Abstract
The baculovirus Autographa californica nucleopolyhedrovirus encodes two proteins with RNA triphosphatase activity. Late expression factor LEF-4, which is an essential gene, is a component of the RNA polymerase and also encodes the RNA capping enzyme guanylyltransferase. PTP/BVP is also an RNA triphosphatase, but is not essential for viral replication, possibly because its activity is redundant to that of LEF-4. To elucidate the role of these proteins in mRNA cap formation, a mutant virus that lacked both RNA triphosphatase activities was constructed. Infection studies revealed that the double-mutant virus was viable and normal with respect to the production of budded virus. Pulse-labeling studies and immunoblot analyses showed that late gene expression in the double mutant was equivalent to that in the wild type, while polyhedrin expression was slightly reduced. Direct analysis of the mRNA cap structure indicated no alteration of cap processing in the double mutant. Together, these results reveal that baculoviruses replicate and express their late genes at normal levels in the absence of its two different types of RNA triphosphatases.
Baculoviruses are unique among eukaryotic DNA viruses in their use of both cellular and viral transcription machinery (19, 25). Viral infection is initiated by the host RNA polymerase II that recognizes and transcribes early promoters which are structurally similar to cellular promoters. Late and very late genes are transcribed by a virus-encoded RNA polymerase composed of four late expression factors (LEFs) called LEF-4, -8, and -9 and P47. These genes were first described for the type species Autographa californica multicapsid nucleopolyhedrovirus (AcMNPV), and homologs have been identified in all the baculovirus genomes that have been sequenced thus far (45). LEF-8 and LEF-9 are believed to form the catalytic pocket, because they contain motifs common to β and β′ RNA polymerases (38, 49, 59). LEF-4 is an RNA capping enzyme with both guanylyltransferase and RNA triphosphatase activities (14, 17, 28), and the function of P47 is unknown.
Late and very late baculovirus mRNAs have a typical methyl-7-guanosine cap structure at their 5′ ends (53). In eukaryotic cells, this cap structure is essential and is required for efficient pre-mRNA splicing, export, stability, and translation initiation (11). Three enzymatic activities are required for cap formation: hydrolysis of the 5′ triphosphate end of the nascent transcript to a diphosphate by RNA triphosphatase, capping of the diphosphate end with GMP by RNA cap guanylyltransferase, and N-7 methylation of guanine cap by RNA cap methyltransferase (11, 57).
Baculoviruses have evolved a novel strategy for capping their late mRNAs. Even though they replicate in the nuclei of infected cells, baculoviruses are unable to use the cellular capping machinery, because these enzymes find their substrates through interactions with the highly repetitive carboxyl-terminal domain of the cellular RNA polymerase II (6, 23, 43). Since the baculovirus RNA polymerase lacks this motif, cellular capping enzymes would not be able to recognize the viral mRNAs. Therefore, baculoviruses must encode their own capping enzymes. This is not uncommon in the virus world, but the incorporation of this activity into a DNA-dependent RNA polymerase is unique to baculoviruses.
The two enzymatic activities of LEF-4 constitute separable and autonomous domains (14, 28, 40, 41). The RNA triphosphatase activity is N-terminal with guanylyltransferase activity at the C terminus. The RNA triphosphatase domain belongs to a family of metal-dependent phosphohydrolases, which also includes RNA capping enzymes of fungi and protozoa (22). These enzymes are defined by two essential glutamate-containing motifs that bind divalent cation. The substitution of any one of these glutamates abolishes enzymatic activity.
As expected for an RNA polymerase subunit, LEF-4 is essential for viral replication, as revealed by studies of a temperature-sensitive virus with a mutation in lef-4 and by RNA silencing experiments (30, 47). LEF-4 is also necessary for transient expression of late and very late genes (48). Recently, we dissected the requirement of LEF-4 for productive infection and showed that the essential function of LEF-4 extends beyond its structural role in transcription (30). In a previous study, a mutant bacmid containing a disruption in the lef-4 gene was unable to produce progeny virus, but the cotransfection of a plasmid encoding wild-type (wt) LEF-4 restored virus production, while cotransfection of a mutant version of LEF-4 with a K255A substitution that abolishes guanylyltransferase activity did not rescue the phenotype (30). This result demonstrated that the capping function of LEF-4 is necessary for viral replication. The question of whether the triphosphatase activity of LEF-4 is also essential was not addressed in that study.
The issue of LEF-4 RNA triphosphatase activity is complicated by the fact that group I baculoviruses, which include AcMNPV, encode another protein with the same enzymatic activity as that of the triphosphatase. This gene was originally named ptp (protein tyrosine phosphatase), due to the presence of a diagnostic P-loop motif that binds the cleavable phosphate (55). Subsequent research, however, showed that its preferred substrate was RNA and that it released phosphates from the 5′ end of RNA (13, 58). It was renamed bvp (baculovirus phosphatase) to reflect its correct function. This name has been adopted outside the baculovirus field, although ptp is more commonly used by baculovirologists. Here we refer to it by the hybrid name ptp/bvp. Crystallographic studies of PTP/BVP showed that its structure was similar to metazoan RNA capping enzymes, thus confirming its specificity for RNA substrates (5). Furthermore, in vivo analyses showed that PTP/BVP could substitute for yeast RNA triphosphatase in the cap synthetic pathway in yeast cells (42). The biological function of PTP/BVP within the context of a baculovirus infection is less well understood. Analyses of mutant viruses showed that ptp/bvp was not required for viral replication in cell culture or in larvae (35). Interestingly, insects infected with ptp/bvp− mutant viruses showed altered behavior, suggesting that it was important in virus transmission (29).
The existence of two baculovirus RNA triphophatases raises questions about their roles in regard to viral mRNA capping. To determine whether the RNA triphosphatase activity of LEF-4 is absolutely essential or whether PTP/BVP performs a backup role, we constructed a mutant virus that lacked both RNA triphosphatase activities by homologous recombination using a bacmid construct in Escherichia coli. Surprisingly, our data show that the double-mutant virus was viable and normal with respect to the production of budded virus and the synthesis of late genes. Furthermore, an analysis of late mRNA cap structure showed no alteration in cap processing in the double-mutant virus.
MATERIALS AND METHODS
Viruses and cells.
Bacmid bMON14272 (Invitrogen Life Technologies) was maintained in DH10B cells as described previously (39). It contains a mini-attTn7 attachment site that allows the generation of recombinant bacmids by the transposition of donor plasmid pFastBac with helper plasmid pMON7124, which encodes the transposase (39). The Sf-9 insect cell line, a clonal isolate of IPLB-Sf21-AE cells derived from the fall armyworm (Spodoptera frugiperda) (60), was cultured in TNMFH medium supplemented with 10% fetal bovine serum, gentamicin (25 μg/ml), and amphotericin (1.25 μg/ml). In all experiments, virus inocula were allowed to adsorb for 1 h at 27°C and then were replaced with fresh medium. Viral DNA was transfected for 4 h before the medium was changed. Time zero was defined as the time when the virus was replaced with fresh medium.
Generation of lef-4 and ptp/bvp double-knockout AcMNPV bacmid.
A double-knockout mutant of AcMNPV was generated in the bacmid bMON14272 by homologous recombination in E. coli using a modification of the phage red recombinase system as described previously (3, 8, 37). A lef-4 single knockout was constructed first. A linear DNA fragment containing the chloramphenicol resistance gene flanked by 50-bp sequences homologous to the 5′ and 3′ ends of lef-4 was generated from pBR325 by PCR using the primer pair lef4Cm-F and lef4Cm-R (Table 1). This fragment was coelectroporated with bacmid bMON14272 DNA into E. coli BW25113-pKD46-competent cells. Potential clones were selected on LB plates containing chloramphenicol, and isolated colonies were screened by PCR with the primer pair A/B, followed by restriction enzyme digestion to confirm the replacement of the lef-4 gene with the chloramphenicol resistance gene. A similar strategy was used to create the double-knockout construct from the lef-4 single-knockout bacmid. To knock out ptp/bvp, primers ptpZeo-F and ptpZeo-R were used to generate a linear Zeocin resistance gene fragment with 50-bp sequences homologous to the 5′ and 3′ regions of ptp/bvp. This fragment was coelectroporated with the lef-4 single-knockout bacmid DNA into E. coli BW25113-pKD46-competent cells. Potential recombinants were selected on LB plates containing Zeocin, and isolated colonies were screened by restriction fragment length polymorphism (RFLP)-PCR with the primer pair C/D to confirm the replacement of ptp/bvp with the Zeocin resistance gene.
TABLE 1.
Primers used in this studya
| Primer name | Position no. | Sequence | Function |
|---|---|---|---|
| lef4Cm-F | 76727-76775 | GTACGACGAAAACGGCTTTCGCACTCGTATACCTATTCAGAGCGCTTGCTGTGTAGGCTGGAGCTGCTTCG | lef-4 KO |
| lef4Cm-R | 77981-77933 | CACGATTCGGTCGCGACGATGTTTCAACACGTTTATTGTCGTGTCCGTGCATATGAATATCCTCCTTA | lef-4 KO |
| ptpZEO-F | 509-558 | CCCGCGCGTTGGCAAACTATTTACAATGCGGCCAAGTTATAAAAGATTCTGTTGACAATTAATCATCGGC | ptp/bvp KO |
| ptpZEO-R | 1080-1031 | GGTTCGCTGGAAGAAGCGCAACAATTTGAAAATAGGATAAAGTATTTGTTCCCGGGAATTCTCAGTCCTGC | ptp/bvp KO |
| A | 373-394 | GTGTGCAAAACATGACATCAGC | Confirm ptp/bvp KO |
| B | 1265-1285 | ACATCCGGTGTTGTGCGTGAC | Confirm ptp/bvp KO |
| C | 76504-76525 | GACACGATGGACGCGAAAATGC | Confirm lef-4 KO |
| D | 78286-78306 | CGCCGCCTTCTCCTATTCCTC | Confirm lef-4 KO |
| ptpR-F | 287-303 | gctactagtGCAAAACATGACATCAG | bvp/ptp repair |
| ptpR-R | 988-1006 | TTATTAAATTAATAAATCTTGAAC | bvp/ptp repair |
| p6.9polyA-F | 86478-86494 | gctactagtGGAAAAACATACCAGCA | Add polyA signal to bvp/ptp |
| p6.9polyA-R | 86695-86711 | atccccgggCAACCAGACATTCCACAC | Add polyA signal to bvp/ptp |
| lef4R-F | 76238-76254 | atcgtcgacCGGCTTGCATCATACTG | lef-4 repair |
| lef4R-R | 77970-77987 | atccccgggATTTGGCACGATTCGGTC | lef-4 repair |
| vp39polyA- F | 75276-75295 | CGCTGCATTTAATCACATCC | Add polyA signal to lef-4 |
| vp39polyA-R | 75513-75534 | gctactagtAAATGGAGTTTGTTAAATTG | Add polyA signal to lef-4 |
| D1/D12 E192/194A-F | GGCCTAATACATCTCTGGcAATAGcATTCACACCTAGAGAC | Construct RTPase− D1/D12 |
Nucleotides added for cloning purposes are shown in lowercase type, restriction enzyme sites are underlined, mutagenic residues are indicated by lowercase bold type. Primers for lef-4 mutagenesis have been described previously (28). KO, knockout.
Bacmid bMON14272 is polyhedrin negative, because most of polyhedrin open reading frame (ORF) was deleted during construction (39). To facilitate the observation of viral infection, polyhedrin (polh) and green fluorescent protein (gfp) genes were inserted into the bacmid genomes. The transfer plasmid pFacT-GFP (a gift from David Theilmann) encoding POLH and GFP was transposed into the polh locus of bMON14272 to generate the bacmid construct vAc-PG, as described previously (7).
The pFacT-GFP plasmid was also used to generate different repair versions containing mutant or wt lef-4 and ptp constructs. The transfer plasmid pFacT-GFP-lef4 was constructed by cloning a 1,777-bp fragment, containing the wt lef-4 ORF with its native promoter, and a 233-bp fragment, containing the p6.9 poly(A) tail region; the fragments were separately amplified by PCR from the AcMNPV genome using primer pairs Lef-4R-F and Lef-4R-R or p6.9polyA-F and p6.9polyA-R, respectively. The lef-4 fragment was digested with SalI and SmaI and then subcloned into pBS (previously digested with SalI and Smal) to yield pBS-lef-4R. The p6.9 poly(A) fragment was digested with SpeI and Smal and then ligated with SpeI- and NotI-digested pBS-lef-4R to construct pBS-lef-4RA. Then, a SalI/SacII-digested fragment from pBS-lef-4RA was cloned into SalI/SmaI-digested pRS416 to give pRS416-lef-4RA. Finally, a XhoI/XbaI-digested fragment from pRS416-lef-4RA was subcloned into pFacT-GFP (previously digested with XhoI and XbaI) to generate pFacT-GFP-lef4.
To construct ptp and lef-4 double-repair viruses, a 744-bp fragment containing the ptp ORF under its native promoter and a 259-bp fragment containing the vp39 poly(A) sequence were amplified from the AcMNPV genome. The two fragments were digested with SpeI and then cloned into pBS (previously digested with SpeI) to give pBS-ptp. The resulting ptp-polyA cassette was then digested with SpeI and subcloned into the XbaI site of pFacT-GFP-lef4, thus generating the double-repair transfer plasmid pFacT-GFP-lef4ptp.
Transfer plasmids encoding mutant versions of lef-4, pFacT-GFP-lef4E2A, and pFacT-GFPE2Aptp were constructed by QuikChange mutagenesis using the primers lef-4/E181,183A (28) and following the recommendations of the manufacturer (Stratagene). The quadruple mutant was made using the double-mutant transfer vector as the template and the primers lef-4/E9,11A (28).
Recombinant bacmids were generated by transposition following the Bac-to-Bac protocol (Invitrogen Life Technologies). The universal M13 reverse primer and an appropriate gene-specific primer were used for PCR screening of potential colonies.
Time course analysis of BV production.
To assess the effect of ptp/bvp or lef-4 deletions on viral infection, a single-step growth curve was performed. Virus stock was prepared by transfecting Sf-9 cells (2.0 ×106 per 35-mm-diameter dish) with 2.0 μg of the appropriate bacmid DNA using Cellfectin (Invitrogen Life Technologies). The supernatant was harvested after 3 days, and titers were determined. Cells were infected in duplicate at a multiplicity of infection (MOI) of 5. One hour after infection, cells were washed twice with fresh medium and replenished with 2 ml of fresh medium supplemented with 10% fetal bovine serum. Virus supernatants were collected at selected time points. The titers of budded virus (BV) were determined by a 50% tissue culture infective dose (TCID50) end point dilution assay on Sf-9 cells (46).
Radiolabeling of proteins and Western blot analyses.
Pulse labeling for virus-infected cells was performed as described previously, with minor modifications (15, 27). Sf-9 cells (4 × 106 per 35-mm-diameter dish) were infected with BV at an MOI of 10 in TNMFH medium with 10% fetal bovine serum. At specific time points, the medium was removed and cells were washed twice with methionine-free Grace's medium and then labeled for 4 h in methionine-free Grace's medium containing 100 μCi of Translabel (ICN Biomedicals, Inc., Irvine, CA; a mixture of about 80% [35S]methionine and 20% [35S]cysteine; specific activity, 1,200 Ci/mmol) per milliliter. Cells were harvested after labeling, and intracellular protein extracts were prepared by treating the cells for 20 min on ice with extraction buffer (50 mM Tris-HCl, pH 8.0-100 mM NaCI-1% Nonidet P-40-1% Empigen BB (Albright & Wilson, Whitehaven, United Kingdom]) and then microcentrifugation for 10 min. The soluble fractions were analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis on 10% gels. Then the gel then was fixed, dried, and exposed to film.
For Western blot analysis, proteins were electrophoretically transferred to nitrocellulose sheets using a semidry apparatus according to the recommendations of the manufacturer (American Bionetics, Hayward, CA). Blots were probed with one of the following four primary antibodies: (i) LEF-4 rabbit antiserum at a 1:1,000 dilution (30), (ii) LEF-3 mouse monoclonal antibody at a 1:1,000 dilution (21), (iii) GP64 mouse monoclonal AcV1 at a 1:250 dilution (61), or (iv) PP31 rabbit antiserum at a 1:500 dilution (16). After incubation with a 1:2,500 dilution of peroxidase-conjugated secondary antibody, signals were detected by enhanced chemiluminescence (ECL; Amersham Bioscience).
Cap marker synthesis and analysis.
To prepare the tetraphosphate cap markers GppppG and m7GppppG, a D1/D12 mutant containing E192/194A substitutions was made with the primer pair D1/D12-E192/194A-F (Table 1) and its complement D1/D12-E192/194A-R on the template pET-14bD1/D12 (56). A QuikChange site-directed mutagenesis kit (Stratagene) was used following the protocol recommended by the manufacturer. Capping enzyme was expressed in bacteria and purified on Talon (Clontech) affinity matrix. mRNA cap markers were prepared as described previously (62). Template RNA was synthesized by in vitro transcription of pBS-SK (Stratagene) previously digested with XbaI. The linearized template was transcribed with T7 RNA polymerase using standard conditions as recommended by the manufacturer (Promega). The resulting 93-nucleotide RNAs were capped using the vaccinia virus capping enzyme D1/D12 or the RNA triphosphatase mutant D1/D12-E192/194A. Capping reactions contained 50 mM Tris (pH 7.9), 5 mM dithiothreitol, 1.25 mM MgCl2, 10 μM GTP, 2.5 μM RNA ends, 0.6 μM capping enzyme, and 10 μCi of [α-32P]GTP with or without 50 μM AdoMet for mRNA cap methylation (56). The resulting cap markers were subsequently purified by two rounds of precipitation with 2.5 M LiCl to remove enzyme and free label and then digested with nuclease P1 (62). Liberated caps were separated by thin-layer chromatography on polyethyleneimine cellulose plates developed in 0.45 M ammonium sulfate. Plates were exposed to a PhosphorImager screen for quantitation and analysis.
In vivo cap labeling and analysis.
Sf-9 cells (5 × 107 cells in suspension culture) were infected with virus at an MOI of 10. At 20 h postinfection, the cells were washed twice with 50 ml of phosphate-free Grace's medium and then incubated in the presence of 5.0 mCi of [32P]orthophosphoric acid (285.5 Ci/mg) diluted in 50 ml of phosphate-free Grace's medium for 4 h at 28°C. After incubation, the cells were harvested by centrifugation and mRNA was extracted with an Oligotex mRNA midi kit (Qiagen) according to the manufacturer's instructions. In vivo mRNA cap analysis was adopted from a procedure described previously by Latner et al. (31). Purified mRNA was digested with nuclease P1 overnight at 37°C, followed by treatment with calf alkaline phosphatase for 3 h at 37°C. Cap markers were also treated with nuclease P1 and calf alkaline phosphatase. Digested samples were then concentrated to approximately 1 μl in an 80°C hot plate and resuspended in 5 μl of sequencing stop buffer (95% formamide, 20 mM EDTA, 0.05% bromophenol blue, 0.05% xylene cyanol FF). Samples were electrophoresed through a 20% polyacrylamide, 8 M urea, 1× Tris-borate-EDTA sequencing gel at 33 W with constant power for 4 h. The excess free-labeled phosphate from this analysis was distributed between the bottom of the gel and the lower buffer reservoir. After electrophoresis, the gel was directly subjected to PhosphorImager analysis.
RESULTS
Analysis of viral replication in Sf-9 cells cotransfected with vAcΔΔRTPase-PG and lef-4 mutant plasmids.
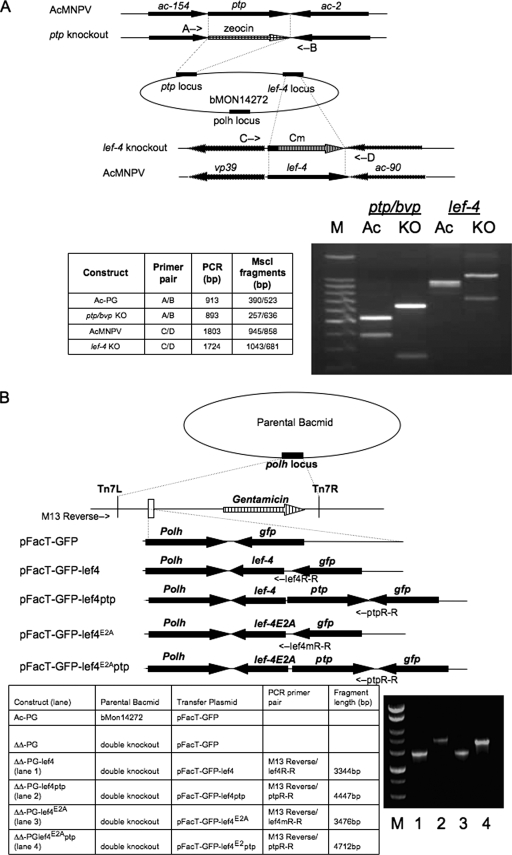
To elucidate the roles of LEF-4 and PTP/BVP RNA triphophatases in the viral life cycle, a double-knockout mutant viral genome was constructed in E. coli. First, the lef-4 gene was deleted by homologous recombination. This knockout was designed so that nearly the entire lef-4 ORF was replaced with the chloramphenicol acetyltransferase (cat) marker. The resulting construct contained only the first 131 bp downstream of the initiating lef-4 AUG codon (Fig. 1A, top panel). It was necessary to leave these residues, because the p39 promoter overlaps the 5′ end of lef-4. The resulting bacmid, Lef-4KO, was confirmed by PCR analysis and restriction enzyme digestions. The primer pair C/D hybridizes to the 5′ and 3′ ends of the lef-4 gene and generated PCR products of 1,803 bp for the parental (wt lef-4 construct) and 1,724 bp for LEF-4KO (data not shown). Because these fragments are close in size, restriction enzyme digestion was used for further confirmation. Sequence analysis predicted that MscI digestion of the wt fragment would produce two bands of 945 and 858 bp, while digestion of LEF-4KO would produce bands of 1,043 and 681 bp (Fig. 1A, lanes 3 and 4, respectively). This PCR-RFLP analysis confirmed that the lef-4 gene had been replaced with the cat gene marker.
FIG. 1.
Construction of double-knockout and rescue bacmids. (A) Strategy for deletion of lef-4 and ptp/bvp genes. Schematic diagram showing location of genes and insertion of antibiotic resistance markers. Positions of primer pairs used to confirm disruption of lef-4 or ptp/bvp are shown by arrows. The table at the lower left summarizes predicted results for PCR-RFLP using appropriate primer pairs for each locus. Actual results are shown on the lower right. (B) Schematic diagrams of vAc-PG and rescue bacmids vAcΔΔRTPase-PGLEF-4, vAcΔΔRTPase-PGLEF-4/PTP, vAcΔΔRTPase-PGLEF-4E2A, and vAcΔΔRTPase-PGLEF-4E2A/PTP, showing the genes inserted into the polyhedrin (polh) locus by Tn7-mediated transposition. PCR analysis (on the bottom right) using the M13-reverse and specific gene primers, whose location is shown on the schematic diagram, to confirm transposition of the rescue constructs. The table shows templates, primers and expected PCR-RFLP sizes. KO, knockout; Ac, AcMNPV; M, 100-bp DNA size markers.
To construct the double-knockout bacmid, the ptp/bvp ORF in the context of the Lef-4KO bacmid was replaced with a Zeocin resistance marker. Construction of the double-knockout bacmid was confirmed by PCR-RFLP. The primer pair A/B hybridizes to the 5′ and 3′ ends of the ptp/bvp gene. PCR generated products of 913 bp for the wt and 893 bp for the knockout construct (data not shown). MscI digestion of the wt fragment produced two bands of 523 and 390 bp, as predicted by the sequence, while the ptp/bvp knockout produced 636- and 257-bp bands (Fig. 1A, lanes 1 and 2, respectively). These results demonstrated that lef-4 and ptp/bvp were successfully replaced by the chloramphenicol and Zeocin resistance markers.
To facilitate the examination of virus infection, polyhedrin and GFP were introduced into the polh locus of the double-knockout bacmid by transposition using the Bac-to-Bac system (Fig. 1B). The resulting construct was named vAcΔΔRTPase-PG. The parental bacmid was also transposed with polyhedrin and gfp for use as a positive control and was named vAc-PG.
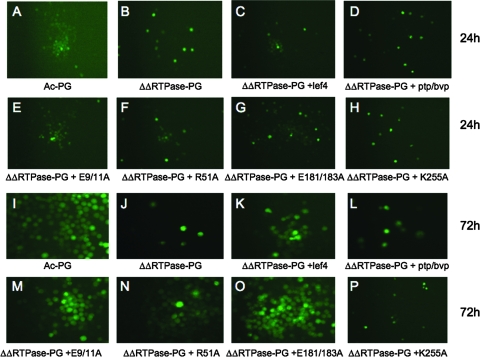
vAcΔΔRTPase-PG bacmid DNA was prepared and transfected into Sf-9 cells alone or cotransfected with plasmids encoding wt PTP/BVP, wt LEF-4, or mutant versions of LEF-4. Fluorescence microscopic analysis revealed similar patterns of GFP in all the samples at 24 h postinfection (Fig. 2), indicating that the transfection efficiencies were equivalent for all DNAs. Three days postinfection, however, the vAcΔΔRTPase-PG transfected cells exhibited no evidence of virus spread. GFP expression was restricted to single cells in a manner similar to that for the 24-h sample. In contrast, the Ac-PG control showed widespread distribution of GFP and polyhedrin formation, as did cells cotransfected with vAcΔΔ RTPase-PG and a wt lef-4 sequence. The LEF-4K255A mutant did not rescue the growth defect. These samples confirm our previous results with BacP+/LEF-4CmR, which demonstrated that the guanylyltransferase activity of lef-4 is essential for viral replication (30).
FIG. 2.
Analysis of viral replication in Sf-9 cells cotransfected with vAcΔΔRTPase-PG and lef-4 or ptp/bvp constructs. Cells were transfected with control bacmid DNA (panels A and I) or the double-knockout RTPase bacmid (ΔΔRTPase-PG) alone (panels B and J) or cotransfected with plasmids encoding wt lef-4 (panels C and K), ptp/bvp (panels D and L), lef-4 mutant E9/11A (panels E and M), lef-4 mutant R51A (panels F and N), lef-4 mutant E181/183 (panels G and O), or lef-4 mutant K255A (panels H and P). Cells were analyzed by fluorescence microscopy at 24 h (panels A to H) and 48 h (panels I to P) posttransfection.
Cotransfection of a plasmid encoding ptp/bvp failed to rescue the double mutant. At 72 h postinfection, these cells showed no evidence of polyhedron formation and GFP was restricted to single cells, similar to the case for the K255A mutant. This result was expected, because the ptp/bvp construct could not restore guanylyltransferase activity.
We then tested whether RNA triphosphatase mutants of lef-4 could rescue the double mutant. Three different mutant versions were analyzed. Previously, we showed that all three of these had low to undetectable RNA triphosphatase activities (28). Two mutants contained alanine substitutions in pairs of conserved glutamic acid residues (E9/11A and E181/183A). Crystallographic analysis of the vaccinia virus ortholog has demonstrated that these residues chelate the essential divalent metal ion (36). Alanine substitution at R51 impairs activity, because this residue contacts the gamma phosphate and positions it for cleavage (28, 36, 41).
Cells cotransfected with vAcΔΔRTPase-PG and one of the three different LEF-4 RNA triphosphatase mutants (E9/11A, R51A, or E181/183A) exhibited widespread patterns of GFP distribution and polyhedron formation, similar to that of vAcΔΔRTPase-PG cotransfected with wt lef-4. The finding that lef-4 RNA triphosphatase mutants could rescue virus production, even in the absence of ptp/bvp, suggests that neither RNA triphosphatase function is required for viral replication.
Construction and analysis of repair bacmids.
To further characterize the extent of the RNA triphosphatase defects on viral replication, repair viruses were constructed containing wt lef-4, mutant lef-4, or ptp/bvp genes inserted at the polyhedrin locus of the double-knockout bacmid by transposition (Fig. 1B). Four different repair bacmids were constructed. One contained only the wt lef-4 gene, which should provide all functions of lef-4, but does not restore ptp/bvp. This virus was named vAcΔΔRTPase-PGLEF-4. The second was an RNA triphosphatase mutant version of lef-4 containing substitutions at E181/183A, called vAcΔΔRTPase-PGLEF-4E2A. This mutant was chosen because it had the lowest RNA triphosphatase activity among the three mutants tested (28). The third bacmid, which restored both wt lef-4 and the ptp/bvp gene, was called vAcΔΔRTPase-PGLEF-4/PTP. Finally, a construct containing the E181/183A mutant version of lef-4 and wt ptp/bvp, called vAcΔΔRTPase-PGLEF-4E2A/PTP, was made. All constructs were confirmed by PCR analysis by using M13 reverse and gene-specific primers. The relative positions of the primers and expected sizes of the PCR products generated from the bacmid constructs are illustrated in Fig. 2B. Results from these analyses were consistent with the expectations, indicating successful transposition of the intended genes.
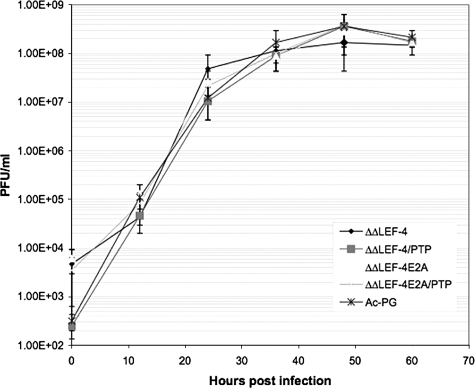
All four repair bacmids and the control vAc-PG bacmid were separately transfected into Sf-9 cells, and BV stocks were produced and their titers were determined. Then a one-step growth curve experiment was performed at an MOI of 5, and infectious virus production was measured at different times postinfection (Fig. 3). The growth curve patterns for all of the repair bacmids were essentially the same as those of the control, even for vAcΔΔRTPase-PGLEF-4E2A, which lacks both virus-encoded RNA triphosphatase activities. The result of this more stringent test of viral replication was consistent with those of the transfection experiments, again indicating that neither RNA triphosphatase function was essential for virus replication.
FIG. 3.
One-step growth curve of vAc-PG, vAcΔΔRTPase-PGLEF-4, vAcΔΔRTPase-PGLEF-4/PTP, vAcΔΔRTPase-PGLEF-4E2A, and vAcΔΔ RTPase-PGLEF-4E2A/PTP. Sf-9 cells were infected with the indicated viruses at an MOI of 5. Cell culture supernatants were harvested at the indicated times postinfection and assayed for the production of infectious virus by the TCID50 assay. Each data point represents the average titer derived from independent duplicate sample. Bars represent standard errors.
Analysis of late gene expression in mutant bacmids.
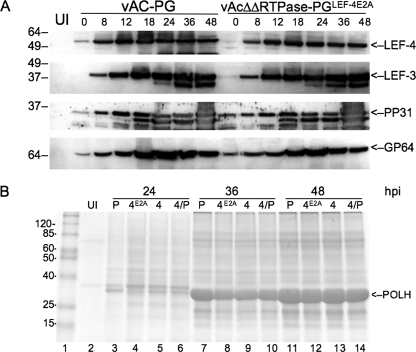
Although there was no apparent effect on production of budded virus, we wanted to analyze the expression of viral late genes to look for more subtle effects. In these experiments, we compared only the control virus vAc-PG and the virus lacking both RNA triphosphatases vAcΔΔRTPaseLEF-4E2A. We first examined LEF-4 expression to confirm that it was expressed at normal levels as expected (Fig. 4A, top panel). Immunoblots with polyclonal LEF-4 antiserum revealed that the lef-4 promoter translocated to the polyhedrin locus drove the expression of LEF-4 at essentially the same levels as those of the native promoter. The expression of LEF-3 was also unaffected by the lef-4 RTPase mutation. This result was expected because lef-3 is a delayed early gene and should not be dependent upon the viral-encoded RNA polymerase (Fig. 4A, second panel). Two genes that are regulated by tandem delayed early and late promoters were analyzed next. These two genes, pp31 and gp64, are expressed during both the early and late phases, but the accumulation of protein is mainly due to the stronger late promoters (4, 18). The expression patterns of both of these proteins in the mutant-infected cells were essentially indistinguishable from that of the control.
FIG. 4.
Western blot analysis of viral protein expression. (A) Sf-9 cells were infected with the double-mutant virus vAcΔΔRTPase-PGLEF-4E2A or the control vAc-PG at an MOI of 5. Cells were harvested at the indicated times, and detergent-soluble protein extracts were separated on SDS gels. Each lane represents 1 × 104 cells. The lane labeled UI represents uninfected cells. Time zero is the end of a 1-h adsorption period. Extracts were probed with antibody against rabbit antiserum raised against LEF-4 or PP31 or mouse monoclonal antibodies against LEF-3 or GP64. (B) Analysis of polyhedrin expression in cells infected with vAC-PG (P), vAcΔΔRTPase-PGLEF-4E2A (4A), vAcΔΔRTPase-PGLEF-4 (4), or vAcΔΔ RTPase-PGLEF-4/PTP (4/P). Cells were harvested at the indicated times and detergent-insoluble extracts were separated on 12% acrylamide gels and stained with Coomassie brilliant blue.
Polyhedrin expression was analyzed by the staining of SDS gels with Coomassie brilliant blue (Fig. 4B). Only the detergent-insoluble fraction was analyzed after the disruption of polyhedra with alkali. For polyhedrin analysis, we also examined two additional repair viruses, because we noticed a decrease in the accumulation of polyhedrin in the vAcΔΔRTPaseLEF-4E2A-infected cells relative to vAc-PG. This decrease was particularly evident in the 24-h sample. vAC-PG cells accumulated sufficient polyhedrin for detection by Coomassie staining, while polyhedrin was not evident at this time in the vAcΔΔRTPaseLEF-4E2A sample (Fig. 4B, compare lanes 3 and 4). Cells infected with the lef-4 wt repair virus vAcΔΔRTPaseLEF-4 showed polyhedrin accumulation at 24 h, as did the cells infected with vAcΔΔRTPaseLEF-4/PTP, although accumulation levels for both of these samples were less than that for the control (Fig. 4B, lanes 3 to 6). By 36 and 48 h postinfection, polyhedrin accumulated to high levels in the double-RTPase mutant, although it was still approximately 20% lower than control levels (Fig. 4B, compare lanes 11 and 12).
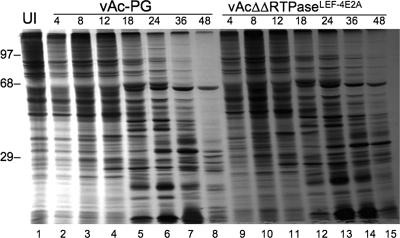
Total viral protein synthesis was also measured by protein pulse labeling and gel analysis of the detergent-soluble fraction of infected cells. Cells infected with mutant virus lacking both RNA triphosphatases appeared to synthesize viral proteins with kinetics similar to those of the control virus (Fig. 5). Although the exact identity of the protein bands is unknown, virus-specific protein bands appeared at the same times in both samples. Some bands, however, persisted longer in the mutant samples than in the control. This persistence was especially evident in the two 48-h samples, where there was only one band in the upper half of the gel in the control sample, while numerous bands were evident in the double-RTPase mutant sample.
FIG. 5.
Pulse-labeling time course of viral protein synthesis. Sf-9 cells were infected with the double-mutant virus vAcΔΔRTPase-PGLEF-4m or the control vAc-PG at an MOI of 5. At different times postinfection, cells were pulse labeled with [35S]methionine-cysteine for 4 h. At the end of the labeling period, detergent-soluble protein extracts were prepared and analyzed on an SDS-10% polyacrylamide gel. The times above each lane represent the harvest time after a 4-h labeling period. Each lane represents 1 × 104 cells. The positions of molecular markers are shown on the left. UI, uninfected cells.
Analysis of viral mRNA caps.
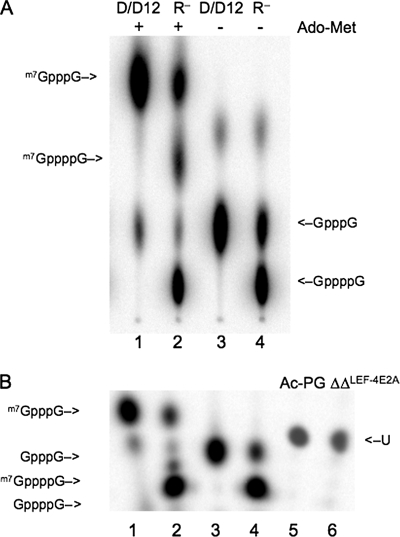
The fact that viral protein synthesis was unaffected by the absence of RNA triphosphatase activity suggested that viral mRNA caps were efficiently formed in the absence of this enzyme. To directly test this suggestion, we radiolabeled RNAs with inorganic 32P from 20 to 24 h postinfection, a time at which viral RNA synthesis is maximal and host RNA synthesis has been suppressed. Viral mRNA was purified on oligo(dT) columns, and caps were liberated by treatment with P1 nuclease and calf intestinal phosphatase. We were unable to efficiently separate the caps from the background of free phosphate on thin-layer chromatograms, because most of the radiolabel in the RNA samples was composed of internal nucleotides and was released as free phosphate. Therefore, we analyzed the samples on 20% acrylamide gels, which yielded cleaner separations. First, however, we wanted to confirm that the gels were capable of separating tetraphosphate caps, which would be generated by the complete absence of RNA triphosphatase activity, from normal triphosphate caps. Triphosphate caps were synthesized using the vaccinia virus capping enzyme D1/D12, a heterodimer that contains RNA triphosphatase, guanylyltransferase, and cap methyltransferase activities (63). Accurate synthesis of the markers was verified by thin-layer chromatograms (Fig. 6A). In the absence of Ado-Met, D1/D12 adds a GMP cap to exogenous RNA, which was synthesized by T7 RNA polymerase. Since the 5′ residue on this RNA is G, the cap structure is GpppG (Fig. 6A, lane 3). In the presence of Ado-Met, the cap is methylated to produce m7GpppG (Fig. 6A, lane 1). Tetraphosphate caps were produced using a mutant version of the vaccinia enzyme containing alanine substitutions at glutamic acid residues 192 and 194 (63), which are equivalent to the LEF-4 residues 181 and 183. In the absence of RNA triphosphatase activity, the major product is a tetraphosphate cap (Fig. 6A, lane 4). A minor amount of triphosphate cap was made, which is likely due to residual triphosphatase activity. The addition of Ado-Met efficiently converts the minor amount of triphosphate cap to methylated cap, but very little tetraphosphate cap is methylated, because it does not fit into the methyltransferase active site (Fig. 6A, lane 2).
FIG. 6.
Cap structures of viral RNAs. (A) Preparation of m7GppppG, m7GpppG, GppppG, and GpppG markers. Capping reactions contained 50 mM Tris (pH 7.9), 5 mM dithiothreitol, 1.25 mM MgCl2, 10 μM GTP, 2.5 μM RNA ends, and 0.6 μM wt (lanes 1 and 3) or mutant (lanes 2 and 4) vaccinia virus D1/D12 and 12.5 μCi of [α-32P]GTP with (+) (lanes 1 and 2) or without (−) (lanes 3 and 4) 50 μM AdoMet. Caps were liberated by nuclease P1 digestion and separated by thin-layer chromatography on polyethyleneimine cellulose plates in 0.45 M ammonium sulfate. The positions of the methylated caps are indicated on the left and nonmethylated caps on the right. (B) Analysis of baculovirus mRNA caps. Sf-9 cells (5 × 107) in a 50-ml spinner flask were infected with vAc-PG (lane 5) or vAcΔΔRTPase-PGLEF-4m (lane 6) at an MOI of 10 and labeled with 5.0 mCi of [32P]orthophosphoric acid (285.5 Ci/mg) from 20 to 24 h postinfection. mRNA was extracted from cells, digested with nuclease P1 and treated with calf alkaline phosphatase. Digested samples were electrophoresed on a 20% polyacrylamide, 8-M urea sequencing gel. The cap markers are the same as those described for panel A. The position of the unknown (U) viral cap is indicated on the right. After electrophoresis, the gel was directly subjected to PhosphorImager analysis.
Acrylamide gel electrophoresis of the same cap markers revealed that this method was also capable of cleanly separating the different species (Fig. 6, compare lanes 1 to 4 of both panels). Cap migration was not affected by treatment of the markers with calf intestinal phosphatase, which was necessary to reduce background in the viral RNAs labeled in vivo (data not shown). A comparison of the viral RNA caps revealed that they migrated faster than did the m7GpppG marker. We were unable to definitely identify the structure of this cap, but believe that it represents GpppNm. Significantly, however, RNAs from the control virus Ac-PG and the double-RNA triphosphatase mutant migrated at exactly the same position (Fig. 6B, lanes 5 and 6), indicating that cap formation was unaffected by the RNA triphosphatase mutations in this virus.
Construction of a LEF-4 quadruple mutant.
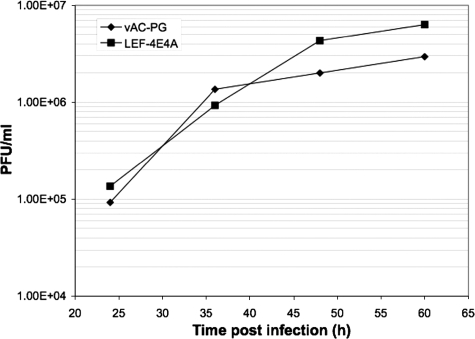
We next considered the possibility that the lack of a phenotype associated with the LEF-4 RTPase E181/183A mutation was due to residual RNA triphosphatase activity. This possibility seemed somewhat unlikely, because mutation of a single one of the four glutamates that chelate metal is known to be lethal in yeast (Saccharomyces cerevisiae) (22). Still, we decided to test this idea by constructing a rescue mutant in which all four chelating glutamates (E9, E11, E181, and E183) were mutated to alanines. This bacmid construct was named vAcΔΔRTPaseLEF-4E4A. The transfection of the mutant bacmid DNA resulted in the production of viable progeny after transfection into insect cells (data not shown). A single-step growth curve showed that it grew at least as well as the parental construct vAC-PG (Fig. 7). The quadruple mutant virus was apparently normal in all respects, except for a defect in the production of polyhedrin (data not shown).
FIG. 7.
One-step growth curve of vAc-PG and vAcΔΔRTPase-PGLEF-4E4A/PTP. Sf-9 cells were infected with the indicated viruses at an MOI of 5. Cell culture supernatants were harvested at the indicated times postinfection and assayed for the production of infectious virus by the TCID50 assay. Each data point represents the average titer derived from an independent duplicate sample.
DISCUSSION
By exploiting a lef-4 and ptp/bvp double-mutant virus, we have been able to probe the biological role of LEF-4 and PTP/BVP in replication of AcMNPV in Sf-9 cells. Our results indicate a surprising conclusion that RNA triphosphatase activity is neither required for viral replication in tissue culture nor necessary for the formation of a 5′ cap structure on late viral mRNAs. In the absence of both RNA triphosphatase functions, the production of double-mutant progeny virus was nearly identical to a virus possessing both of these functions. In addition, protein pulse labeling and Western blotting experiments showed that there was little difference between the double-mutant and control viruses with respect to the expression of late viral genes, although the expression of polyhedrin was affected. Moreover, mRNA cap structure analysis indicated that the double mutant had a 5′ cap structure that was the same as that of the control virus.
The 5′ m7GpppN cap plays an essential role in the life cycle of eukaryotic cellular and viral mRNA and is required for efficient pre-mRNA splicing, export, stability, and translation. Cellular enzymes find their RNA substrates through interactions with RNA polymerase II (6, 43). RNA viruses, which utilize their own RNA-dependent RNA polymerases, have evolved diverse strategies to contend with the capping problem. Vesicular stomatitis virus (1, 32, 33), alphavirus (2), and Sindbis virus (54) encode their own enzymes that specifically cap and methylate their positive-strand transcripts, while influenza virus steals caps from cellular transcripts (34, 52). Picornaviruses and hepatitis C virus adopt unique RNA structures that bypass the cap requirement for stability and allow internal initiation of mRNA translation (26, 50). DNA viruses, on the other hand, employ somewhat less adventurous strategies. The mRNAs of retroviruses and most nuclear DNA viruses (e.g., papovaviruses, adenoviruses, and herpesviruses) are transcribed by cellular RNA polymerase II and are capped by the cellular capping and methylating enzymes. A few DNA virus families, however, encode their own RNA polymerases and mRNA capping enzymes; these include poxviruses, which replicate entirely in the cytoplasm (44), and African swine fever virus, which has a cytoplasmic replication phase (51). Among all the DNA viruses, baculoviruses are unique in that they employ host RNA polymerase II for early gene transcription and encode their own RNA polymerases for late mRNA synthesis.
The notion that baculoviruses encode their own enzymes for mRNA capping gained currency in light of the in vitro RNA triphosphatase and guanylyltransferase activities of LEF-4, a subunit of viral RNA polymerase. The presence of ptp/bvp with RNA triphosphatase activity challenged the potential role of LEF-4 as the sole RNA triphosphatase involved in the capping of viral late mRNAs. Crystallography (5) and domain swap studies (42) between baculovirus and mammalian enzymes suggested that this enzyme might provide a backup mechanism for catalyzing the RNA triphosphatase step in the capping pathway. After our initial results with a lef-4 single-knockout virus with three different versions of LEF-4 triphosphatase mutants indicated that its triphosphatase activity is not required for viral replication (unpublished observations), we assumed that PTP/BVP was supplying the essential triphosphatase function. To our surprise, however, we found that the deletion of the ptp/bvp gene, even in the context of a LEF-4 RNA triphosphatase mutation, had no effect on viral replication or on the RNA cap structure.
This intriguing result raises the question of which enzyme is responsible for the hydrolysis of γ-phosphate from triphosphate-terminated nascent RNA strands. We considered the possibility that residual activity from the E181/183A mutant was sufficient for mRNA capping, although it had undetectable levels of RNA triphosphatase when assayed in vitro (28). Figure 6 shows the effect of similar mutations on the vaccinia virus RNA capping enzyme. In the absence of RTPase activity, tetraphosphate caps were formed, and these were inefficiently methylated (63). In S. cerevisiae, alanine substitution of even a single residue corresponding to the conserved LEF-4 residues E9, E11, R51, E181, or E183 was lethal (22). In our experiments, we tested mutant versions containing pairs of glutamic acid residue substitutions that should have even lower residual activities, but these still supported growth. To further impair the RTPase activity of LEF-4, however, we constructed a mutant version containing all four essential glutamates mutated to alanine and found that viral replication was still supported.
Two possible explanations remain for our observations that cap structure and viral replication were unaffected by the complete absence of known RNA triphosphatases. One is that baculoviruses encode another RNA triphosphatase gene that has not been identified and the other is that the cellular enzyme provides this necessary function. While plausible, the first explanation seems unlikely. Baculoviruses are the only known genomes that encode both types of RNA triphosphatases known to be involved in cap formation. Metal-dependent enzymes like LEF-4 are found in some DNA viruses and fungi, while metal-independent cysteine phosphatases are used in plants and metazoans. Conserved residues for both of these families are now well defined, and we have not been able to identify additional orthologs in AcMNPV.
A host enzyme could provide RNA triphosphatase activity. Although host protein synthesis is shut down during the late phase of infection, it is likely that the enzyme persists. However, the fact that cellular capping enzymes are directed to their substrates through interactions with the C-terminal domain of RNA polymerase II has been well documented (6, 43). Baculovirus RNA polymerases lack this feature, which we presume is why they have evolved their own capping enzymes. The structure of baculovirus RNA polymerase is unknown, but it seems reasonable to propose that the active sites of LEF-4 are oriented so that RNA is capped as it leaves the polymerase tunnel. In other systems, capping has been shown to occur cotranscriptionally, and cellular enzymes cannot cap exogenous RNAs (12, 20). Indeed, we have been unable to demonstrate capping of exogenous RNAs with either purified polymerase or the single-subunit LEF-4 (J. Jin and L. Guarino, unpublished data). The vaccinia virus capping enzyme D1/D12 is an exception to this rule, and that is why it has become a useful tool for capping RNAs after in vitro transcription. We expect, however, that the insect cell capping enzymes would not be attracted to the viral RNA polymerase and may not have sufficient access to the nascent RNA to dephosporylate it cotranscriptionally. We attempted to test whether cellular capping enzymes were involved in late RNA synthesis through RNA silencing studies, but this analysis is complicated by the fact that early viral RNAs require the host capping enzymes. This experiment would require the ability to rapidly inactivate the host enzyme as the virus enters the late phase, for example, with a temperature-sensitive mutant. We doubt that RNA silencing, which lacks the ability to fine-tune the timing of the effect, would give a clean answer.
Unlike the case for S. cerevisiae, the Candida albicans RNA triphosphatase gene is nonessential (9). Cells with null alleles were viable, although they grew more slowly. Cap structures were also affected. While the altered structures were not fully characterized, their migration appeared to be consistent with an unmethylated tetraphosphate cap. While this result suggests that RNA triphosphatases are not always essential, it differs from the baculovirus case in that growth of the viruses was not affected, nor was the cap structure altered.
We were unable to identify the cap structure on viral RNAs. It migrated faster than the m7GpppG marker, and only slightly slower than the unmethylated marker GpppG. Its migration suggests that it may correspond to GpppNm. Ribose methylation has a lesser effect on the mobility of cap structures than does N7 methylation, because the former alters mass but not charge. The methylation on both sites (m7GpppGm) results in a mobility that is slower than that of the m7GpppG marker (31). If correct, this cap structure would be consistent with the presence of mtase-1 [an RNA cap (2′-O)-methyltransferase] in the genome of AcMNPV, but there are no good candidates for an RNA cap (guanine-7)-methyltransferase enzyme. These two cap methylations are catalyzed by enzymes with different active site structures (10, 24), and it is unlikely that mtase-1 could catalyze both reactions.
Still unresolved by our work is the question of why group I baculoviruses encode the RNA triphosphatase gene ptp/bvp. The gene is not essential for viral replication in cell culture, but the work of Kamita et al. (29) suggests that it plays an important role in the virus lifestyle. Viruses with deletions in this gene were unable to induce wandering, a behavior that results in infected insects crawling to the tops of vegetation and aids in virus dispersion. The mechanism for its action is unknown, but the role of the protein for ptp/bvp in RNA processing suggests that it may be mediated through RNA regulatory mechanisms.
Acknowledgments
We thank Xiaojiang Dai and David Theilmann for assistance with bacmid constructs, and Wen Dong in our lab for assistance with virus titers.
This material is based upon work supported by the National Science Foundation under grant no. MCB-0416484.
Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation.
Footnotes
Published ahead of print on 2 April 2008.
REFERENCES
- 1.Abraham, G., D. P. Rhodes, and A. K. Banerjee. 1975. The 5′ terminal structure of the methylated mRNA synthesized in vitro by vesicular stomatitis virus. Cell 551-58. [DOI] [PubMed] [Google Scholar]
- 2.Ahola, T., and L. Kaariainen. 1995. Reaction in alphavirus mRNA capping: formation of a covalent complex of nonstructural protein nsP1 with 7-methyl-GMP. Proc. Natl. Acad. Sci. USA 92507-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bideshi, D. K., and B. A. Federici. 2000. The Trichoplusia ni granulovirus helicase is unable to support replication of Autographa californica multicapsid nucleopolyhedrovirus in cells and larvae of T. ni. J. Gen. Virol. 811593-1599. [DOI] [PubMed] [Google Scholar]
- 4.Blissard, G. W., and G. F. Rohrmann. 1989. Location, sequence, transcriptional mapping, and temporal expression of the gp64 envelope glycoprotein gene of the Orgyia pseudotsugata multicapsid nuclear polyhedrosis virus. Virology 170537-555. [DOI] [PubMed] [Google Scholar]
- 5.Changela, A., A. Martins, S. Shuman, and A. Mondragon. 2005. Crystal structure of baculovirus RNA triphosphatase complexed with phosphate. J. Biol. Chem. 28017848-17856. [DOI] [PubMed] [Google Scholar]
- 6.Cho, E. J., T. Takagi, C. R. Moore, and S. Buratowski. 1997. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 113319-3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dai, X., T. M. Stewart, J. A. Pathakamuri, Q. Li, and D. A. Theilmann. 2004. Autographa californica multiple nucleopolyhedrovirus exon0 (orf141), which encodes a RING finger protein, is required for efficient production of budded virus. J. Virol. 789633-9644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dunyak, D. S., D. S. Everdeen, J. G. Albanese, and C. L. Quinn. 2002. Deletion of individual mRNA capping genes is unexpectedly not lethal to Candida albicans and results in modified mRNA cap structures. Eukaryot. Cell 11010-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fabrega, C., S. Hausmann, V. Shen, S. Shuman, and C. D. Lima. 2004. Structure and mechanism of mRNA cap (guanine-N7) methyltransferase. Mol. Cell 1377-89. [DOI] [PubMed] [Google Scholar]
- 11.Furuichi, Y., and A. J. Shatkin. 2000. Viral and cellular mRNA capping: past and prospects. Adv. Virus Res. 55135-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gillian-Daniel, D. L., N. K. Gray, J. Astrom, A. Barkoff, and M. Wickens. 1998. Modifications of the 5′ cap of mRNAs during Xenopus oocyte maturation: independence from changes in poly(A) length and impact on translation. Mol. Cell. Biol. 186152-6163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gross, C. H., and S. Shuman. 1998. Characterization of a baculovirus-encoded RNA 5′-triphosphatase. J. Virol. 727057-7063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gross, C. H., and S. Shuman. 1998. RNA 5′-triphosphatase, nucleoside triphosphatase, and guanylyltransferase activities of baculovirus LEF-4 protein. J. Virol. 7210020-10028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guarino, L. A. 1990. Identification of a viral gene encoding a ubiquitin-like protein. Proc. Natl. Acad. Sci. USA 87409-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guarino, L. A., W. Dong, B. Xu, D. R. Broussard, R. W. Davis, and D. L. Jarvis. 1992. Baculovirus phosphoprotein pp31 is associated with virogenic stroma. J. Virol. 667113-7120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guarino, L. A., J. Jin, and W. Dong. 1998. Guanylyltransferase activity of the LEF-4 subunit of baculovirus RNA polymerase. J. Virol. 7210003-10010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guarino, L. A., and M. Smith. 1992. Regulation of delayed-early gene transcription by dual TATA boxes. J. Virol. 663733-3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guarino, L. A., B. Xu, J. Jin, and W. Dong. 1998. A virus-encoded RNA polymerase purified from baculovirus-infected cells. J. Virol. 727985-7991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hagler, J., and S. Shuman. 1992. A freeze-frame view of eukaryotic transcription during elongation and capping of nascent mRNA. Science 255983-986. [DOI] [PubMed] [Google Scholar]
- 21.Hang, X., W. Dong, and L. A. Guarino. 1995. The lef-3 gene of Autographa californica nuclear polyhedrosis virus encodes a single-stranded DNA-binding protein. J. Virol. 693924-3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ho, C. K., Y. Pei, and S. Shuman. 1998. Yeast and viral RNA 5′ triphosphatases comprise a new nucleoside triphosphatase family. J. Biol. Chem. 27334151-34156. [DOI] [PubMed] [Google Scholar]
- 23.Ho, C. K., V. Sriskanda, S. McCracken, D. Bentley, B. Schwer, and S. Shuman. 1998. The guanylyltransferase domain of mammalian mRNA capping enzyme binds to the phosphorylated carboxyl-terminal domain of RNA polymerase II. J. Biol. Chem. 2739577-9585. [DOI] [PubMed] [Google Scholar]
- 24.Hodel, A. E., P. D. Gershon, X. Shi, and F. A. Quiocho. 1996. The 1.85 Å structure of vaccinia protein VP39: a bifunctional enzyme that participates in the modification of both mRNA ends. Cell 85247-256. [DOI] [PubMed] [Google Scholar]
- 25.Hoopes, R. R., Jr., and G. F. Rohrmann. 1991. In vitro transcription of baculovirus immediate early genes: accurate mRNA initiation by nuclear extracts from both insect and human cells. Proc. Natl. Acad. Sci. USA 884513-4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jang, S. K. 2006. Internal initiation: IRES elements of picornaviruses and hepatitis C virus. Virus Res. 1192-15. [DOI] [PubMed] [Google Scholar]
- 27.Jarvis, D. L., and M. D. Summers. 1989. Glycosylation and secretion of human tissue plasminogen activator in recombinant baculovirus-infected insect cells. Mol. Cell. Biol. 9214-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin, J., W. Dong, and L. A. Guarino. 1998. The LEF-4 subunit of baculovirus RNA polymerase has RNA 5′-triphosphatase and ATPase activities. J. Virol. 7210011-10019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kamita, S. G., K. Nagasaka, J. W. Chua, T. Shimada, K. Mita, M. Kobayashi, S. Maeda, and B. D. Hammock. 2005. A baculovirus-encoded protein tyrosine phosphatase gene induces enhanced locomotory activity in a lepidopteran host. Proc. Natl. Acad. Sci. USA 1022584-2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Knebel-Morsdorf, D., I. Quadt, Y. Li, L. Montier, and L. A. Guarino. 2006. Expression of baculovirus late and very late genes depends on LEF-4, a component of the viral RNA polymerase whose guanyltransferase function is essential. J. Virol. 804168-4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Latner, D. R., J. M. Thompson, P. D. Gershon, C. Storrs, and R. C. Condit. 2002. The positive transcription elongation factor activity of the vaccinia virus J3 protein is independent from its (nucleoside-2′-O-) methyltransferase and poly(A) polymerase stimulatory functions. Virology 30164-80. [DOI] [PubMed] [Google Scholar]
- 32.Li, J., E. C. Fontaine-Rodriguez, and S. P. Whelan. 2005. Amino acid residues within conserved domain VI of the vesicular stomatitis virus large polymerase protein essential for mRNA cap methyltransferase activity. J. Virol. 7913373-13384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li, J., J. T. Wang, and S. P. Whelan. 2006. A unique strategy for mRNA cap methylation used by vesicular stomatitis virus. Proc. Natl. Acad. Sci. USA 1038493-8498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li, M. L., P. Rao, and R. M. Krug. 2001. The active sites of the influenza cap-dependent endonuclease are on different polymerase subunits. EMBO J. 202078-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li, Y., and L. K. Miller. 1995. Properties of a baculovirus mutant defective in the protein phosphatase gene. J. Virol. 694533-4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lima, C. D., L. K. Wang, and S. Shuman. 1999. Structure and mechanism of yeast RNA triphosphatase: an essential component of the mRNA capping apparatus. Cell 99533-543. [DOI] [PubMed] [Google Scholar]
- 37.Lin, G., and G. W. Blissard. 2002. Analysis of an Autographa californica nucleopolyhedrovirus lef-11 knockout: lef-11 is essential for viral DNA replication. J. Virol. 762770-2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu, A., and L. K. Miller. 1994. Identification of three late expression factor genes within the 33.8- to 43.4-map-unit region of Autographa californica nuclear polyhedrosis virus. J. Virol. 686710-6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luckow, V. A., S. C. Lee, G. F. Barry, and P. O. Olins. 1993. Efficient generation of infectious recombinant baculoviruses by site-specific transposon-mediated insertion of foreign genes into a baculovirus genome propagated in Escherichia coli. J. Virol. 674566-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martins, A., and S. Shuman. 2003. Mapping the triphosphatase active site of baculovirus mRNA capping enzyme LEF4 and evidence for a two-metal mechanism. Nucleic Acids Res. 311455-1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martins, A., and S. Shuman. 2001. Mutational analysis of baculovirus capping enzyme Lef4 delineates an autonomous triphosphatase domain and structural determinants of divalent cation specificity. J. Biol. Chem. 27645522-45529. [DOI] [PubMed] [Google Scholar]
- 42.Martins, A., and S. Shuman. 2002. The domain order of mammalian capping enzyme can be inverted and baculovirus phosphatase can function in cap formation in vivo. Virology 304167-175. [DOI] [PubMed] [Google Scholar]
- 43.McCracken, S., N. Fong, E. Rosonina, K. Yankulov, G. Brothers, D. Siderovski, A. Hessel, S. Foster, S. Shuman, and D. L. Bentley. 1997. 5′-Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes Dev. 113306-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moss, B. 1996. Poxviridae: the viruses and their replication, p. 2637-2671. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven, Philadelphia, PA.
- 45.Okano, K., A. L. Vanarsdall, V. S. Mikhailov, and G. F. Rohrmann. 2006. Conserved molecular systems of the Baculoviridae. Virology 34477-87. [DOI] [PubMed] [Google Scholar]
- 46.O'Reilly, D. R., L. K. Miller, and V. A. Luckow. 1992. Baculovirus expression vectors: a laboratory manual. W. H. Freeman & Co., New York, NY.
- 47.Partington, S., H. Yu, A. Lu, and E. B. Carstens. 1990. Isolation of temperature sensitive mutants of Autographa californica nuclear polyhedrosis virus: phenotype characterization of baculovirus mutants defective in very late gene expression. Virology 17591-102. [DOI] [PubMed] [Google Scholar]
- 48.Passarelli, A. L., and L. K. Miller. 1993. Identification of genes encoding late expression factors located between 56.0 and 65.4 map units of the Autographa californica nuclear polyhedrosis virus genome. Virology 197704-714. [DOI] [PubMed] [Google Scholar]
- 49.Passarelli, A. L., J. W. Todd, and L. K. Miller. 1994. A baculovirus gene involved in late gene expression predicts a large polypeptide with a conserved motif of RNA polymerases. J. Virol. 684673-4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pelletier, J., G. Kaplan, V. R. Racaniello, and N. Sonenberg. 1988. Translational efficiency of poliovirus mRNA: mapping inhibitory cis-acting elements within the 5′ noncoding region. J. Virol. 622219-2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pena, L., R. J. Yanez, Y. Revilla, E. Vinuela, and M. L. Salas. 1993. African swine fever virus guanylyltransferase. Virology 193319-328. [DOI] [PubMed] [Google Scholar]
- 52.Plotch, S. J., M. Bouloy, I. Ulmanen, and R. M. Krug. 1981. A unique cap(m7GpppXm)-dependent influenza virion endonuclease cleaves capped RNAs to generate the primers that initiate viral RNA transcription. Cell 23847-858. [DOI] [PubMed] [Google Scholar]
- 53.Qin, J. C., and R. F. Weaver. 1982. Capping of viral RNA in cultured Spodoptera frugiperda cells infected with Autographa californica nuclear polyhedrosis virus. J. Virol. 43234-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rozanov, M. N., E. V. Koonin, and A. E. Gorbalenya. 1992. Conservation of the putative methyltransferase domain: a hallmark of the ‘Sindbis-like’ supergroup of positive-strand RNA viruses. J. Gen. Virol. 732129-2134. [DOI] [PubMed] [Google Scholar]
- 55.Sheng, Z., and H. Charbonneau. 1993. The baculovirus Autographa californica encodes a protein tyrosine phosphatase. J. Biol. Chem. 2684728-4733. [PubMed] [Google Scholar]
- 56.Shuman, S. 1990. Catalytic activity of vaccinia mRNA capping enzyme subunits coexpressed in Escherichia coli. J. Biol. Chem. 26511960-11966. [PubMed] [Google Scholar]
- 57.Shuman, S. 2001. Structure, mechanism, and evolution of the mRNA capping apparatus. Prog. Nucleic Acid Res. Mol. Biol. 661-40. [DOI] [PubMed] [Google Scholar]
- 58.Takagi, T., G. S. Taylor, T. Kusakabe, H. Charbonneau, and S. Buratowski. 1998. A protein tyrosine phosphatase-like protein from baculovirus has RNA 5′-triphosphatase and diphosphatase activities. Proc. Natl. Acad. Sci. USA 959808-9812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Titterington, J. S., T. K. Nun, and A. L. Passarelli. 2003. Functional dissection of the baculovirus late expression factor-8 gene: sequence requirements for late gene promoter activation. J. Gen. Virol. 841817-1826. [DOI] [PubMed] [Google Scholar]
- 60.Vaughn, J. L., R. H. Goodwin, G. J. Tompkins, and P. McCawley. 1977. The establishment of two cell lines from the insect Spodoptera frugiperda (Lepidoptera; Noctuidae). In Vitro 13213-217. [DOI] [PubMed] [Google Scholar]
- 61.Volkman, L. E., and P. A. Goldsmith. 1988. Resistance of the 64K protein of budded Autographa californica nuclear polyhedrosis virus to functional inactivation by proteolysis. Virology 166285-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu, X., and L. A. Guarino. 2003. Autographa californica nucleopolyhedrovirus orf69 encodes an RNA cap (nucleoside-2′-O)-methyltransferase. J. Virol. 773430-3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yu, L., A. Martins, L. Deng, and S. Shuman. 1997. Structure-function analysis of the triphosphatase component of vaccinia virus mRNA capping enzyme. J. Virol. 719837-9843. [DOI] [PMC free article] [PubMed] [Google Scholar]