ∼3 × 107 clones, which is well in the range of the four previously described AAV-2 counterparts (with estimated diversities of 1.2 × 107 to 1.7 × 108 clones) (48, 50, 58, 76). Analyses of 24 individual clones confirmed the complete randomization of the inserted oligonucleotides and the absence of repetitive sequences in our library (data not shown). Of the 168 triplets in these clones, only three carried the single possible stop codon TAG, representing a 1.8% frequency in our plasmid library (see Fig. S6 in the supplemental material). This level was even below the theoretical prediction for the (NNB)7 design (2.1%) and also slightly below previous results with an AAV-2 (NNK)7 library (about 2%, with a theoretical possibility of 3.1%) (50, 76). Very similar data (not shown) were obtained for an AAV-2-based library that we generated in parallel, by following the same rules, as a control for the AAV-DJ library. Both plasmid libraries were then packaged into virions (in 2 × 109 cells, to maintain library diversity), resulting in large and complex viral libraries with titers of 5 × 1012 (AAV-2) and 5.8 × 1012 (AAV-DJ) particles/ml, matching or in fact slightly exceeding (in the case of AAV-DJ) the highest numbers reported previously (5.5 × 1012) (76).
In vivo biopanning of peptide-modified AAV capsids in mouse lung tissue.
Critical to the success of AAV capsid evolution is to find an ideal multiplicity of infection giving minimal target cell transduction with a single or only a few particles, which are subsequently amplified by the helper virus. This approach will avoid the uncoupling of genotypes and phenotypes, which can occur if cells are initially infected with excess AAV particles and which will hamper the selection process. Likewise crucial is to find a helper virus dose resulting in efficient yet slow infection, giving the AAVs enough time for productive amplification. To optimize these parameters, we nasally coinfected mouse lungs with various multiplicities of infection of the AAV-2- and AAV-DJ-based libraries, ranging from 109 to 1011 physical particles, together with escalating volumes (0.2, 1, and 5 μl) of human or murine adenovirus stocks (from the ATCC). Our aim was to find the virus dose combination that would allow PCR detection of amplified AAV particles in helper virus-infected mice but not in controls lacking the adenovirus. The mice were maintained for 4 or 7 days before their lungs were harvested. At the highest dose of murine adenovirus (5 μl of ATCC stock VR-550), all lungs had macroscopic evidence of tissue inflammation. In contrast, mice inoculated with the human adenovirus lacked any gross signs of infection, and their lungs looked normal under all conditions (doses and time points).
A comparison of the results with various AAV doses showed that inoculation with 1010 AAV-DJ library particles gave the ideal result, i.e., a clear band for the murine helper virus-treated mice but no or very faint signals in the absence of adenovirus (data not shown). Conversely, we did not obtain bands with the same dose of the AAV-2 library, regardless of the absence or presence of helper virus, and there was also no detectable AAV amplification in any mice infected with the human helper virus. Together, these findings confirmed that the AAV-DJ particles had successfully replicated their genomes in the mouse lungs in the presence of the appropriate murine helper. Moreover, the fact that we observed AAV-2 amplification only from a 10-fold-higher starting dose of 1011 particles validated our hypothesis that using AAV-DJ as a parental capsid would increase the in vivo fitness of an AAV library compared to that of a library based on the conventionally used serotype 2.
We next cloned the amplified AAV-DJ genomes from three mice and sequenced 15 to 24 clones per mouse. We were able to read 46 full sequences, whose translation and alignment are shown in Table 5 (for amino acid compositions, see Fig. S6 in the supplemental material). Clearly, the most notable observation was the enrichment with peptides NSSRDLG and NDVRAVS in the first mouse. This result was curious, as the identical peptides had previously been isolated independently from an AAV-2-based library after in vitro passaging on two other cell types, coronary arterial and venous endothelial cells (50, 76). Several other findings were also interesting. Firstly, we noted the recurrence of particular peptide motifs composed of the amino acids D-V-R and S-R-D-G in different arrangements among all three mice (Table 5). The two lead peptides from mouse 1 were also enriched with S-R-D(-G-V) residues in distinct permutations. Secondly, we discovered an elevated frequency (∼3-fold over that predicted) of arginines (R) in positions 4 of the peptides, especially apparent again in the two lead candidates. Thirdly, the frequency of glycine (G) residues was also markedly increased (from a predicted 6% to 15 to 20%) in multiple positions, particularly 1 and 7 (the start and end points of the peptides), but also 4. As a result of the G/R increase in positions 4, the overall frequency of hydrophilic amino acids in this spot was also elevated (from a predicted 55 to 61%), like that of positively charged residues (from a predicted 15 to 26%). A last remarkable finding was a drop in negatively charged polar amino acid frequency from a predicted 10% to 2 and 4% for positions 6 and 7, respectively, and a concurrent mild increase in the frequency of noncharged polar residues (from 30% to 33 and 35%). Conversely, the frequency of hydrophilic amino acids was lowest (46%, down from a predicted 55%) in positions 6.
TABLE 5.
Peptide sequences from in vivo biopanning in murine lungsa
| Mouse (no. of clones) | Peptide (frequency) | Charge pattern | Hydrophobicity pattern |
|---|---|---|---|
| 1 (19) | NSSRDLG (4) | xxx+−yy | −−−−−++ |
| NDVRAVS (3) | x−y+yyx | −−+−++− | |
| NWLLDSG | xyxx−xy | −+++−−+ | |
| NTQDVNK | xxx−yx+ | −−−−+−− | |
| GLEGSSN | yy−yxxx | ++−+−−− | |
| GEQSSFG | y−xxxyy | +−−−−++ | |
| SADRQGP | xy−+xyy | −+−−−++ | |
| DELQGCT | −−yxyxx | −−+−+−− | |
| VLDGMCR | yy−yyx+ | ++−++−− | |
| DNNGLVV | −xxyyyy | −−−++++ | |
| ADWLCRA | y−yyx+y | +−++−−+ | |
| GKEKDTI | y+−+−xy | +−−−−−+ | |
| HCMVRPC | +xyy+yx | −−++−+− | |
| KMEGIFN | +y−yyyx | −+−+++− | |
| 2 (16) | YGGSRSN | xyyx+xx | −++−−−− |
| DLRVQGC | −y+xxyx | −+−+−+− | |
| DFSVSFV | −yxyxyy | −+−+−++ | |
| HDVRWAV | +yy+yyy | −−+−+++ | |
| WVRDVML | yy+−yyy | ++−−+++ | |
| ARATDRV | y+yxy+y | +−+−−−+ | |
| VALGRES | yyyy+−x | ++++−−− | |
| FVLTVGG | yyyxyyy | +++−+++ | |
| SYRVQTS | xx+yxxx | −−−+−−− | |
| ICEGHSR | yx−y+x+ | +−−+−−− | |
| GLRQPFS | yy+xyyx | ++−−++− | |
| ASSVYWY | yxxyyyy | +−−+−+− | |
| VIISPTS | yyyxyxx | +++−+−− | |
| KDIVRKV | +−yy++y | −−++−−+ | |
| KPGGNQL | +yyyxxy | −+++−−+ | |
| GTGTQSD | yxyxxx- | +−+−−−− | |
| 3 (11) | RDVDVGR | +−y−yy+ | −−+−++− |
| DVGRIRD | −yy+y+− | −++−+−− | |
| GDVDSNK | y−y-xx+ | +−+−−−− | |
| GKFGSGP | y+yyxyy | +−++−++ | |
| SMEPRLC | xy−y+yx | −+−+−+− | |
| DGSTLTR | −yxxyx+ | −+−−+−− | |
| LYKDSGY | yx+−xyy | +−−−−+− | |
| EAERGRY | −y−+y+x | −+−−+−− | |
| IDGGWMI | y−yyyyy | +−+++++ | |
| CWWQEYN | xyyx−xx | −++−−−− | |
| LKASVLW | y+yxyyy | +−+−+++ |
Shown are 46 peptide sequences from AAV genomes isolated from mouse lungs (from three individual animals) after in vivo biopanning of the AAV-DJ-based peptide display library. The peptides NSSRDLG and NDVRAVS were isolated four and three times, respectively, from the same mouse (all others were isolated once). Residues or short sequence motifs that were enriched in all three animals (see the text) are in bold or underlined. Also shown is the pattern of amino acid side chains, with the following classifications: +, positively charged; −, negatively charged; x, uncharged polar; and y, nonpolar. Moreover, their classification according to hydrophobicity is listed, in which + represents hydrophobic and − represents hydrophilic.
Generally, we noted high-level diversity among the peptide sequences from different mice or even among those from the same animal. This finding was not surprising, in light of identical and consistent prior results from in vitro biopannings of AAV-2 libraries on cultured cells (48, 50, 58, 76). As discussed later, there are several possible explanations for this sequence diversity, especially the very complex lung architecture, with multiple cells and receptors. Another important factor was that the sequences in Table 5 were obtained after a single infection round, implying that consecutive in vivo reamplification may result in further enrichment with specific peptides. However, we actually failed to detect AAV DNA signals in new animals infected with whole-lung protein extracts. The most likely reason was the generation of neutralizing antibodies in the primary immunocompetent mice, and this possibility was indeed corroborated by the lack of signs of adenovirus infection in secondary animals, even after prolonged incubation. We therefore pooled the extracts from mice 1 and 2 from the first round and depleted the extracts of murine Igs by using a commercial kit. The cleared eluates were then used for reinfection, together with freshly added murine helper virus. This approach resulted in the expected lung infection and inflammation at day 7 and enabled us to PCR amplify and clone replicated AAV genomes from total lung DNA. Surprisingly, we found all 24 analyzed clones from the second round to be identical and to encode the peptide MVNNFEW. This sequence was unique and had been observed neither in the original unselected library nor in the three primary mice.
Analyses of lung cell tropisms of peptide-modified AAV-DJ vectors.
Our next goal was to study whether the isolated peptides would confer new tropism on the AAV-DJ capsid in murine lungs in vivo. We therefore engineered luciferase-expressing recombinant AAV-DJ vectors to display the NSSRDLG peptide (the peptide most frequently recovered in the first round) (Table 5) or the MVNNFEW peptide (the peptide from the second round) and infused them by nasal aspiration at a dose of 1011 particles per mouse. As controls, we applied the identical vector genomes corresponding to unmodified DJ capsids, the DJ-2/8 HBD mutant capsid, or the wild-type AAV-8 capsid. In vivo luciferase expression was monitored starting at day 1 and continuing over 4 weeks, before all major tissues including lungs were extracted and imaged separately.
The representative mice shown in Fig. 10A exemplify an approximate twofold variation in overall lung transduction efficacies among the five vectors, with the NSSRDLG mutant peptide (and the AAV-8 capsid) typically yielding slightly better results than the other three DJ variant forms. This finding was corroborated by the imaging of isolated lungs (Fig. 10B). Notably, analyses of all other main tissues gave no evidence for vector spillover beyond the lung (data not shown).
FIG. 10.
Analyses of in vivo transduction with peptide-displaying AAV-DJ capsid variants. (A) Wild-type FVB mice were nasally infected with luciferase-expressing AAV vectors as described in the text. Shown are representative examples of lung-directed luciferase expression (numbers below the image are photon counts) 7 days after the inoculation. The black arrow highlights an example of very occasionally observed vector spillover into the stomach (an artifact from the installation procedure). DJ, parental AAV-DJ; NSS, NSSRDLG mutant; MVN, MVNNFEW mutant; ΔHBD, HBD-negative AAV-DJ/8 mutant; 8, wild-type AAV-8. (B) In vitro imaging of isolated lungs confirmed the similarity of luciferase expression levels among all vectors and the mild trend toward higher numbers with the NSSRDLG variant. C, control. (C) Representative examples of results for wild-type FVB mice 7 days after peripheral injection (via the tail vein) with all luciferase-expressing vectors as described in the text. Note the slight (two- to threefold) drop in expression from the MVNNFEW mutant compared to that from the other vectors. (D) Histological analyses (by X-Gal staining) of murine lungs 2 weeks after nasal infection with β-galactosidase-expressing AAV-DJ variants (5 × 1010 particles per mouse). Shown are various representative examples of different sections at different magnifications (×100 or ×200) for each mouse. Black arrows highlight the predominantly transduced cell type for each capsid: (putative) alveolar type II cells for the NSSRDLG variant and alveolar macrophages for the MVNNFEW peptide. The arrows in the images corresponding to the AAV-DJ/8 mutant-infected mouse (labeled ΔHBD) point at occasionally observed positive alveolar cells, while the main transduction target was pulmonary endothelial or smooth muscle cells (top frame), identical to the target of the parental AAV-DJ capsid (the middle frame corresponding to the AAV-DJ-infected mouse shows a vessel cross-section). Sections from an uninfected control mouse (rightmost frames) showed no background signals, with the exception of very faint macrophage staining that was also found in sections from the other mice (not visible here, except in those from the MVNNFEW mutant-infected mouse, which had stronger and more clustered signals than sections from the other mice).
In a parallel biodistribution study, we systemically infused all vectors at an intermediate dose of 2 × 1011 particles via peripheral tail vein injection (Fig. 10C). Whole-body imaging revealed comparable levels of liver transduction for most DJ vectors and AAV-8, corroborating our hFIX expression data (see above) (Fig. 7). The only exception was the MVNNFEW mutant vector, which consistently gave two- to threefold-lower luciferase levels than the others. The imaging of all other major tissues gave no clear evidence for the retargeting of the two peptide mutants to a particular organ, including the lungs, from this injection route (data not shown). Intriguingly, this finding seems to contradict the results of a prior study in which the NSSRDLG peptide mediated systemic AAV-2 detargeting from the liver and retargeting to the heart (50). As discussed later, these discrepancies may be due to several crucial differences between the two studies.
The luciferase data had validated lung transduction with all vectors but gave no information on the infected cell types. We thus nasally infused a new set of vectors based on all four DJ variants, expressing β-galactosidase from a CMV promoter. Two weeks later, the lungs were perfused with agarose, sectioned, and X-Gal stained. A number of salient observations are depicted in Fig. 10D. Firstly, both the unmodified DJ and the HBD mutant (a control lacking a peptide insert) strongly transduced cells of the vasculature (likely pulmonary endothelial cells or smooth muscle cells), whereas these particular cells were entirely negative with the two peptide mutants. Secondly, the HBD mutant but not wild-type DJ additionally infected occasional alveolar cells, which by size, morphology, and location appeared to be type II cells. Thirdly, these specific cells were in fact the main and only distinct target of the NSSRDLG mutant. This obvious contrast to DJ or the HBD mutant highlights the retargeting effect (from endothelial to alveolar cells) mediated by this particular peptide. Yet, both endothelial and alveolar cells in lungs infected with the MVNNFEW mutant were conspicuously negative. Instead, we observed a mild increase in the staining of yet another dispersed cell type, alveolar macrophages. In fact, these cells stained faintly positive in all mice (not visible in Fig. 10D), including an uninfected control (right panels) which was otherwise entirely negative. Even so, the apparent increase in alveolar macrophage transduction with the MVNNFEW mutant may well explain the origin of luciferase expression depicted in Fig. 10A and B, as well as the loss of liver signals (Fig. 10C). Moreover, the evolution of macrophage tropism during AAV passaging in the presence of helper virus would be highly reasonable, as these cells are a main target for adenovirus infection in the lung.
AAV capsid library biopanning in all major organs.
Encouraged by the results with lung tissue, we performed a pilot study to assess the potential of our libraries for biopanning in all major tissues of a mouse. We therefore injected two mice each with 5 × 1011 particles from the original shuffled library or the AAV-DJ-derived peptide display library. A week later, we harvested nine major tissue types and screened total genomic DNA for the presence of AAV viral DNA via PCR. As seen in Fig. 11, we could clearly detect AAV cap genes in all samples, although we noted differences among the individual tissues and animals (note that the PCR was not quantitative). Strong signals were consistently observed in liver, heart, kidney, and spleen tissues, the typical AAV targets (compare Fig. 8). However, we also obtained good bands for all four brain DNA samples, exemplifying the potential of our unselected libraries to infect tissues that are otherwise inaccessible to the majority of AAVs (in particular, from peripheral virus application).
FIG. 11.
Biodistribution of AAV capsid libraries following peripheral delivery (via tail vein injection). Wild-type FVB mice were infused with 5 × 1011 particles of the shuffled (A) or AAV-DJ-based peptide display (B) library, and 1 week later, all major organs were harvested for the preparation of total genomic DNA. AAV DNA genomes were detected via PCR using primers flanking the entire cap gene (∼2.2 kb; arrows). Numbers (in kilobases) on the left refer to a DNA size marker. Shown are results from two representative mice per injection protocol. Note that AAV DNA signals could be detected in all analyzed tissues, including brain tissue, highlighting the potential for the biopanning and evolution of AAV capsids in all major organs in vivo.
Our next question was whether we would also be able to enrich optimized capsids or further targeting peptides from these organs. For proof of the concept, we performed the following injections (route and tissue harvested are shown in parentheses): (i) shuffled library (intranasally, lung), (ii) shuffled library (intravenously, liver), and (iii) AAV-2 peptide library (intravenously, liver). We chose the AAV-2-based library since AAV-DJ had already been evolved on liver cells, lowering the chances of recovering hepatotropic peptides. We injected one mouse each for injections i and ii (shuffled libraries) with 5 × 1011 particles, as well as one mouse for injection iii (AAV-2 peptide library) with 2 × 1012 particles. Because of the overall lower level of in vivo fitness of the AAV-2 library, we used a four-times-higher dose than that used before in the biodistribution study with the AAV-DJ counterpart (Fig. 11) in order to permit efficient PCR recovery of viral genomes. All three mice were additionally coinfected with murine adenovirus (5 μl of ATCC stock VR-550) to help AAV genome amplification.
One week after virus installation, the two lungs and the liver from each mouse were harvested, and potentially replicated and enriched AAV genomes were cloned and sequenced (from the 3′ end over ∼500 nucleotides, including those corresponding to the HBD and peptide insertion, respectively). Of 20 clones per mouse, we obtained readable sequences for 14 from injection i (shuffled-library particles in lung tissue), 18 from injection ii (shuffled-library particles in liver tissue), and 19 from injection iii (AAV-2 peptide particles in liver tissue). We first performed alignments of the 32 sequences from the shuffled library with the 8 parental viral genomes. Although the numbers of mice and clones per group were too low for comprehensive statistical analyses, we observed markedly different trends between the lung and liver samples. In lung samples, 6 of 14 clones had retained the AAV-2 HBD, whereas the others were homologous in this region to AAV-8 (1 of 14), AAV-9 (1 of 14), or avian AAV (6 of 14) and thus lacked heparin binding capability. Conversely, only 2 of 18 liver clones had the AAV-2 HBD region, while 6 of 18 carried the corresponding sequence from AAV-8, 8 of 18 had that from AAV-9, and 2 of 18 carried the sequence from avian AAV. In summary, ∼43% of lung clones contained an HBD, as opposed to only ∼11% from the liver tissue. Moreover, it was noteworthy that we unambiguously recovered avian AAV sequences in 8 of the 32 clones, although this serotype was lost in our in vitro selection protocols. The opposite was observed for serotypes 4 and 5, which were present in clones after in vitro selection but missing in the 32 (partial) sequences from lung and liver tissues. Finally, it was interesting that in all eight avian AAV sequence-containing clones, the 3′ ends (bases 2069 to 2208) were identical and were from AAV-2.
Curious results were also obtained with the 19 clones from the AAV-2 peptide library from liver tissue (Table 6). As in our prior findings with lung tissue, we recovered a specific sequence twice, NRGYGAE. We also noted a significant increase in glycine but also asparagine residues (G, 22.6% versus 6.3% predicted by the NNB design; N, 9.0% versus 4.2%), as well as enrichment with a G-V motif in half of the clones (9 of 19). The glycine residues accumulated particularly in positions 1 and 3 to 5 of the peptides, whereas positions 1 were enriched with N. This pattern was reminiscent of our lead peptides recovered from lung tissue before and also similar to that of many clones previously isolated by others from various cell types (48, 50, 76), perhaps suggesting a general benefit from asparagine in this spot (see Discussion). Concurrent with the glycine accumulation in the center of the peptide, we found a marked increase in hydrophobic nonpolar residues in positions 3 and 4 to about 80 and 60%, respectively (normal would be 45%).
TABLE 6.
Peptide sequences from in vivo biopanning in murine liver tissuea
| Peptide (frequency) | Charge pattern | Hydrophobicity pattern |
|---|---|---|
| NRGYGAE (2) | x+yxyy− | −−++++− |
| NLTSGVY | xyxxyyx | −+−−++− |
| NSLGSTV | xxyyxxy | −−++−−+ |
| NEARLQG | x−y+yxy | −−+−+−+ |
| NQVGKGT | xxyy+yx | −−++−+− |
| GPGSYRE | yyyxx+− | +++−−−− |
| GSPVGQR | yxyyyx+ | +−+++−− |
| GRAPSST | y+yyxxx | +−++−−− |
| GSDGNMS | yx−yxyx | +−−+−+− |
| GVGVGGI | yyyyyyy | +++++++ |
| RGGDGSW | +yy−yxy | −++−+−+ |
| DRMVGEG | −+yyy−y | −−+++−+ |
| STVGERV | xxyy−+y | −−++−−+ |
| MDGNHRM | y−yx++y | +−+−−−+ |
| LRIGVAN | y+yyyyx | +−++++− |
| KNGVNAG | +xyyxyy | −−++−++ |
| FNSAMSL | yxxyyxy | +−−++−+ |
| HIRSQMA | +y+xxyy | −+−−−++ |
Shown are 19 peptide sequences from AAV genomes isolated from murine liver tissue (from one animal, injected with 2 × 1012 particles of the AAV-2-based peptide display library). The peptide NRGYGAE was isolated twice, and all others were isolated once. Residues or short sequence motifs that were enriched are underlined. See Table 5 for the definition of symbols indicating amino acid classifications.
In analogy to our lung work before, we finally tried to further amplify the potentially enriched and replicated AAV genomes in the liver extract from a second mouse. Yet, this time we consistently failed to detect AAV signals in tissue from the reinfected mouse, regardless of identical attempts to purify anti-AAV and -adenovirus IgGs from the tissue extract. Our most likely explanation is that unlike that in the lung, the adenovirus used for coinfection had not been able to exert the full helper function for the AAV particles in the liver. Interestingly, the mouse had been visibly sick at the time of organ harvesting, and the liver was pale, both observations indicative of successful adenovirus infection. Thus, the inability to reamplify AAVs from the liver but not the lung implies a tissue-specific block in the life cycle of AAV or adenovirus (or both), preventing the production of infectious AAV progeny. Alternatively or in addition, it is possible that the infection conditions for the first mouse (doses, ratios of adenovirus and AAV, and time points) require further fine-tuning to increase the amount of amplified AAV for reinoculation.
DISCUSSION
Our long-standing goal, to optimize the AAV vector system for therapeutic liver transduction, has now led us to devise novel methods for directed AAV diversification from a multispecies set of wild-type viruses. The potent molecular evolution technologies reported here, DNA family shuffling and in vivo biopanning, should significantly complement the current strategies. Indeed, DNA family shuffling provides numerous benefits, as is evident from a comparison of three key parameters: diversity, complexity, and versatility. First and foremost, the diversity in our library, before and after selection, was markedly enhanced over that described in all prior reports. Individual capsids from our unselected library had as little as ∼50% homology to the AAV-2 prototype, and our lead candidate (AAV-DJ) differed from its closest natural relative, AAV-2, by 60 residues (of 737 [∼8.1%]). This high degree of diversity resulted from the recombination of largely different parental viruses and was further increased by sporadic point mutations. Also, data for wild-type AAVs (11, 18, 44) imply that individual clones can recombine during library growth on cells, further enhancing diversity and explaining the similarity of our pools A and B. In striking contrast, the highest level of diversity achieved before was a difference of only seven residues, in the case of peptide display libraries (50, 58, 76). Moreover, since all insertions occured at the same location, diversification by continuous recombination was unlikely. A maximum of six mutations prior to selection was the upper limit for the second library type, created by error-prone PCR amplification of the AAV-2 capsid gene (46, 59). Postselection, respective lead candidates were ∼99.9% identical to wild-type AAV-2, differing by only one or two residues. Hence, the sum of all previous attempts at directed AAV evolution, including four libraries and over 370 mutants from multiple groups (also counting site-directed mutagenesis strategies [42]), did not change more than 1% of capsid residues, at best.
DNA family shuffling also appears to be superior to all nonlibrary strategies for the creation of AAV hybrids, including transcapsidation, marker rescue, and rational domain swapping (8, 30, 62, 64). From therapeutic and technical standpoints, a drawback of the first two approaches is the poor reproducibility, as the genetic templates are not recovered. This problem is solved by the domain-cloning strategy, yet this method can yield hybrids not only identical in phenotype to the parental viruses but also sometimes being noninfectious altogether (64, 85). Rational domain swapping also remains hampered by our limited knowledge of the structure-function relationships for most non-2 serotypes. The same issue continues to restrict the expansion of peptide display into alternative serotypes, with only a few reports of such expansion to date (4, 38). In this respect, a benefit of the AAV-2 HBD in AAV-DJ was that it allowed the straightforward adaptation of peptide display methodology. To our knowledge, we are in fact the first to utilize a synthetic AAV capsid for peptide display and also for library biopanning in vivo. Error-prone PCR is theoretically easier to adapt than peptide-based strategies, as it does not rely on sequence knowledge. Yet, the inherently low level of diversity will pertain as a drawback regardless of the parental virus. Thus, we believe that at this point, DNA family shuffling is the only available method able to yield highly variable libraries whose generation is neither serotype restricted nor limited by our knowledge (or lack thereof) of capsid structures and functions and which truly represent the viral diversity found in nature.
Complexity, i.e., the number of diverse capsids in the library, is a second crucial aspect of evolution methods. A good approximation is the number of bacterial colonies in the plasmid DNA library. Yet, equally important factors that are often ignored in the literature are the degree of interclonal diversity and technical aspects. For example, the simplicity of peptide insertions that require only direct cloning facilitates the creation of large bacterial libraries with up to 108 clones (50, 58, 76). This high level of efficacy was readily reproduced with our own peptide libraries and their complexities of ∼3 × 107 clones. PCR randomization is also undemanding, well established, and adaptable, thus easily yielding 106 to 107 capsid mutants when used for AAV-2 diversification (46, 59). Yet, such numbers may overestimate the true functional complexity, due to contamination with wild-type AAV-2 or phenocopies that falsely raise the library titer. Also, many resulting plasmids do not yield functional virions and are lost during selection. A good measure for vitality is particle yield per transfected cell during viral library production. Our high final titer of 8.2 × 1011 particles/ml, or 3.3 × 1012 total virions, is thus remarkable, as it translates into 3,300 full capsids/cell. These numbers match or exceed the best prior results, indicative of the high degree of vitality of our shuffled library. This vitality was expected, as the principle of DNA family shuffling is homology-based, in-frame recombination of functional sequences. Even so, we cannot technically rule out that our library contained noninfectious clones, similar to observations with domain swapping (64). Notably, our typical AAV-DJ vector yields (including those of all mutants) of >1013 particles/ml also exceeded those of earlier chimeras (e.g., reference 76), further supporting the high degrees of vitality and stability of the chimeric AAV sequences in our library. Accordingly, we are pleased with the complexity of our pilot shuffled plasmid library of ∼7 × 105 clones, especially as we had used extreme parameters, i.e., eight parental AAVs with as little as ∼50% homology. We can readily anticipate that future libraries based on fewer and/or more-related serotypes will have even higher levels of complexity. It will also always be feasible to increase the titer and complexity by upscaling library production.
Together, diversity and complexity determine a third parameter, versatility. DNA family shuffling also excels here and expands on existing technologies. We define versatility as the sum of viral properties, particularly within a single capsid, that can be molecularly evolved. Peptide display libraries are limited in this regard, as their main purpose is to improve a single parameter, tropism. Some inserts may diminish antibody recognition (33) or affect intracellular processing, but it remains a restriction that only receptor binding is subjected directly to selection. Also, as the capsid gene itself remains unaltered, there is no evolution pressure on the underlying AAV DNA per se. In contrast, error-prone capsid gene mutation via PCR can theoretically alter any residue. Yet, the random and inefficient nature of this method limits library diversity, complexity, and vitality and, thus, versatility. A main drawback is that the technique will typically yield only one or two viable mutations per capsid. Although single residue switches can alter the AAV phenotype (42, 84), such events are probably rare and hard to recap with such libraries. Considering the structure of AAV capsids, with 60 subunits and up to ∼735 residues each, it is statistically unlikely that all viral properties can be enhanced by random point mutation. In fact, the only two applications reported to date increased AAV-2 capsid resistance to neutralizing antibodies (46, 58). This result was expected, as prior work had already shown the role of specific residues in antibody recognition (42, 84). Yet, it is unclear how the same technique could be used to improve other properties, or even multiple features at once, especially ones relying on several dispersed residues or domains. The versatility of classical libraries is further restricted if they are derived from AAV-2 (as are almost all to date), as the remaining >99% identity suggests that all capsids will share some of the adverse features that hamper AAV-2 use in humans, including susceptibility to the prevalent immunity.
In conclusion, DNA family shuffling may be the most powerful and potent library-based AAV evolution method to date. With its hallmark of recombination of functional genes, it is an ideal tool to molecularly breed novel viruses merging multiple properties in a single capsid. This capacity is exemplified by our lead candidate, AAV-DJ, which is best characterized as a magnified AAV-2. It merges AAV-2 assets—high-level and broad-range in vitro efficiency, binding to the AAV-2 receptor, and relatively restricted in vivo biodistribution—with those of AAV-8 and AAV-9—superb liver performance, plus the ability to evade preexisting human immunity. By actually surpassing the in vitro efficacy of the best parental wild-type AAVs, AAV-DJ also demonstrates the great potential of DNA family shuffling to create de novo gain-of-function phenotypes. At this point, we are unaware of another synthetic capsid combining a similar extent of valuable assets in a single sequence or any other evolution method providing the same potential to concurrently create high degrees of diversity and versatility (while maintaining reasonable complexity and high-level vitality).
In addition, DNA family shuffling is a potent reverse-genetics tool to study AAV biology. We have provided examples of gain- and loss-of-function phenotypes that it can create and that give insights into diverse viral properties, such as antibody binding and protease cleavage. We also indirectly confirmed key residues constituting the HBD (previously identified via mutagenesis; R484-487-585-588 and K532 [37, 56]), based on conservation in all our selected clones and the critical role of the HBD in vitro. Many dispersed amino acids earlier identified as antigenic determinants in AAV-2 were also recognized in AAV-DJ by alignment with clones evolved without IVIG. Examples are AAV-2 R471, N705, and V708, which are critical for IVIG resistance (42), and R459, the mutation of which permits escape from anti-AAV-2 antibodies (59). Our alignments also confirmed many of the HVRs in the capsid gene (11, 16) and identified various new areas of diversity. We recently reported the first AAV-8 receptor (LamR) (1), plus an endosomal protease (cathepsin B) involved in intracellular capsid processing (2), and had proposed before that rapid nuclear uncoating is key to potent AAV-8 transduction (73). Future comparisons of shuffled and wild-type capsids will help to further unravel the sequences and mechanisms underlying such viral properties.
Equally crucial for an AAV evolution approach is particle selection and its stringency and clinical relevance, and both were maximized in our study. We particularly consider our use of pooled human antisera (IVIG) for selection an advance in stringency and clinical relevance over approaches described in prior reports, which relied solely on single antisera. These single antisera do not represent the assortment of anti-AAV antibodies in the human population, leaving the usefulness of the evolved particles unclear. The necessity to use pools is validated by a report by Huttner et al., who compared the activities of 65 human serum samples toward AAV-2 mutants (33). Depending on the peptide insertion site, they found substantial differences between individual sera in the ability to bind and/or to neutralize AAV-2 capsids, implying that monoselection is insufficient. As further evidence, AAV-2 variants with point mutations evolved with a single antiserum varied fourfold in their capacities for escape from seven other sera (59).
A second benefit from IVIG was that it forced the evolution of a single hybrid, AAV-DJ, that was not recovered under less stringent conditions. Our key conclusion is that library growth on cells already permissive for one or more parental AAVs is insufficient to select individual capsids. Instead, we obtained pools of related AAVs with homology to AAV-2 and interspersed regions from other AAVs. Notably, we also isolated similarly related derivatives of serotypes 2, 4, 5, 8, and 9 but no single lead candidates from human embryonic kidney cells (293 cells) and mouse fibroblast (NIH 3T3) or liver (Hep1A) cells (data not shown). Most likely key to our success, and likely imperative for future attempts, was to combine stringent positive pressure (growth on cells) and negative pressure (IVIG). Under these conditions, IVIG exerted dual functions. It forced the elimination of immunogenic residues and favored the infectious capsids best able to rapidly escape from neutralization. This conclusion is supported by our result that IVIG was active in culture for >12 h, maintaining pressure on the replicating AAVs. It also explains the superiority of AAV-DJ in vitro, because we inadvertently selected for a capsid that was extremely robust at transducing cells. Some prior evolution attempts also yielded capsids more infectious than that of AAV-2, but usually the increase was lower than that with AAV-DJ (about twofold) (e.g., reference 59). These capsids were also not tested thoroughly and mostly not in vivo. As of now, we do not fully understand all the properties of the AAV-DJ capsid, but most likely, they are determined by synergistic or additive contributions from the parental strains, such as the juxtaposition of multiple peptide motifs potentially involved in cell binding, viral uptake, and subsequent steps (Fig. 12; also see below). Indeed, less-chimeric capsids (obtained without IVIG pressure, e.g., those in pools A and B in Fig. 3) yielded lower levels of gene expression in mice, more like those yielded by AAV-2 (data not shown).
Another key observation was that library growth on cultured cells invariably led to enrichment with the AAV-2 HBD. This bias is likely inherent in any in vitro selection strategy (provided that the library contains AAV-2), considering the role of the HBD in culture. Whether or not its presence is ultimately useful depends on the application. With the liver as our main target, the presence of the HBD was highly rewarding due to the multifaceted role of the HBD in vector biodistribution. Restricted dissemination after intravenous delivery and minimal brain transduction (unlike the transduction patterns occuring with AAV-8 and AAV-9) are significant benefits for human liver gene therapy. More work is needed to elucidate the underlying mechanism(s), but an HBD may affect capsid stability in the blood or the particles' ability to traverse the endothelial cell lining, two prerequisites to transducing remote organs. Notably, a homologous domain was recently associated with capsid-specific T-cell responses in nonhuman primates injected intramuscularly with AAV-2 vectors (74). Yet, it remains unclear whether and to what extent these monkey studies can explain findings in a clinical trial in which one patient injected intraportally with an AAV-2 vector developed cytolytic T cells against capsid-bearing hepatocytes. The authors of the study attributed the incident to two immunogenic peptides in the AAV-2 capsid (47). These are absent in AAV-DJ (Fig. 2D); instead, this capsid part is identical to AAV-8, which did not elicit a T-cell response in the primate study. Yet, recent findings suggest that non-2 serotypes may also be able to elicit T-cell responses, depending on AAV-specific CD8+ memory T cells (49). Clearly, more work and in vivo data are needed to truly understand the anti-AAV cellular immune response and the potential role of the HBD. Importantly, as the constituting residues are known, it is trivial to mutate lead capsids should heparin binding be undesired for an application. Yet, another asset will then also be lost, i.e., the usefulness of the HBD for heparin affinity purification, a scalable and thus superior method to CsCl gradient centrifugation.
Our preliminary data from in vivo biopanning in liver and lung tissues revealed a set of crucial principles which may be broadly applicable to other AAV evolution strategies. Firstly, we noted that the outcomes of in vitro and in vivo evolution can largely differ. For instance, we frequently recovered avian AAV DNA in vivo, despite its absence after in vitro selection, and vice versa for AAV-4 and AAV-5. This result also underscores the diversity and complexity of our initial library, which evidently contained functional capsids derived from all parents. Secondly, the dependencies on heparin binding differed in vitro and in vivo and in distinct tissues. Our pilot data suggest that heparin binding, which is critical for in vitro transduction, may be more important in lung tissue than in liver tissue. This possibility is in line with our results that at higher doses, the HBD-deficient AAV-DJ mutants, as well as AAV-8 and AAV-9, transduced liver tissue better than AAV-DJ. We also noted a much higher proportion of AAV-8 and AAV-9 sequences in liver tissue (14 of 18 sequences; 78%) than in lung tissue (2 of 14 sequences; 14%), corroborating the fact that AAV-8 and AAV-9 are the best-known AAVs for the liver. It is also in line with the high percentage of AAV-8 and AAV-9 sequences in AAV-DJ (Table 2). On the other hand, the inhibition of heparin binding hampered liver transduction with the less efficient AAV-2 (Fig. 7C), and adding an HBD to AAV-8 was also disruptive. Thus, the addition or deletion of an HBD must be considered carefully for each target (cell or tissue type), application (ex or in vivo), and capsid.
A third set of key principles was evident from comparisons to data in the literature. A salient finding was that our two major peptides from lung tissue had previously been selected from an AAV-2 peptide library on arterial or venous endothelial cells (50, 76). Similar sequences were also isolated from acute myeloid leukemia cells in a third study (48). We envision multiple explanations for this phenomenon. One is that identical peptides confer distinct effects, e.g., on tropisms, upon exposure on various AAV backbones. This factor may also explain why in the AAV-DJ context, the NSSRDLG peptide did not alter liver transduction while it detargeted AAV-2 (50). Albeit unlikely, we also cannot technically rule out that our lung sample was marginally contaminated with heart tissue and that the lung tropisms were secondary. Yet, a more probable, captivating possibility is that the NSSRDLG peptide and related motifs bind to a common receptor present on all cells from which these sequences have been isolated to date: alveolar, coronary arterial, venous endothelial, and acute myeloid leukemia cells. Indeed, we noted several conspicuous similarities between our clones and many prior lead peptides, supporting the idea of affinity for the same or related receptors. These similarities include the observation that many peptides were generally enriched with the amino acids A, D, G, L, N, R, S, and V. Likewise, an N in the first position and an R in the fourth position were noted suspiciously frequently in peptides from lung tissue (also in our lead peptide from liver tissue, NRGYGAE) as well as earlier. A third, indirect piece of evidence for binding to widely present receptors is that some prior lead peptides mediated good transduction levels across a panel of cell lines (76).
Curious in this regard is the recent identification of a new integrin αVβ1 binding motif in AAV-2 and a few other serotypes (5). Intriguingly, this motif (NGR) is part of a larger sequence with striking similarity to the NSSRDLG peptide, i.e., NGRDSL. The latter is fully conserved in AAV-2 and AAV-DJ (and serotypes 3 and 10) and located on the capsid exterior near the HBD, in line with its putative role in integrin binding. Moreover, we identified two further intriguing motifs within AAV-DJ VP, another one similar to NSSRDLG and one resembling the second lead peptide identified in the present study and that by Müller et al., NDVRAVS (50). Both motifs are also displayed on the capsid (Fig. 12B); in fact, all four (including the HBD) are located very close to one another and near the threefold symmetry axis (Fig. 12A). Together with our findings described above, these results make it tempting to speculate that these three additional motifs contribute to the binding of AAV receptors, e.g., integrins. An appealing ensuing idea is that AAV-DJ's superior efficacy on multiple cell types is due in part to the fact that AAV-DJ combines all four motifs in one sequence, unlike any wild-type AAV. The observation that the binding of AAV-DJ to cultured cells is in fact inferior to that of AAV-2 would suggest that the putative additional receptors act as entry, but not attachment, molecules. This idea is in line with the roles of different integrins, which cooperatively act as secondary entry receptors for many viruses, including αVβ1/αVβ5 for AAV-2 (5). The high level of efficacy of AAV-DJ may then indeed result from synergism from the juxtaposition of multiple parental properties: binding to potent primary attachment and secondary entry receptors and efficient intracellular processing. Further investigations into the true nature of the receptors recognized by selected peptides will certainly be very exciting and important.
In the future, the optimization of in vivo evolution schemes should become a top priority, starting with the AAV helper virus, for multiple reasons. Obvious factors are toxicity and safety issues, but equally critical is the influence of the helper virus on the outcome of selection. AAV libraries can amplify only in coinfected cells, permitting the helper virus to dictate and restrict the tropism of the evolving AAV particles. This property is shown by our findings with lung tissue, in which our lead candidates mimicked adenoviral biodistribution. An intriguing possible solution may be to clone and express the adenoviral helper genes (E1, E2A, E4orf6, and VA genes) from a second AAV library in parallel. Other promising steps toward routine in vivo evolution would be the use of immunodeficient animals or mice with humanized tissues. It remains to be tested whether this approach will help to overcome another hurdle seen in our pilot studies, the block in the AAV or adenoviral life cycle in the liver (and maybe elsewhere). For adenovirus, this problem may also be solved by the expression of the relevant genes from AAV.
Optimized selection schemes will then be useful to resolve many key aspects of AAV vector evolution. For example, parallel in vivo screening of libraries based on different capsids will clarify the role of the hosting virus and the locations and flanking sequences for the displayed peptides. The use of AAV-2-based libraries may be challenging due to the relatively low level of in vivo fitness of the underlying virus, yet our data for liver tissue already show the potential of this approach. Better animal models will also help us to unravel the function of our other lead peptide, MVNNFEW. Thus far, our data imply tropism for alveolar macrophages, albeit this requires validation due to the background staining of alveolar macrophages from even naïve mice. Enrichment with this peptide after IgG depletion suggests an alternative function, i.e., to reduce the recognition of the hosting capsid by neutralizing antibodies. This phenomenon was noted anecdotally before (33) and may be due to sterical or conformational effects, masking immunogenic residues or domains in the capsid. In our case, it is possible that during IgG depletion of our primary lung extract, we also eliminated all capsids still attached to the antibodies, perhaps excluding the MVNNFEW virus which could escape from this procedure. Alternatively, multiple capsids may have evaded depletion but only the MVNNFEW clone could also reamplify in the secondary lung. To distinguish these conceivable possibilites, we will now rescreen our libraries in immunodeficient animals and will purify selected target cells prior to AAV rescue and reamplification.
Finally, essential is the potential clinical usefulness of the AAV clones evolved in this work. For our in vivo-selected capsids, our proof-of-concept data may be too limited to suggest specific uses, as more factors remain to be studied, including biodistribution, doses, time points, delivery routes, and comparisons to more wild-type AAVs. Further in vitro work analogous to and beyond our experiments presented in Fig. 6 will also aim to unravel their mechanisms of transduction. On the other hand, we can envision multiple ex or in vivo gene therapy applications for AAV-DJ vectors. Their high-level efficacy at low doses, from a clinically feasible administration route, implies a use for systemic therapies of hepatodeficiencies such as the hemophilias. The slower kinetics at extreme doses are irrelevant because of the high-level efficiency, with a minimal dose of 5 × 1010 particles per mouse, or 2.5 × 1012 particles/kg of body weight, already sufficing for the expression of 400% of normal hFIX levels. This dose equals the highest particle load in the clinical trial, although the inferior AAV-2 capsid produced only 12% of normal hFIX levels (still in the therapeutic range) (47). Despite the blunted response, the hFIX expression level of ∼300 μg/ml from the highest AAV-DJ dose exceeds most reported data for mice. Also, the extreme dose at which AAV-DJ transduction became saturated would correspond to a level of >2 × 1016 particles in humans, which is technically impossible to produce or administer. Finally, combining potent AAV capsids with efficient self-complementary genomes will allow effective particle doses to be lowered further by at least an order of magnitude (20, 26, 28, 54). Hence, we doubt that dosing will ultimately limit therapies in humans and rather assume that issues such as biodistribution will prevail. This conclusion already justifies the further preclinical evaluation of AAV-DJ, despite the strong competition from wild types 8 and 9. It was no surprise that the in vivo efficacy of AAV-DJ did not exceed their in vivo efficacy, considering that they have evolved to outperform most AAVs in mammalian tissues (19, 52). It is still essential to have another similarly efficient AAV at our disposal, due to the escalating evidence that findings with mice are not inevitably predictive for higher species (17, 32, 54, 75). Consequently, the next challenge will be to translate AAV-DJ for use in appropriate large animals and verify our results in a context more relevant to humans.
Of note, AAV-DJ may prove very useful for an emerging new clinical application, liver RNAi to treat hepatocellular carcinoma or infection with hepatitis viruses (25, 28). We recently evaluated AAV-8 vectors for this purpose and observed lethalities in mice, caused by rapid high-level expression of short hairpin RNAs (shRNAs) (28). Due to its more gradual onset of gene expression at high doses, the use of AAV-DJ for shRNA expression may minimize the risk of overloading the cellular RNAi machinery (28) while yielding sufficient shRNAs to achieve therapeutic effects. The reduced vector dissemination would further limit toxicity and enhance safety, especially in combination with our latest liver-specific shRNA expression cassettes (Giering et al., submitted for publication).
The ultimate task remains to engineer AAV capsids to merge specificity with efficiency and safety and tailor them to patient or disease profiles. We anticipate that DNA family shuffling will play a major role in the future, as it is applicable to all natural AAVs and can readily be adapted for directed vector evolution toward many applications. With the rapidly progressing discovery of multispecies AAVs, expanding our repertoire of capsid genes, it should soon be embraced as the most potent technology for AAV diversification. Moreover, our promising pilot efforts toward in vivo biopanning may help to pave the way for future evolution attempts under physiologically and clinically pertinent conditions. The key technologies and principles established here raise optimism that our work will bring us several steps closer to all these critical goals.
Supplementary Material
Acknowledgments
We thank Brian Garrison for expert help with the cultivation of the human hepatocytes and Kusum Pandey for excellent technical assistance. We are very grateful to James Wilson for providing AAV-8 and AAV-9 helper plasmids and express our gratitude to Jay Chiorini and Rob Kotin for their kind gifts of plasmids harboring the avian or bovine AAV capsid genes.
This work was supported by NIH grant HL64274 and the California Institute for Regenerative Medicine.
Footnotes
Published ahead of print on 9 April 2008.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Akache, B., D. Grimm, K. Pandey, S. R. Yant, H. Xu, and M. A. Kay. 2006. The 37/67-kilodalton laminin receptor is a receptor for adeno-associated virus serotypes 8, 2, 3, and 9. J. Virol. 809831-9836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akache, B., D. Grimm, X. Shen, S. Fuess, S. R. Yant, D. S. Glazer, J. Park, and M. A. Kay. 2007. A two-hybrid screen identifies cathepsins B and L as uncoating factors for adeno-associated virus 2 and 8. Mol. Ther. 15330-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arbetman, A. E., M. Lochrie, S. Zhou, J. Wellman, C. Scallan, M. M. Doroudchi, B. Randlev, S. Patarroyo-White, T. Liu, P. Smith, H. Lehmkuhl, L. A. Hobbs, G. F. Pierce, and P. Colosi. 2005. Novel caprine adeno-associated virus (AAV) capsid (AAV-Go.1) is closely related to the primate AAV-5 and has unique tropism and neutralization properties. J. Virol. 7915238-15245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arnold, G. S., A. K. Sasser, M. D. Stachler, and J. S. Bartlett. 2006. Metabolic biotinylation provides a unique platform for the purification and targeting of multiple AAV vector serotypes. Mol. Ther. 1497-106. [DOI] [PubMed] [Google Scholar]
- 5.Asokan, A., J. B. Hamra, L. Govindasamy, M. Agbandje-McKenna, and R. J. Samulski. 2006. Adeno-associated virus type 2 contains an integrin α5β1 binding domain essential for viral cell entry. J. Virol. 808961-8969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartlett, J. S., J. Kleinschmidt, R. C. Boucher, and R. J. Samulski. 1999. Targeted adeno-associated virus vector transduction of nonpermissive cells mediated by a bispecific F(ab'γ)2 antibody. Nat. Biotechnol. 17181-186. [DOI] [PubMed] [Google Scholar]
- 7.Bleker, S., F. Sonntag, and J. A. Kleinschmidt. 2005. Mutational analysis of narrow pores at the fivefold symmetry axes of adeno-associated virus type 2 capsids reveals a dual role in genome packaging and activation of phospholipase A2 activity. J. Virol. 792528-2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bowles, D. E., J. E. Rabinowitz, and R. J. Samulski. 2003. Marker rescue of adeno-associated virus (AAV) capsid mutants: a novel approach for chimeric AAV production. J. Virol. 77423-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buning, H., M. U. Ried, L. Perabo, F. M. Gerner, N. A. Huttner, J. Enssle, and M. Hallek. 2003. Receptor targeting of adeno-associated virus vectors. Gene Ther. 101142-1151. [DOI] [PubMed] [Google Scholar]
- 10.Chang, C. C., T. T. Chen, B. W. Cox, G. N. Dawes, W. P. Stemmer, J. Punnonen, and P. A. Patten. 1999. Evolution of a cytokine using DNA family shuffling. Nat. Biotechnol. 17793-797. [DOI] [PubMed] [Google Scholar]
- 11.Chen, C. L., R. L. Jensen, B. C. Schnepp, M. J. Connell, R. Shell, T. J. Sferra, J. S. Bartlett, K. R. Clark, and P. R. Johnson. 2005. Molecular characterization of adeno-associated viruses infecting children. J. Virol. 7914781-14792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi, V. W., D. M. McCarty, and R. J. Samulski. 2005. AAV hybrid serotypes: improved vectors for gene delivery. Curr. Gene Ther. 5299-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christians, F. C., L. Scapozza, A. Crameri, G. Folkers, and W. P. Stemmer. 1999. Directed evolution of thymidine kinase for AZT phosphorylation using DNA family shuffling. Nat. Biotechnol. 17259-264. [DOI] [PubMed] [Google Scholar]
- 14.Crameri, A., S. A. Raillard, E. Bermudez, and W. P. Stemmer. 1998. DNA shuffling of a family of genes from diverse species accelerates directed evolution. Nature 391288-291. [DOI] [PubMed] [Google Scholar]
- 15.Crameri, A., E. A. Whitehorn, E. Tate, and W. P. Stemmer. 1996. Improved green fluorescent protein by molecular evolution using DNA shuffling. Nat. Biotechnol. 14315-319. [DOI] [PubMed] [Google Scholar]
- 16.Gao, G., M. R. Alvira, S. Somanathan, Y. Lu, L. H. Vandenberghe, J. J. Rux, R. Calcedo, J. Sanmiguel, Z. Abbas, and J. M. Wilson. 2003. Adeno-associated viruses undergo substantial evolution in primates during natural infections. Proc. Natl. Acad. Sci. USA 1006081-6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao, G., Y. Lu, R. Calcedo, R. L. Grant, P. Bell, L. Wang, J. Figueredo, M. Lock, and J. M. Wilson. 2006. Biology of AAV serotype vectors in liver-directed gene transfer to nonhuman primates. Mol. Ther. 1377-87. [DOI] [PubMed] [Google Scholar]
- 18.Gao, G., L. H. Vandenberghe, M. R. Alvira, Y. Lu, R. Calcedo, X. Zhou, and J. M. Wilson. 2004. Clades of adeno-associated viruses are widely disseminated in human tissues. J. Virol. 786381-6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao, G. P., M. R. Alvira, L. Wang, R. Calcedo, J. Johnston, and J. M. Wilson. 2002. Novel adeno-associated viruses from rhesus monkeys as vectors for human gene therapy. Proc. Natl. Acad. Sci. USA 9911854-11859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao, G. P., Y. Lu, X. Sun, J. Johnston, R. Calcedo, R. Grant, and J. M. Wilson. 2006. High-level transgene expression in nonhuman primate liver with novel adeno-associated virus serotypes containing self-complementary genomes. J. Virol. 806192-6194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Girod, A., M. Ried, C. Wobus, H. Lahm, K. Leike, J. Kleinschmidt, G. Deleage, and M. Hallek. 1999. Genetic capsid modifications allow efficient re-targeting of adeno-associated virus type 2. Nat. Med. 51052-1056. [DOI] [PubMed] [Google Scholar]
- 22.Grifman, M., M. Trepel, P. Speece, L. B. Gilbert, W. Arap, R. Pasqualini, and M. D. Weitzman. 2001. Incorporation of tumor-targeting peptides into recombinant adeno-associated virus capsids. Mol. Ther. 3964-975. [DOI] [PubMed] [Google Scholar]
- 23.Grimm, D. 2002. Production methods for gene transfer vectors based on adeno-associated virus serotypes. Methods 28146-157. [DOI] [PubMed] [Google Scholar]
- 24.Grimm, D., and M. A. Kay. 2003. From virus evolution to vector revolution: use of naturally occurring serotypes of adeno-associated virus (AAV) as novel vectors for human gene therapy. Curr. Gene Ther. 3281-304. [DOI] [PubMed] [Google Scholar]
- 25.Grimm, D., and M. A. Kay. 2006. Therapeutic short hairpin RNA expression in the liver: viral targets and vectors. Gene Ther. 13563-575. [DOI] [PubMed] [Google Scholar]
- 26.Grimm, D., K. Pandey, and M. A. Kay. 2005. Adeno-associated virus vectors for short hairpin RNA expression. Methods Enzymol. 392381-405. [DOI] [PubMed] [Google Scholar]
- 27.Grimm, D., K. Pandey, H. Nakai, T. A. Storm, and M. A. Kay. 2006. Liver transduction with recombinant adeno-associated virus is primarily restricted by capsid serotype not vector genotype. J. Virol. 80426-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grimm, D., K. L. Streetz, C. L. Jopling, T. A. Storm, K. Pandey, C. R. Davis, P. Marion, F. Salazar, and M. A. Kay. 2006. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature 441537-541. [DOI] [PubMed] [Google Scholar]
- 29.Grimm, D., S. Zhou, H. Nakai, C. E. Thomas, T. A. Storm, S. Fuess, T. Matsushita, J. Allen, R. Surosky, M. Lochrie, L. Meuse, A. McClelland, P. Colosi, and M. A. Kay. 2003. Preclinical in vivo evaluation of pseudotyped adeno-associated virus vectors for liver gene therapy. Blood 1022412-2419. [DOI] [PubMed] [Google Scholar]
- 30.Hauck, B., L. Chen, and W. Xiao. 2003. Generation and characterization of chimeric recombinant AAV vectors. Mol. Ther. 7419-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hauck, B., and W. Xiao. 2003. Characterization of tissue tropism determinants of adeno-associated virus type 1. J. Virol. 772768-2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herzog, R. W. 2007. Immune responses to AAV capsid: are mice not humans after all? Mol. Ther. 15649-650. [DOI] [PubMed] [Google Scholar]
- 33.Huttner, N. A., A. Girod, L. Perabo, D. Edbauer, J. A. Kleinschmidt, H. Buning, and M. Hallek. 2003. Genetic modifications of the adeno-associated virus type 2 capsid reduce the affinity and the neutralizing effects of human serum antibodies. Gene Ther. 102139-2147. [DOI] [PubMed] [Google Scholar]
- 34.Inagaki, K., S. Fuess, T. A. Storm, G. A. Gibson, C. F. McTiernan, M. A. Kay, and H. Nakai. 2006. Robust systemic transduction with AAV9 vectors in mice: efficient global cardiac gene transfer superior to that of AAV8. Mol. Ther. 1445-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jang, J. H., K. I. Lim, and D. V. Schaffer. 2007. Library selection and directed evolution approaches to engineering targeted viral vectors. Biotechnol. Bioeng. 98515-524. [DOI] [PubMed] [Google Scholar]
- 36.Kay, M. A., C. S. Manno, M. V. Ragni, P. J. Larson, L. B. Couto, A. McClelland, B. Glader, A. J. Chew, S. J. Tai, R. W. Herzog, V. Arruda, F. Johnson, C. Scallan, E. Skarsgard, A. W. Flake, and K. A. High. 2000. Evidence for gene transfer and expression of factor IX in haemophilia B patients treated with an AAV vector. Nat. Genet. 24257-261. [DOI] [PubMed] [Google Scholar]
- 37.Kern, A., K. Schmidt, C. Leder, O. J. Muller, C. E. Wobus, K. Bettinger, C. W. Von der Lieth, J. A. King, and J. A. Kleinschmidt. 2003. Identification of a heparin-binding motif on adeno-associated virus type 2 capsids. J. Virol. 7711072-11081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koerber, J. T., J. H. Jang, J. H. Yu, R. S. Kane, and D. V. Schaffer. 2007. Engineering adeno-associated virus for one-step purification via immobilized metal affinity chromatography. Hum. Gene Ther. 18367-378. [DOI] [PubMed] [Google Scholar]
- 39.Kolkman, J. A., and W. P. Stemmer. 2001. Directed evolution of proteins by exon shuffling. Nat. Biotechnol. 19423-428. [DOI] [PubMed] [Google Scholar]
- 40.Kronenberg, S., B. Bottcher, C. W. von der Lieth, S. Bleker, and J. A. Kleinschmidt. 2005. A conformational change in the adeno-associated virus type 2 capsid leads to the exposure of hidden VP1 N termini. J. Virol. 795296-5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kwon, I., and D. V. Schaffer. 1 September 2007, posting date. Designer gene delivery vectors: molecular engineering and evolution of adeno-associated viral vectors for enhanced gene transfer. Pharm. Res. doi: 10.1007/s11095-007-9431-0. [DOI] [PMC free article] [PubMed]
- 42.Lochrie, M. A., G. P. Tatsuno, B. Christie, J. W. McDonnell, S. Zhou, R. Surosky, G. F. Pierce, and P. Colosi. 2006. Mutations on the external surfaces of adeno-associated virus type 2 capsids that affect transduction and neutralization. J. Virol. 80821-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Loiler, S. A., T. J. Conlon, S. Song, Q. Tang, K. H. Warrington, A. Agarwal, M. Kapturczak, C. Li, C. Ricordi, M. A. Atkinson, N. Muzyczka, and T. R. Flotte. 2003. Targeting recombinant adeno-associated virus vectors to enhance gene transfer to pancreatic islets and liver. Gene Ther. 101551-1558. [DOI] [PubMed] [Google Scholar]
- 44.Lukashov, V. V., and J. Goudsmit. 2001. Evolutionary relationships among parvoviruses: virus-host coevolution among autonomous primate parvoviruses and links between adeno-associated and avian parvoviruses. J. Virol. 752729-2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lux, K., N. Goerlitz, S. Schlemminger, L. Perabo, D. Goldnau, J. Endell, K. Leike, D. M. Kofler, S. Finke, M. Hallek, and H. Buning. 2005. Green fluorescent protein-tagged adeno-associated virus particles allow the study of cytosolic and nuclear trafficking. J. Virol. 7911776-11787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maheshri, N., J. T. Koerber, B. K. Kaspar, and D. V. Schaffer. 2006. Directed evolution of adeno-associated virus yields enhanced gene delivery vectors. Nat. Biotechnol. 24198-204. [DOI] [PubMed] [Google Scholar]
- 47.Manno, C. S., G. F. Pierce, V. R. Arruda, B. Glader, M. Ragni, J. J. Rasko, M. C. Ozelo, K. Hoots, P. Blatt, B. Konkle, M. Dake, R. Kaye, M. Razavi, A. Zajko, J. Zehnder, P. K. Rustagi, H. Nakai, A. Chew, D. Leonard, J. F. Wright, R. R. Lessard, J. M. Sommer, M. Tigges, D. Sabatino, A. Luk, H. Jiang, F. Mingozzi, L. Couto, H. C. Ertl, K. A. High, and M. A. Kay. 2006. Successful transduction of liver in hemophilia by AAV-factor IX and limitations imposed by the host immune response. Nat. Med. 12342-347. [DOI] [PubMed] [Google Scholar]
- 48.Michelfelder, S., M. K. Lee, E. DeLima-Hahn, T. Wilmes, F. Kaul, O. Muller, J. A. Kleinschmidt, and M. Trepel. 2007. Vectors selected from adeno-associated viral display peptide libraries for leukemia cell-targeted cytotoxic gene therapy. Exp. Hematol. 351766-1776. [DOI] [PubMed] [Google Scholar]
- 49.Mingozzi, F., and K. A. High. 2007. Immune responses to AAV in clinical trials. Curr. Gene Ther. 7316-324. [DOI] [PubMed] [Google Scholar]
- 50.Müller, O. J., F. Kaul, M. D. Weitzman, R. Pasqualini, W. Arap, J. A. Kleinschmidt, and M. Trepel. 2003. Random peptide libraries displayed on adeno-associated virus to select for targeted gene therapy vectors. Nat. Biotechnol. 211040-1046. [DOI] [PubMed] [Google Scholar]
- 51.Muzyczka, N., and K. H. Warrington, Jr. 2005. Custom adeno-associated virus capsids: the next generation of recombinant vectors with novel tropism. Hum. Gene Ther. 16408-416. [DOI] [PubMed] [Google Scholar]
- 52.Nakai, H., S. Fuess, T. A. Storm, S. Muramatsu, Y. Nara, and M. A. Kay. 2005. Unrestricted hepatocyte transduction with adeno-associated virus serotype 8 vectors in mice. J. Virol. 79214-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nakai, H., C. E. Thomas, T. A. Storm, S. Fuess, S. Powell, J. F. Wright, and M. A. Kay. 2002. A limited number of transducible hepatocytes restricts a wide-range linear vector dose response in recombinant adeno-associated virus-mediated liver transduction. J. Virol. 7611343-11349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nathwani, A. C., J. T. Gray, C. Y. Ng, J. Zhou, Y. Spence, S. N. Waddington, E. G. Tuddenham, G. Kemball-Cook, J. McIntosh, M. Boon-Spijker, K. Mertens, and A. M. Davidoff. 2006. Self-complementary adeno-associated virus vectors containing a novel liver-specific human factor IX expression cassette enable highly efficient transduction of murine and nonhuman primate liver. Blood 1072653-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nicklin, S. A., H. Buening, K. L. Dishart, M. de Alwis, A. Girod, U. Hacker, A. J. Thrasher, R. R. Ali, M. Hallek, and A. H. Baker. 2001. Efficient and selective AAV2-mediated gene transfer directed to human vascular endothelial cells. Mol. Ther. 4174-181. [DOI] [PubMed] [Google Scholar]
- 56.Opie, S. R., K. H. Warrington, Jr., M. Agbandje-McKenna, S. Zolotukhin, and N. Muzyczka. 2003. Identification of amino acid residues in the capsid proteins of adeno-associated virus type 2 that contribute to heparan sulfate proteoglycan binding. J. Virol. 776995-7006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pekrun, K., R. Shibata, T. Igarashi, M. Reed, L. Sheppard, P. A. Patten, W. P. Stemmer, M. A. Martin, and N. W. Soong. 2002. Evolution of a human immunodeficiency virus type 1 variant with enhanced replication in pig-tailed macaque cells by DNA shuffling. J. Virol. 762924-2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Perabo, L., H. Buning, D. M. Kofler, M. U. Ried, A. Girod, C. M. Wendtner, J. Enssle, and M. Hallek. 2003. In vitro selection of viral vectors with modified tropism: the adeno-associated virus display. Mol. Ther. 8151-157. [DOI] [PubMed] [Google Scholar]
- 59.Perabo, L., J. Endell, S. King, K. Lux, D. Goldnau, M. Hallek, and H. Buning. 2006. Combinatorial engineering of a gene therapy vector: directed evolution of adeno-associated virus. J. Gene Med. 8155-162. [DOI] [PubMed] [Google Scholar]
- 60.Perabo, L., D. Goldnau, K. White, J. Endell, J. Boucas, S. Humme, L. M. Work, H. Janicki, M. Hallek, A. H. Baker, and H. Buning. 2006. Heparan sulfate proteoglycan binding properties of adeno-associated virus retargeting mutants and consequences for their in vivo tropism. J. Virol. 807265-7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Powell, S. K., M. A. Kaloss, A. Pinkstaff, R. McKee, I. Burimski, M. Pensiero, E. Otto, W. P. Stemmer, and N. W. Soong. 2000. Breeding of retroviruses by DNA shuffling for improved stability and processing yields. Nat. Biotechnol. 181279-1282. [DOI] [PubMed] [Google Scholar]
- 62.Rabinowitz, J. E., D. E. Bowles, S. M. Faust, J. G. Ledford, S. E. Cunningham, and R. J. Samulski. 2004. Cross-dressing the virion: the transcapsidation of adeno-associated virus serotypes functionally defines subgroups. J. Virol. 784421-4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Scallan, C. D., H. Jiang, T. Liu, S. Patarroyo-White, J. M. Sommer, S. Zhou, L. B. Couto, and G. F. Pierce. 2006. Human immunoglobulin inhibits liver transduction by AAV vectors at low AAV2 neutralizing titers in SCID mice. Blood 1071810-1817. [DOI] [PubMed] [Google Scholar]
- 64.Shen, X., T. Storm, and M. A. Kay. 2007. Characterization of the relationship of AAV capsid domain swapping to liver transduction efficiency. Mol. Ther. 151955-1962. [DOI] [PubMed] [Google Scholar]
- 65.Shi, W., and J. S. Bartlett. 2003. RGD inclusion in VP3 provides adeno-associated virus type 2 (AAV2)-based vectors with a heparan sulfate-independent cell entry mechanism. Mol. Ther. 7515-525. [DOI] [PubMed] [Google Scholar]
- 66.Shi, W., A. Hemminki, and J. S. Bartlett. 2006. Capsid modifications overcome low heterogeneous expression of heparan sulfate proteoglycan that limits AAV2-mediated gene transfer and therapeutic efficacy in human ovarian carcinoma. Gynecol. Oncol. 1031054-1062. [DOI] [PubMed] [Google Scholar]
- 67.Snyder, R. O., C. H. Miao, G. A. Patijn, S. K. Spratt, O. Danos, D. Nagy, A. M. Gown, B. Winther, L. Meuse, L. K. Cohen, A. R. Thompson, and M. A. Kay. 1997. Persistent and therapeutic concentrations of human factor IX in mice after hepatic gene transfer of recombinant AAV vectors. Nat. Genet. 16270-276. [DOI] [PubMed] [Google Scholar]
- 68.Sonntag, F., S. Bleker, B. Leuchs, R. Fischer, and J. A. Kleinschmidt. 2006. Adeno-associated virus type 2 capsids with externalized VP1/VP2 trafficking domains are generated prior to passage through the cytoplasm and are maintained until uncoating occurs in the nucleus. J. Virol. 8011040-11054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Soong, N. W., L. Nomura, K. Pekrun, M. Reed, L. Sheppard, G. Dawes, and W. P. Stemmer. 2000. Molecular breeding of viruses. Nat. Genet. 25436-439. [DOI] [PubMed] [Google Scholar]
- 70.Stemmer, W. P. 1994. DNA shuffling by random fragmentation and reassembly: in vitro recombination for molecular evolution. Proc. Natl. Acad. Sci. USA 9110747-10751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stemmer, W. P. 1994. Rapid evolution of a protein in vitro by DNA shuffling. Nature 370389-391. [DOI] [PubMed] [Google Scholar]
- 72.Summerford, C., and R. J. Samulski. 1998. Membrane-associated heparan sulfate proteoglycan is a receptor for adeno-associated virus type 2 virions. J. Virol. 721438-1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Thomas, C. E., T. A. Storm, Z. Huang, and M. A. Kay. 2004. Rapid uncoating of vector genomes is the key to efficient liver transduction with pseudotyped adeno-associated virus vectors. J. Virol. 783110-3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vandenberghe, L. H., L. Wang, S. Somanathan, Y. Zhi, J. Figueredo, R. Calcedo, J. Sanmiguel, R. A. Desai, C. S. Chen, J. Johnston, R. L. Grant, G. Gao, and J. M. Wilson. 2006. Heparin binding directs activation of T cells against adeno-associated virus serotype 2 capsid. Nat. Med. 12967-971. [DOI] [PubMed] [Google Scholar]
- 75.Wang, L., R. Calcedo, T. C. Nichols, D. A. Bellinger, A. Dillow, I. M. Verma, and J. M. Wilson. 2005. Sustained correction of disease in naive and AAV2-pretreated hemophilia B dogs: AAV2/8-mediated, liver-directed gene therapy. Blood 1053079-3086. [DOI] [PubMed] [Google Scholar]
- 76.Waterkamp, D. A., O. J. Muller, Y. Ying, M. Trepel, and J. A. Kleinschmidt. 2006. Isolation of targeted AAV2 vectors from novel virus display libraries. J. Gene Med. 81307-1319. [DOI] [PubMed] [Google Scholar]
- 77.White, S. J., S. A. Nicklin, H. Buning, M. J. Brosnan, K. Leike, E. D. Papadakis, M. Hallek, and A. H. Baker. 2004. Targeted gene delivery to vascular tissue in vivo by tropism-modified adeno-associated virus vectors. Circulation 109513-519. [DOI] [PubMed] [Google Scholar]
- 78.Wistuba, A., A. Kern, S. Weger, D. Grimm, and J. A. Kleinschmidt. 1997. Subcellular compartmentalization of adeno-associated virus type 2 assembly. J. Virol. 711341-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wistuba, A., S. Weger, A. Kern, and J. A. Kleinschmidt. 1995. Intermediates of adeno-associated virus type 2 assembly: identification of soluble complexes containing Rep and Cap proteins. J. Virol. 695311-5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wobus, C. E., B. Hugle-Dorr, A. Girod, G. Petersen, M. Hallek, and J. A. Kleinschmidt. 2000. Monoclonal antibodies against the adeno-associated virus type 2 (AAV-2) capsid: epitope mapping and identification of capsid domains involved in AAV-2-cell interaction and neutralization of AAV-2 infection. J. Virol. 749281-9293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Work, L. M., H. Buning, E. Hunt, S. A. Nicklin, L. Denby, N. Britton, K. Leike, M. Odenthal, U. Drebber, M. Hallek, and A. H. Baker. 2006. Vascular bed-targeted in vivo gene delivery using tropism-modified adeno-associated viruses. Mol. Ther. 13683-693. [DOI] [PubMed] [Google Scholar]
- 82.Work, L. M., S. A. Nicklin, N. J. Brain, K. L. Dishart, D. J. Von Seggern, M. Hallek, H. Buning, and A. H. Baker. 2004. Development of efficient viral vectors selective for vascular smooth muscle cells. Mol. Ther. 9198-208. [DOI] [PubMed] [Google Scholar]
- 83.Wu, P., W. Xiao, T. Conlon, J. Hughes, M. Agbandje-McKenna, T. Ferkol, T. Flotte, and N. Muzyczka. 2000. Mutational analysis of the adeno-associated virus type 2 (AAV2) capsid gene and construction of AAV2 vectors with altered tropism. J. Virol. 748635-8647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wu, Z., A. Asokan, J. C. Grieger, L. Govindasamy, M. Agbandje-McKenna, and R. J. Samulski. 2006. Single amino acid changes can influence titer, heparin binding, and tissue tropism in different adeno-associated virus serotypes. J. Virol. 8011393-11397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wu, Z., A. Asokan, and R. J. Samulski. 2006. Adeno-associated virus serotypes: vector toolkit for human gene therapy. Mol. Ther. 14316-327. [DOI] [PubMed] [Google Scholar]
- 86.Zadori, Z., J. Szelei, M. C. Lacoste, Y. Li, S. Gariepy, P. Raymond, M. Allaire, I. R. Nabi, and P. Tijssen. 2001. A viral phospholipase A2 is required for parvovirus infectivity. Dev. Cell 1291-302. [DOI] [PubMed] [Google Scholar]
- 87.Zhong, L., W. Li, Y. Li, W. Zhao, J. Wu, B. Li, N. Maina, D. Bischof, K. Qing, K. A. Weigel-Kelley, I. Zolotukhin, K. H. Warrington, Jr., X. Li, W. B. Slayton, M. C. Yoder, and A. Srivastava. 2006. Evaluation of primitive murine hematopoietic stem and progenitor cell transduction in vitro and in vivo by recombinant adeno-associated virus vector serotypes 1 through 5. Hum. Gene Ther. 17321-333. [DOI] [PubMed] [Google Scholar]
- 88.Zolotukhin, S., M. Potter, W. W. Hauswirth, J. Guy, and N. Muzyczka. 1996. A “humanized” green fluorescent protein cDNA adapted for high-level expression in mammalian cells. J. Virol. 704646-4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.