Abstract
The transcription factor (TF) Foxp2 has been shown to partially repress surfactant protein C (SP-C) transcription, presumably through interaction of an independent repressor domain with a conserved Foxp2 consensus site in the SP-C promoter. We explored the role of interactions between Foxp2 and the homeodomain TF Nkx2.1 that may contribute to the marked reduction in SP-C expression accompanying phenotypic transition of alveolar epithelial type II (AT2) to type I (AT1) cells. Foxp2 dose-dependently inhibited Nkx2.1-mediated activation of SP-C in MLE-15 cells. While electrophoretic mobility shift assays and chromatin immunoprecipitations revealed an interaction between Foxp2 and the conserved consensus motif in the SP-C promoter, Nkx2.1-mediated activation of the 318-bp proximal SP-C promoter (which lacks a Foxp2 consensus) was attenuated by increasing amounts of Foxp2. Co-immunoprecipitation and mammalian two-hybrid assays confirmed a physical interaction between Nkx2.1 and Foxp2 mediated through the Nkx2.1 homeodomain. Formation of an Nkx2.1 complex with an SP-C oligonucleotide was inhibited dose-dependently by recombinant Foxp2. These findings demonstrate that direct interaction between Foxp2 and Nkx2.1 inhibits Nkx2.1 DNA-binding and transcriptional activity and suggest a mechanism for down-regulation of SP-C (and probably other AT2 cell genes) during transition of AT2 cells to an AT1 cell phenotype.
Keywords: alveolar epithelium, transcriptional regulation, forkhead box, Nkx2.1, differentiation
CLINICAL RELEVANCE
The research provides insight into transcriptional mechanisms that regulate transitions between alveolar epithelial type II and type I cells during normal maintenance of the epithelium and repair after injury.
Alveolar epithelium is composed of two morphologically distinct cell types, type I (AT1) and type II (AT2) cells (1). AT2 cells are believed to serve as progenitors for replacement of both AT2 and AT1 cells in vivo during development and after lung injury, while AT1 cells have been presumed to be terminally differentiated (2–4). In culture, primary AT2 cells lose their characteristic phenotypic hallmarks (e.g., lamellar bodies and expression of surfactant apoproteins) (5, 6), and gradually acquire AT1 cell features, including morphologic characteristics and expression of phenotypic markers (e.g., aquaporin-5 and T1α). These observations suggest that, similar to development/repair in vivo, AT2 cells transdifferentiate in vitro toward the AT1 cell phenotype (6–8). However, transcriptional mechanisms that govern activation/repression of differentiation-related genes during transitions between AT2 and AT1 cell phenotypes in adult lung are poorly understood.
Forkhead box (Fox) proteins constitute a family of transcription factors (TF) that play important roles in the regulation of cell fate during morphogenesis and differentiation (9). Fox TF share a conserved winged-helix/forkhead DNA-binding domain (10) and, while they mostly function as transcriptional activators, they can also repress gene transcription (11, 12). Several Fox genes are expressed in lung, where they have been implicated as important regulators of lung-specific promoters (e.g., surfactant protein [SP]-B, Clara cell secretory protein [CC10], and Nkx2.1/thyroid transcription factor-1 [TTF-1]) (11, 13, 14). FOXP (FOXP1-FOXP4) TF constitute a novel Fox subfamily characterized by a highly divergent forkhead DNA-binding domain (15). Mutations in this DNA-binding domain are implicated in a number of developmental disorders (16–19).
Foxp1, Foxp2, and Foxp4 are expressed at high levels in embryonic and adult mouse lung and function as sequence-specific transcriptional repressors (20). Foxp1 and Foxp4 are expressed in both proximal and distal airway epithelium, while Foxp2 becomes progressively restricted to distal lung epithelium during development (11, 20). Foxp2 has been shown to repress the promoters of lung-enriched genes (CC10 and SP-C) through a homologous N-terminal repression domain that is shared by Foxp1 and Foxp4 (11), suggesting a role in restriction of expression to specific cell types within the respiratory epithelium. Foxp2 has also been shown to interact synergistically with the co-repressor CtBP-1 to inhibit transcription (21). However, mutation of the CtBP-1 site in Foxp1 and Foxp2 did not alter their ability to repress transcription, suggesting that CtBP-1 is not essential for transcriptional repression. Additional mechanisms underlying transcriptional repression mediated by Foxp TF, including possible DNA-independent interactions with other TF, have not been investigated.
TTF-1 is a member of the Nkx2 class of homeodomain-containing TF that is essential for lung morphogenesis and a critical determinant of lung epithelium–specific gene expression (22, 23). Nkx2.1 is found primarily in AT2 cells and is not expressed in AT1 cells (24–26). Nkx2.1 transactivates the AT2 cell–specific SP-C gene through a conserved critical proximal binding site located −186/−163 bp upstream of the transcription start site (27, 28). Although transcriptional activation of SP-C by Nkx2.1 and interacting co-activators (e.g., GATA-6 [29], NFI [30], TAZ [31] and Erm [32]) has been extensively investigated, mechanisms involved in down-regulation of SP-C (such as during transition of alveolar epithelial cells [AEC] from the AT2 to the AT1 cell phenotype) are largely unknown. In particular, potential interactions between Nkx2.1 and transcriptional repressors (e.g., FoxP TF) in this process have not been examined.
Fox TF have been shown to interact with a number of homeodomain proteins to regulate transcription (33, 34). Although interactions between Fox and homeodomain TF are usually stimulatory, this interaction has been reported in some instances to be inhibitory (e.g., between Foxa2/engrailed and Foxa2/Otx2) (34, 35), possibly by preventing binding of the interacting partner with DNA. In this regard, Foxa2 has recently been shown to interact with Nkx2.1 to repress expression of SP-C through a DNA-independent mechanism (36). In the current study, we investigated the role of Foxp2 in regulating expression of SP-C through potential inhibitory interactions with Nkx2.1.
MATERIALS AND METHODS
Cell Isolation and Preparation of AEC Monolayers
AT2 cells were isolated from adult male Sprague-Dawley rats by disaggregation with elastase (2.0–2.5 U/ml; Worthington Biochemical, Freehold, NJ), followed by differential adherence on IgG-coated bacteriological plates (37). Freshly isolated AT2 cells were plated in minimal defined serum-free medium (MDSF) consisting of Dulbecco's modified Eagle's medium and Ham's F12 nutrient mixture in a 1:1 ratio (DME-F12), supplemented with 1.25 mg/ml bovine serum albumin (BSA), 10 mM HEPES, 0.1 mM nonessential amino acids, 2.0 mM L-glutamine, 100 U/ml sodium penicillin G, and 100 μg/ml streptomycin. Cells were seeded onto 0.4-μm pore polycarbonate filter cups (Transwell; Corning, Corning, NY) at 1 × 106 cells/cm2. AT2 cell purity (> 90%) was assessed by staining freshly isolated cells with an antibody (Ab) to p180 lamellar membrane protein (Covance Research Products, Berkeley, CA). MLE-15 cells (J. Whitsett, University of Cincinnati [38]) were cultivated in HITES medium ((RPMI 1640 medium; Invitrogen, Carlsbad, CA) supplemented with 10 nM hydrocortisone, 5 μg/ml insulin, 5 μg/ml human transferrin, 10 nM β-estradiol, 5 μg/ml selenium, 2 mM L-glutamine, 10 mM HEPES, 100 U/ml penicillin, 100 μg/ml streptomycin; and 4% fetal bovine serum (FBS) (Atlanta Biologicals, Lawrenceville, GA)).
Plasmids
SP-C-3.7-Luc contains the 3.7-kb human SP-C promoter in pGL2Basic (Promega, Madison, WI). pRC/CMV/Nkx2.1 contains the 2.3-kb human Nkx2.1 cDNA in pRC/CMV (Invitrogen). p318 muSP-C-Luc contains -318 to +18 of the murine SP-C promoter cloned into SmaI/XhoI sites of pGL2Basic. pM3/Nkx2.1 contains the full-length 2.3-kb human Nkx2.1 cDNA inserted in pM (Clontech, Mountain View, CA). pGEX-2T-Nkx2.1 contains Nkx2.1 cDNA (either full-length or specific domains) inserted in pGEX-2T (Amersham Pharmacia Biotech, Piscataway, NJ). pCMV/Tag2B/Foxp2 contains the mouse Foxp2 cDNA. pRC/CMV/Foxp2 was generated by inserting Foxp2 cDNA from pCMV/Tag2B/Foxp2 into BamHI/ApaI sites of pRC/CMV. pVP16/Foxp2 contains Foxp2 cDNA excised from pCMV/Tag2B/Foxp2 and inserted in-frame into the EcoRI site of pVP16 (Clontech). pGEX-4T-1-Foxp2 contains Foxp2 cDNA inserted in-frame into the EcoRI site of pGEX-4T-1 (Amersham Pharmacia Biotech). pFR-Luc is from Stratagene (La Jolla, CA).
Co-Transfection Assays
Transient transfections in MLE-15 cells were performed using SuperFect reagent (Qiagen, Valencia, CA). MLE-15 cells were seeded at 6 × 104 cells/well in 24-well plates 1 day before transfection. Cells were transfected with 0.75 μg of SP-C-3.7-Luc reporter and pRC/CMV/Nkx2.1 and pCMV/Tag2B/Foxp2 in equal amounts (0.15 μg) or corresponding empty expression vectors. To evaluate dose-dependence of Foxp2 repression on trans-activation of the 3.7-kb SP-C promoter by Nkx2.1, SP-C-3.7-Luc was co-transfected with 0.25 μg of pRC/CMV/Nkx2.1 and increasing amounts of pCMV/Tag2B/Foxp2 (12.5–200 ng). To determine whether Nkx2.1 could overcome repression by Foxp2, SP-C-3.7-Luc was co-transfected with 0.15 μg of pCMV/Tag2B/Foxp2 and increasing amounts of pRC/CMV/Nkx2.1 (3.125–50 ng). To evaluate dose-dependence of Foxp2 repression on trans-activation of the 318-bp SP-C promoter by Nkx2.1, p318 muSP-C-Luc was co-transfected with 0.6 μg of pRC/CMV/Nkx2.1 and increasing amounts of pCMV/Tag2B/Foxp2 (0–600 ng). After 24 hours, cells were harvested and firefly luciferase activity was determined with the Dual-Luciferase reporter assay system (Promega). Firefly luciferase was normalized to Renilla luciferase activity.
Preparation of Nuclear Extracts for Electrophoretic Mobility Shift Assay
NIH3T3 cells grown in 100-mm culture plates were transfected with 10 μg pCMV/Tag2B/Foxp2 expression vector using Superfect reagent (Qiagen) (39). After 48 hours, cells were harvested in PBS. After centrifugation, pellets (from four plates) were resuspended in 1 ml hypotonic buffer (10 mM HEPES pH 7.5, 10 mM KCl, 3 mM MgCl2, 0.05% Nonidet P-40, 1 mM EDTA pH 8.0, 10 mM NaF, and 0.1 mM Na3VO4) with 5 μl/ml protease inhibitor cocktail III (Calbiochem, San Diego, CA) and incubated on ice for 20 minutes, followed by vortexing for 10 seconds and centrifugation at 500 × g for 10 minutes at 4°C. Nuclear pellets were washed twice with hypotonic buffer and lysed in 600 μl lysis buffer (100 mM HEPES [pH 7.5], 0.5 M KCl, 5 mM MgCl2, 28% glycerol, and protease inhibitor cocktail III) for 30 minutes on ice with shaking. After centrifugation at 20,000 × g for 30 minutes to remove debris, extracts were stored at −70°C. Preparation of nuclear extracts from MLE-15 cells was performed using a kit from Panomics (Redwood City, CA).
Electrophoretic Mobility Shift Assay
Double-stranded oligonucleotides (5′-GAGGCTTAGGCAAATATTTAAGGGGGCA-3′) spanning the conserved FoxP2 binding site in the SP-C promoter were radioactively labeled using 33P-ATP and polynucleotide kinase (Invitrogen). Nuclear extracts (∼ 4 μg) from NIH3T3 cells transfected with Foxp2 expression plasmid and approximately 2 × 105 cpm of labeled oligonucleotide were incubated in electrophoretic mobility shift assay (EMSA) buffer (20 mM Tris pH 7.5, 2 mM NaCl, 2 mM EDTA, 10% glycerol, 2 mM DTT [freshly added] and 0.2 μg poly [dI/dC]) (Amersham Pharmacia Biotech) for 20 minutes at 4°C. DNA–protein complexes were resolved from free probe by electrophoresis on 5% nondenaturing polyacrylamide gels in 0.5× TBE (1× TBE: 89 mM Tris, 49 mM boric acid, 2 mM EDTA). Two mutant oligonucleotide probes (5′-GAGGCTTAGGCGCGTATTTAAGGGGCA-3′ and 5′-GAGGCTTAGGCAAGCGCTTAAGGGGCA-3′) were used for competition. For supershift, nuclear extracts were pre-incubated with Foxp2 antibody at room temperature for 20 minutes before addition of radiolabeled probe. For competitive EMSA, 5 to 10 μg of nuclear extract was incubated in buffer C (12 mM HEPES, pH 7.9, 25 mM KCl, 5 mM MgCl, 1 mM EDTA, 50 ng/ml poly [dI/dC] [Roche Applied Science, Indianapolis, IN], 10% glycerol, 1 mM dithiothreitol, and 0.5 mM fresh phenylmethylsulfonyl fluoride) for 10 minutes on ice. To test the ability of Foxp2 to compete with Nkx2.1 binding to its cognate binding site in the SP-C promoter, nuclear extracts were first incubated with increasing amounts of unlabeled purified glutathione-S-transferase (GST)-Foxp2 (1, 2, 4, and 6 μg) or GST alone for 20 minutes. 33P-labeled oligonucleotide (5′-TAGGCCAAGGGCCTTGGGGCTCT-3′) containing the Nkx2.1 binding site of the SP-C promoter (-186/-183) (27) was added and incubated for 15 minutes on ice.
Mammalian Two-Hybrid Assay
The luciferase reporter pFR-luc was co-transfected with pM3/Nkx2.1 and pVP16/Foxp2 into MLE-15 cells using SuperFect reagent (Qiagen) (36). Transfection controls included combinations of pM3+pVP16, pM3+pVP16/Foxp2, and pM3/Nkx2.1+pVP16. Cells were harvested 48 hours after transfection, and luciferase activity was analyzed with the Galacto-Light kit (TROPIX, Bedford, MA) and normalized to β-galactosidase activity.
Co-Immunoprecipitation
Nuclear extracts harvested from MLE-15 cells were precleared with IgG and Protein A/G beads. One hundred fifty microliters of Protein A/G plus-agarose beads (Santa Cruz Biotechnology, Santa Cruz, CA) were prewashed with 750 μl IP Low Wash Buffer (Active Motif, Carlsbad, CA) three times and resuspended in 120 μl IP Low Wash Buffer. For co-immunoprecipitation (co-IP) of Foxp2 with Nkx2.1, nuclear extracts were incubated with anti-Nkx2.1 monoclonal antibody (Lab Vision, Fremont, CA) or with mouse IgG (control) in IP Low Wash Buffer overnight at 4°C. After adding 150 μl prewashed Protein A/G beads, incubation was continued for an additional 4 hours at 4°C followed by centrifugation at 3,000 rpm for 30 seconds. Pelleted beads were washed and resuspended in 30 μl of 4× sodium dodecyl sulfate (SDS) loading buffer (240 mM Tris pH 6.8, 26% glycerol, 0.1% DTT, 8% SDS, and 0.06% bromophenol blue) followed by boiling for 5 minutes. Supernatants were collected and applied to 7.5% gels for electrophoresis and subsequent Western analysis with polyclonal anti-Foxp2 Ab (Abcam, Cambridge, MA). For co-IP of Nkx2.1 with Foxp2, nuclear extracts were incubated with rabbit polyclonal anti-Foxp2 Ab or rabbit IgG. Western blotting was performed using anti-Nkx2.1 monoclonal antibody.
Purification of GST Fusion Proteins
Escherichia coli L21 bacteria harboring pGEX-2T-Nkx2.1 (either full-length or specific domains) or pGEX-4T-Foxp2 were grown in 15 ml of LB media overnight. Five milliliters of bacteria were transferred to 250 ml LB medium and grown to an optical density of 0.7 to 0.8 followed by addition of isopropyl β-D-1-thiogalactopyranoside (IPTG) (1 mM final) and growth at 28°C for 2 hours to induce fusion protein expression. Bacteria were pelleted, resuspended in 35 ml phosphate buffered saline (PBS, pH 7.2), and sonicated (2 min × 2), followed by addition of 1.5 ml 20% Triton-X 100 and incubation for 30 minutes at 4°C. After centrifugation for 10 minutes at 12,000 × g, supernatants were transferred to a new 50-ml tube followed by addition of 50% glutathione-sepharose 4 Fast Flow Beads (Amersham Pharmacia Biotech) and rotation for 30 minutes at room temperature. The mixture was transferred to Poly-Prep Chromatography Columns (Bio-Rad, Hercules, CA). GST fusion proteins were eluted with elution buffer (10 mM glutathione in 50 mM Tris-HCl at pH 8.0) after washing the column three times with 2 ml PBS, and dialyzed in PBS at 4°C overnight.
GST Pull-Down Assay
Nkx2.1 and Foxp2 proteins were synthesized by in vitro translation using plasmids pRC/CMV/Nkx2.1 and pRC/CMV/Foxp2 in the presence of methionine using TNT T7 Quik Coupled Transcription/Translation Reaction kit (Promega) (36). Translated Nkx2.1 and Foxp2 were precleared by incubation with glutathione-sepharose. To evaluate the interaction between Nkx2.1 and Foxp2, in vitro translated Nkx2.1 was incubated with GST-Foxp2 fusion protein–adsorbed glutathione-sepharose in binding buffer containing 50 mM Tris-HCl, 120 mM NaCl, 2 mM EDTA, and 0.1% NP-40 for 1 hour at room temperature. After extensive washing, adsorbed protein complexes were boiled and analyzed by Western blotting using anti-Nkx2.1 Ab (Lab Vision). GST-adsorbed glutathione-sepharose was used as control. To determine which domain of Nkx2.1 interacts with Foxp2, in vitro translated Foxp2 was incubated with GST-Nkx2.1 full-length, GST-N-terminal, GST-homeodomain, or GST-C-terminal Nkx2.1 fusion protein–adsorbed glutathione-sepharose in binding buffer for 1 hour at room temperature. After extensive washing, adsorbed protein complexes were boiled and analyzed by Western blotting using anti-Foxp2 Ab (Abcam).
Chromatin Immunoprecipitation
Day 1 (D1) and D8 rat AEC cultivated in MDSF were directly crosslinked with formaldehyde (1% final concentration) by incubating for 10 minutes at room temperature, followed by addition of glycine to 0.125 M for an additional 5 minutes (40). Cell pellets were collected by centrifugation after rinsing twice with PBS, resuspended in hypotonic buffer (10 mM HEPES pH 7.8, 10 mM KCl, 1.5 mM MgCl2, and 5 μl/ml protease inhibitor cocktail III), and incubated on ice for 10 minutes. Nuclear pellets were collected by centrifugation and resuspended in nuclear lysis buffer (1% SDS, 50 mM Tris-Cl pH 8.0, 10 mM EDTA, and 5 μl/ml protease inhibitor cocktail III). DNA was sheared by sonication to yield soluble chromatin. For immunoprecipitation (IP), 2 μg goat anti-Foxp2 Ab (Novus Biologicals, Littleton, CO), or 2 μg goat IgG (Santa Cruz Biotechnology), and 40 μg chromatin were incubated in IP dilution buffer (0.01% SDS, 20 mM Tris-Cl pH 8.0, 1.1% Triton X-100, 167 mM NaCl, and 1.2 mM EDTA) at 4°C overnight. Complexes were recovered by incubating with 20 μl protein A/G Plus-agarose beads (Santa Cruz Biotechnology) preblocked with sonicated salmon sperm DNA and BSA, at 4°C for 2 hours. After repeated washes, immunocomplexes were eluted from beads by incubation with 150 μl IP elution buffer (1% SDS with 0.1 M NaHCO3). NaCl (0.3 M final) was added to eluted samples and heated at 65°C overnight to reverse DNA–protein crosslinks. DNA was recovered by phenol-chloroform extraction and ethanol precipitation and resuspended in 30 μl Tris-EDTA. PCR was performed using primers (forward 5′-GAAAACTAGCTCCCCTCTCC-3′ and reverse 5′-CAGCATGGCATCTGAGATG-3′) spanning the conserved FoxP2 binding site in the rat SP-C promoter. For input DNA, the chromatin preparation not incubated with antibodies was subjected to PCR. PCR reactions for amplifying the FoxP2 binding site were performed in a volume of 50 μl containing 50 mM Tris-HCl pH 8.3, 0.15 mM MgCl2, 1× TaqMaster, 200 μm dNTPs, 0.5 μl Taq polymerase (Eppendorf, Westbury, NY) and 1 μM primer for 30 cycles (45 s denaturation [95°C], 45 s annealing [57°C], and 1 min extension [72°C]). PCR products were resolved and visualized with ethidium bromide. The expected size of the PCR product is 150 bp.
Western Analysis
Total protein was harvested in SDS sample buffer (2% SDS, 10% glycerol, 5% β-mercaptoethanol, pH 6.8) from AEC monolayers in MDSF on D0, D1, D3, D5, and D8 and protein concentrations were measured using the DC Protein Analysis System (Bio-Rad). Samples were resolved by SDS-PAGE and electrophoretically blotted onto Immuno-Blot polyvinylidene fluoride (PVDF) membranes (Bio-Rad). Membranes were blocked in 5% nonfat dry milk for Nkx2.1 and pro–SP-C immunoblotting or 1% BSA for FoxP2 immunoblotting followed by incubation with primary Abs (1:1,000 for anti-Nkx2.1 [NovoCastra, Norwell, MA], 1:500 for anti-pro-SP-C [Millipore, Billerica, MA], and 1:250 for anti-Foxp2 [20]) at 4°C overnight. After washing with TBS-T (20 mM Tris-7.5, 0.5 M NaCl, 0.01% Tween-20), blots were incubated with horseradish peroxidase–linked anti-IgG conjugates (Promega) for 45 minutes at room temperature. Complexes were visualized by SuperSignal West Femto Maximum Sensitivity Substrate (Pierce, Rockford, IL) for pro–SP-C and FoxP2, and enhanced chemiluminescence (ECL) (Amersham Pharmacia Biotech) for Nkx2.1 with an Alpha Ease Imaging System (Alpha Innotech, San Leandro, CA).
Statistical Analysis
Data are shown as mean ± SEM, where (n) is the number of observations. We used z-tests to determine if the ratiometric data (i.e., normalized against control) were different from controls. We performed one-way ANOVA followed by post hoc comparisons of group means using the Newman-Keuls-Student procedure. P < 0.05 was considered significant.
RESULTS
Foxp2 Represses Nkx2.1-Induced SP-C Promoter Activity
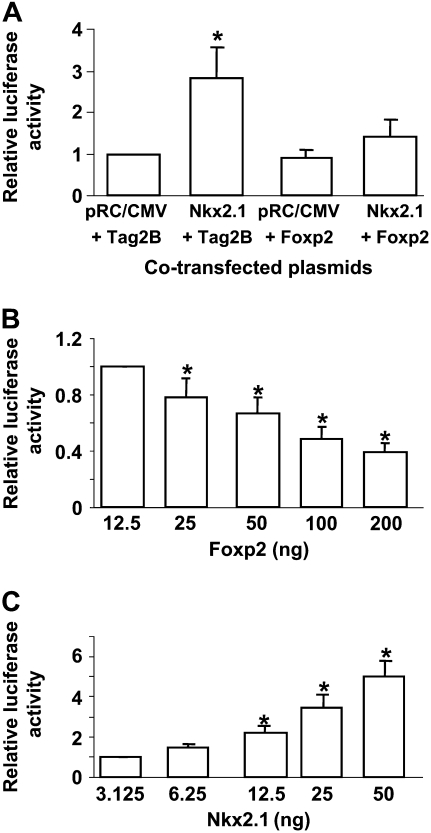
Nkx2.1 is known to activate SP-C through a proximal Nkx2.1-binding site in both human and mouse SP-C promoters (27, 28). To investigate potential interactions between Foxp2 and Nkx2.1 in regulating expression of SP-C, MLE-15 cells were co-transfected with a luciferase reporter construct containing the 3.7-kb human SP-C promoter (3.7-SP-C-Luc) together with combinations of Foxp2 and Nkx2.1 expression plasmids. As shown in Figure 1A, co-transfection of Nkx2.1 alone activates 3.7-SP-C-Luc approximately 3-fold compared with empty vector. Co-transfection of Foxp2 alone has only a minimal effect on basal reporter activity, although higher amounts of Foxp2 (0.75 μg) reduced reporter activity by approximately 60% as previously reported (11). When co-transfected together with Nkx2.1, Foxp2 blunted the Nkx2.1-mediated stimulation by approximately 70%. Furthermore, transfection of increasing amounts of Foxp2 expression plasmid causes a dose-dependent reduction in trans-activation of SP-C by Nkx2.1 (Figure 1B), which could be rescued by transfection of increasing amounts of Nkx2.1 (Figure 1C). Of note, overexpression of Foxp2 did not alter endogenous Nkx2.1 protein levels (data not shown).
Figure 1.
Foxp2 represses transcriptional activation of surfactant protein (SP)-C promoter by Nkx2.1 in a dose-dependent fashion. The 3.7-kb human SP-C–luciferase construct was co-transfected into MLE-15 cells with combinations of Foxp2 (pCMV/Tag2B/Foxp2) and Nkx2.1 (pRC/CMV/Nkx2.1) expression plasmids or empty vector as control (A), 50 ng pRC/CMV/Nkx2.1 and increasing amounts of pCMV/Tag2B/Foxp2 (12.5–200 ng) (B), and 150 ng pCMV/Tag2B/Foxp2 and increasing amounts of pRC/CMV/Nkx2.1 (3.125–50 ng) (C). Firefly luciferase was measured 48 hours after transfection and normalized to Renilla luciferase activity. Data represent mean ± SEM, with n = 3 for A and B and n = 4 for C. *P < 0.05 compared with all other conditions (A), compared with 12.5 ng Foxp2 (B), and compared with 3.125 ng Nkx2.1 (C).
Foxp2 Represses Nkx2.1-Mediated Stimulation of the Mouse 318-bp SP-C Proximal Promoter
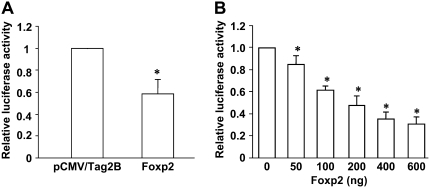
To further evaluate whether Foxp2-mediated attenuation of SP-C stimulation by Nkx2.1 was dependent on direct Foxp2 binding to the SP-C promoter, we transfected MLE-15 cells with a 318-bp SP-C-luciferase construct (which does not contain a Foxp2 binding site). Cells were co-transfected with pRC/CMV/Nkx2.1 and increasing amounts of pCMV/Tag2B/Foxp2. As shown in Figure 2A, Foxp2 alone decreased transcriptional activity of the 318-bp SP-C-Luc construct approximately 40%. Furthermore, Foxp2 antagonized Nkx2.1-mediated trans-activation of the 318-bp SP-C promoter in a dose-dependent fashion, with approximately 3-fold repression observed with the 600-ng dose (Figure 2B). These results suggest that Foxp2 attenuates Nkx2.1-mediated activation of SP-C via direct interaction with Nkx2.1 and without a requirement for binding to a Foxp2 DNA motif.
Figure 2.
Foxp2 represses transcriptional activation of mouse 318-bp SP-C promoter by Nkx2.1. The 318-bp mouse SP-C luciferase construct was co-transfected into MLE-15 cells with Foxp2 (pCMV/Tag2B/Foxp2) and empty vector pCMV/Tag2B (A) or 600 ng pRC/CMV/Nkx2.1 and increasing amounts of pCMV/Tag2B/Foxp2 (0–600 ng) (B). Firefly luciferase was measured 24 hours after transfection and normalized to Renilla luciferase activity. Data represent mean ± SEM, with n = 3. *P < 0.05 compared with empty vector (A) or no Foxp2 (B).
Interactions between Nkx2.1 and Foxp2 by Mammalian Two-Hybrid Assay and Co-IP
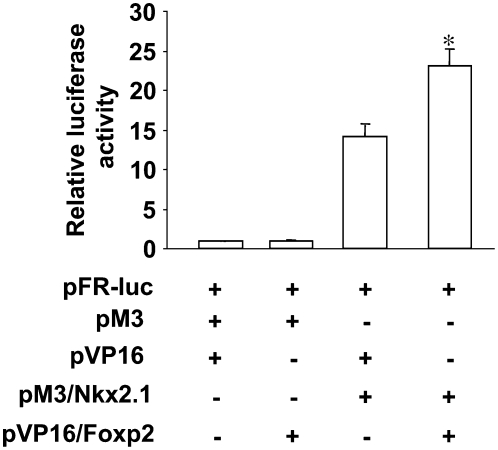
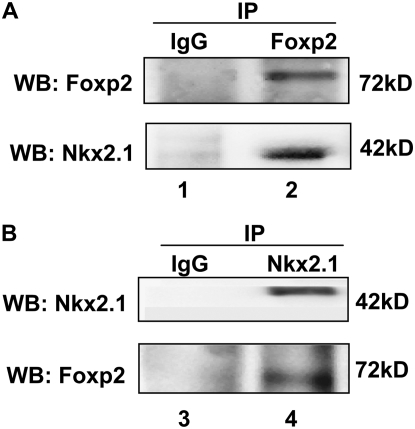
To investigate potential interactions between Nkx2.1 and Foxp2, mammalian two-hybrid assay was performed in MLE-15 cells. The luciferase reporter pFR-luc was co-transfected with pM3/Nkx2.1 and pVP16/Foxp2, a Gal4 DNA-binding domain fusion protein and VP16 DNA trans-activation domain fusion protein, respectively. As shown in Figure 3, luciferase activity increased significantly after transfection of pM3/Nkx2.1 and pVP16/Foxp2 compared with controls, suggesting interaction between Nkx2.1 and Foxp2. Consistent with these results, co-IP assays using either anti-Nkx2.1 Ab or anti-Foxp2 Ab for IP demonstrated association between endogenous Nkx2.1 and Foxp2 in MLE-15 cells (Figure 4).
Figure 3.
Interaction between Nkx2.1 and Foxp2 by mammalian two-hybrid assay. The luciferase reporter pFR-luc (1 μg) was co-transfected with pM3/Nkx2.1 (0.05 μg) and pVP16/Foxp2 (1.5 μg) into MLE-15 cells. Transfection controls included pM3+pVP16, pM3+pVP16/Foxp2, and pM3/Nkx2.1+pVP16. Firefly luciferase activity was measured 48 hours after transfection and normalized to pSV-β-gal. Data represent mean ± SEM, with n = 6. *P < 0.05 compared with all other conditions.
Figure 4.
Co-immunoprecipitation of Nkx2.1 and Foxp2. IP was performed with nuclear extracts harvested from MLE-15 cells using either anti-Foxp2 Ab (A) or anti-Nkx2.1 Ab (B) and analyzed by Western blotting. Representative Western blots (n = 3) demonstrate that Nkx2.1 is detected in protein complexes immunoprecitated by anti-Foxp2 Ab (lane 2), and that Foxp2 can be detected in protein complexes immunopreciptated by anti-Nkx2.1 Ab (lane 4), demonstrating an association between Foxp2 and Nkx2.1. Lanes 1 and 3 are negative controls (IgG at an equivalent concentration to the respective precipitating Ab).
Foxp2 Interacts Directly with the Nkx2.1 Homeodomain
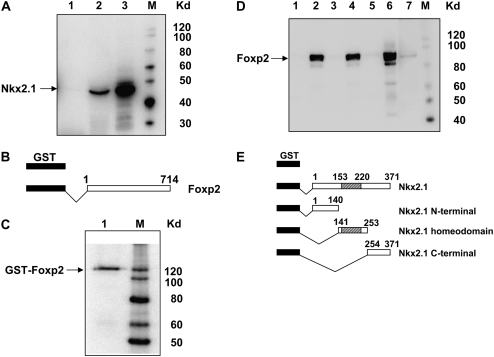
To test whether Nkx2.1 and Foxp2 interact directly, in vitro GST pull-down assays were performed. In vitro-translated Nkx2.1 was incubated with immobilized GST-Foxp2 or GST as control, and the presence of Nkx2.1 after pull-down with GST-Foxp2 was tested by Western analysis. As shown in Figure 5A, full-length Nkx2.1 interacts with GST-Foxp2 but not with GST alone. Western analysis (Figure 5C) demonstrates full-length GST-Foxp2 used as input. These results suggest that Foxp2 could inhibit Nkx2.1 activity upon target genes by directly interacting with Nkx2.1 domains that otherwise interact with DNA or with co-activators, thereby disrupting the function of Nkx2.1. To address these possibilities, pull-down assays were performed with in vitro-translated Foxp2 and GST fusion proteins containing different domains of Nkx2.1. Results demonstrate that Foxp2 interacts directly with the Nkx2.1 homeodomain but not with the C- or N-terminal domains (Figure 5D). GST-fusion proteins encompassing different Nkx2.1 domains used as input (Figure 5E) are as previously described (36).
Figure 5.
Physical interactions between Nkx2.1 and Foxp2 in GST pull-down assay. (A) In vitro translated Nkx2.1 was incubated with GST (lane 1) or GST-Foxp2 (lane 2) or proteins coupled to glutathione-sepharose. Bound Nkx2.1 was visualized by Western blotting using anti-Nkx2.1 Ab. Lane 3 is positive control for Nkx2.1. (B) Diagram of constructs used for pull-down of Nkx2.1. (C) Input of GST-Foxp2 fusion protein detected by anti-Foxp2 antibody. Arrow denotes full-length GST-Foxp2. (D) In vitro translated Foxp2 was incubated with GST-Nkx2.1 fusion proteins encompassing full-length Nkx2.1 protein (lane 2), N-terminal domain (lane 3), homeodomain (lane 4), and C-terminal domain (lane 5) of Nkx2.1. Lane 1 is GST alone, lane 6 is in vitro translated Foxp2 protein, and lane 7 represents nuclear extract from NIH3T3 cells transfected with pCMV/Tag2B/Foxp2. Bound Foxp2 was visualized by Western blotting using anti-Foxp2 Ab. (E) Diagram of constructs used for pull-down of FoxP2.
Foxp2 Inhibits Nkx2.1 DNA-Binding Activity
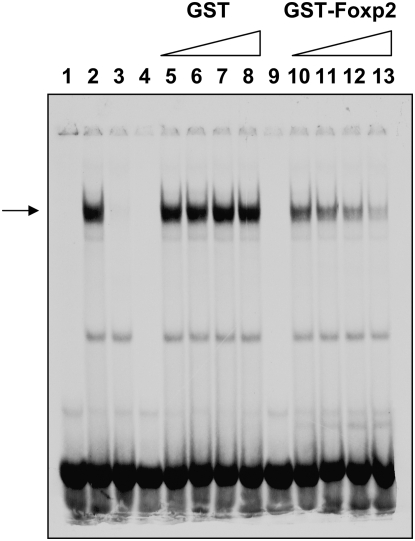
Because Foxp2 interacts with the DNA-binding homeodomain of Nkx2.1, we tested by competitive EMSA (Figure 6) whether Foxp2 inhibits Nkx2.1 DNA-binding activity. As previously reported (27), nuclear extracts from MLE-15 cells (which strongly express Nkx2.1) form a complex with a −186/−163 SP-C promoter probe spanning the consensus Nkx2.1 motif (Figure 6, lane 2). Increasing amounts of recombinant GST-Foxp2 fusion protein inhibited formation of the Nkx2.1-DNA complexes in a dose-dependent manner (Figure 6, lanes 10–13), while the same amounts of GST alone had no effect on complex formation (Figure 6, lanes 5–8). These results suggest that Foxp2 suppresses SP-C transcription by preventing Nkx2.1 from binding to the SP-C proximal promoter.
Figure 6.
Foxp2 interferes with binding of Nkx2.1 to the SP-C promoter. Electrophoretic mobility shift assay (EMSA) was performed with nuclear extracts from MLE-15 cells and 33P-labeled oligonucleotides encompassing the Nkx2.1 DNA-binding site on the SP-C promoter. The nucleoprotein complexes formed (lane 2) can be competed by cold competitor (40×) (lane 3). Addition of increasing amounts (1, 2, 4, 6 μg) of GST-Foxp2 proteins (lanes 10–13), but not GST alone (lanes 5–8), interferes with nucleoprotein complex formation in a dose-dependent fashion. GST-Foxp2 (lane 4) and GST (lane 9) proteins do not form a complex with the oligonucleotide probe. Lane 1 is probe only.
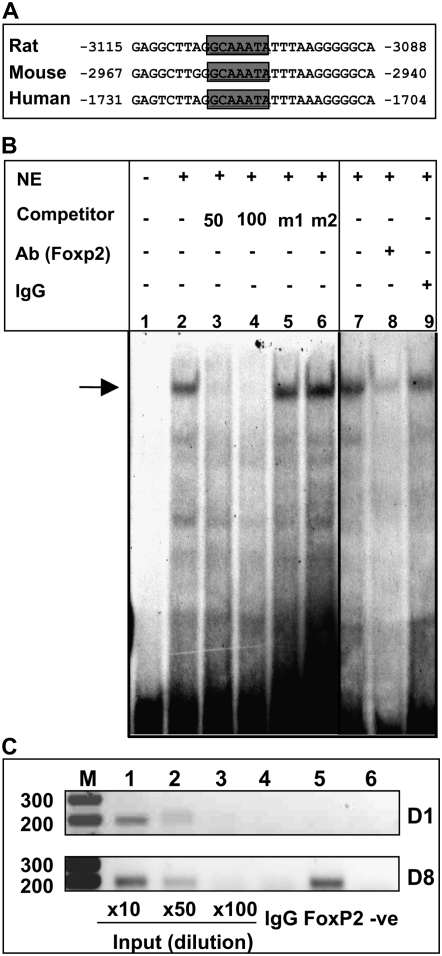
FoxP2 Directly Binds to Conserved Consensus Binding Site in the SP-C Promoter
To further explore the mechanisms underlying effects of Foxp2 on SP-C expression, we examined interactions of Foxp2 with the SP-C promoter. Analysis of the 3.7-kb sequence of the human SP-C promoter identified a putative Foxp2-binding site (5′-GCAAATA-3′) located at nucleotide −1724 to −1718, which is conserved among human, rat, and mouse (Figure 7A). EMSA and ChIP assays were performed to determine whether Foxp2 interacted with this site using nuclear extracts from NIH3T3 cells transfected with a Foxp2 expression plasmid in vitro or cross-linked chromatin from AT1-like cells in vivo, respectively. These nuclear extracts formed a shifted DNA–protein complex with a labeled oligonucleotide encompassing the conserved Foxp2 site from the SP-C promoter (Figure 7B, lanes 2 and 7). The complex could be competed with unlabeled probe (Figure 7B, lanes 3 and 4) and by anti-Foxp2 Ab (lane 8), but not by mutated probe (lanes 5 and 6) or IgG (lane 9). Furthermore, ChIP assays showed that Foxp2 occupies its conserved motif at the SP-C promoter in primary AEC cultures. This occupancy is not observed in AT2 cells on D1, but is clearly present on D8 after transdifferentiation to the AT1 cell-like phenotype (Figure 7C).
Figure 7.
Foxp2 binds to a conserved consensus site in the SP-C promoter in vitro. (A) Conserved Foxp2 consensus binding site among rat, mouse, and human. (B) EMSA was performed using nuclear extracts (NE) from NIH3T3 cells transfected with pCMV/Tag2B/Foxp2 and a 33P-labeled oligonucleotide probe spanning the conserved FoxP2 binding site in the rat SP-C promoter. Arrow indicates formation of a shifted complex between Foxp2 and the labeled probe (lanes 2 and 7), which can be attenuated by 50- and 100-fold excess cold competitor (lanes 3 and 4) and FoxP2 Ab (lane 8), but not by mutated oligonuceotides (m1 and m2) (lanes 5 and 6) or IgG (lane 9). Lane 1 is probe only. (C) FoxP2 interacts with putative binding site in the SP-C promoter by chromatin immunoprecipitation (ChIP). ChIP assays were performed with soluble chromatin harvested from AEC on D1 and D8. PCR was performed after IP with either a polyclonal anti-Foxp2 Ab (lane 5) or control rabbit IgG (lane 4) with primers amplifying the FoxP2 consensus binding sequence in the rat SP-C promoter. Input designates PCR product derived from chromatin that was not subjected to IP. Different dilutions of input DNA demonstrate that PCR products are within the dynamic range (lanes 1–3). Lane 6 indicates PCR without template DNA. Results are representative of three separate experiments.
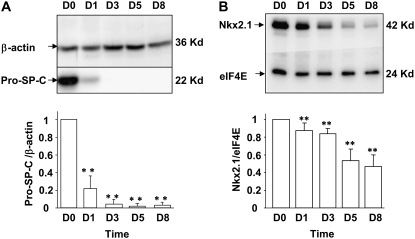
Reciprocal Relationship of FoxP2 and Nkx2.1 during AT2 to AT1 Cell Transition In Vitro
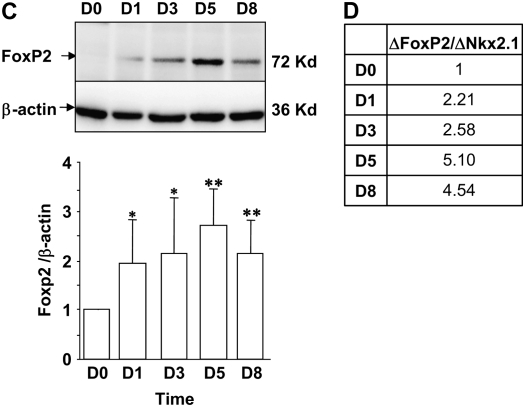
We examined changes in expression of FoxP2 and Nkx2.1 accompanying transition of rat AT2 cells toward the AT1 cell phenotype (5, 6) by Western analysis. As shown in Figure 8A, pro–SP-C is expressed at high levels in freshly isolated AT2 cells and declines dramatically by D1. Nkx2.1 protein is abundantly expressed in freshly isolated AT2 cells and declines gradually over time in culture through D8 (Figure 8B). In contrast, FoxP2 protein is not expressed on D0, increases markedly by D1, and remains elevated through D8 (Figure 8C). FoxP2 increases 2.21-fold between D0 and D1 relative to the decline in Nkx2.1 over this period, and this relative ratio increases to approximately 5-fold on D5 and D8 (Figure 8D). Together with the results of EMSA and ChIP, these findings suggest that relative levels of and interactions between FoxP2 and Nkx2.1 may be particularly important for down-regulation of SP-C early in the transition from AT2 to AT1 cell phenotype (between D0 and D3), while direct binding of FoxP2 to its cognate binding site may predominate to repress SP-C on later days in AT1-like cells once Nkx2.1 levels have already declined.
Figure 8.
Expression of SP-C, FoxP2, and Nkx2.1 during transdifferentiation of AEC in vitro. Total protein was harvested from freshly isolated AT2 cells (D0) or from AEC on D1, D3, D5, and D8 in primary culture and analyzed by Western blotting. Representative Western blots from at least three different experiments and densitometric analyses are shown for SP-C (A), Nkx2.1 (B), and FoxP2 (C). D shows the ratio of the change in FoxP2 relative to the change in Nkx2.1 expression levels from D0 to D1, D3, D5, and D8. Data represent mean ± SEM of more than three different experiments. *P < 0.05 and **P < 0.01 compared with D0.
DISCUSSION
To elucidate transcriptional mechanisms involved in down-regulation of SP-C during phenotypic transition of AT2 to AT1 cells, we examined the involvement of the forkhead/winged helix TF Foxp2 and its interactions with the homeodomain protein Nkx2.1, the major transcriptional activator of SP-C. Foxp2 repressed Nkx2.1-mediated activation of the 3.7-kb SP-C promoter in a dose-dependent fashion in transient transfection assays, suggesting that these TF interact. In addition, Foxp2 inhibited Nkx2.1-mediated transactivation of a transiently transfected 318-bp proximal SP-C promoter that lacks a putative Foxp2 consensus site, further suggesting that Foxp2 and Nkx2.1 interact. Using several complementary approaches, we demonstrated that Foxp2 and Nkx2.1 associate through a direct interaction between Foxp2 and the Nkx2.1 homeodomain. Competitive EMSA demonstrated that a GST-Foxp2 fusion protein prevented formation of Nkx2.1–DNA complexes in a dose-dependent manner. However, examination of the human SP-C promoter revealed a putative consensus FoxP binding site at −1724/−1718 that is conserved among rat, mouse, and human, leading us to further examine whether Foxp2 also interacts with this site in vitro and in vivo. Binding of Foxp2 to this site was demonstrated by EMSA as well as ChIP assay in primary AT1 cell-like cultures on D8, suggesting that repression of SP-C by FoxP2 may also involve direct binding to DNA. Western analysis demonstrated reciprocal changes in expression of Nkx2.1 and Foxp2 concurrent with down-regulation of SP-C during transition of AT2 cells toward the AT1 cell-like phenotype in vitro. These results suggest that attenuation of SP-C transcription by FoxP2 over time in culture is likely mediated by both DNA-dependent and -independent mechanisms and leads to a model in which the relative balance between FoxP2 and Nkx2.1 determines cell-specific effects. In this model, in AT2 cells containing abundant Nxk2.1 but low levels of FoxP2, SP-C is transcriptionally activated by Nkx2.1. As cells transition toward the AT1 cell phenotype, increasing levels of FoxP2 lead to greater interaction with Nkx2.1, preventing interaction of the Nkx2.1 homeodomain with DNA and leading to down-regulation of SP-C. Reciprocally, in AT1 cells that express abundant FoxP2 and low or no Nkx2.1, binding of FoxP2 to its cognate DNA-binding site predominates, leading to direct repression of SP-C.
Although Fox proteins generally act as transcriptional activators/co-activators, members of the Foxp subfamily have been shown to function as transcriptional repressors of CC10 and SP-C promoters (11, 20). However, mechanisms underlying repression of SP-C by Foxp2 were not fully elucidated. In the current study, we demonstrate a novel interaction between Foxp2 and Nkx2.1 that disrupts Nkx2.1 binding and attenuates Nkx2.1-mediated transactivation of SP-C. Since Foxp2 does not bind to the Nkx2.1 oligonucleotide used in EMSA, these results suggest that interaction between Foxp2 and Nkx2.1 leads to dissociation of the Nkx2.1 homeodomain from DNA, implicating more than one mechanism of inhibition of SP-C transcription by Foxp2. In a recent study, deletion of Foxp2 in the mouse resulted in defects in postnatal alveolarization, although levels of SP-C were unchanged at E18.5 (41). Lack of an effect on SP-C expression during development in Foxp2−/− mice may reflect functional redundancy among Foxp and possibly other Fox TF that interact with Nkx2.1 (e.g., Foxa1 and FKHR) (36, 42). Indeed, loss of one allele of Foxp1 in addition to a complete loss of Foxp2 leads to increased severity of lung epithelial defects in compound mutants, including respiratory failure at birth (41).
Foxp2 consists of a winged-helix DNA-binding domain and two functionally and structurally independent transcriptional repression subdomains located in the N-terminal region and containing various protein–protein interaction motifs, including a zinc finger and a putative leucine zipper, suggesting the potential for interaction with other TF with either activator or repressor functions (21). Co-regulation of target genes by Fox TF and homeodomain proteins have been shown to play an important role in regulation of tissue-specific gene expression and morphogenesis (43, 44). Interactions among these TF may be cooperative, resulting in enhancement, or antagonistic, leading to attenuation, of DNA binding/transcriptional activity. Occasionally, the same TF can function as either activator or repressor, depending on the specific cellular context (38, 45–47). In this regard, Foxa1 was recently shown to differentially regulate the CC10 and SP-C promoters based on the presence or absence of functional Fox-binding sites in the respective promoters (36). These findings demonstrate that Fox TF can either inhibit or activate transcription depending on the specific cellular context, interacting partner, and target gene.
Nkx2.1 is characterized by a 60–amino acid region involved in DNA binding (the homeodomain), an N-terminal domain and the C-terminal Nk-2–specific domain (NK-2-SD) (48). All three domains have been implicated in recruiting transcriptional regulators that function as co-factors to regulate target gene expression in both a gene- and cell-specific manner (29, 31, 49). Interactions of homeodomain proteins in general, and of Nkx2.1 in particular, with co-factors generally lead to transcriptional activation (30–32). However, two recent reports demonstrate interactions between Nkx2.1 and Smad3 or FOXA1 that attenuate SP-B and SP-C gene transcription, respectively (36, 50). In the current study, we have described a direct interaction between Nkx2.1 and Foxp2 that attenuates transcriptional activity of Nkx2.1, indicating that the particular interacting partner determines gene expression in a cell-specific context. Interaction between Foxp2 and Nkx2.1 was evaluated in several different ways, including co-immunopreciptation, mammalian two-hybrid assay and GST pull-down. Studies demonstrating interactions between Foxp2 and Nkx2.1 were for the most part performed in the MLE-15 cell line, which is of mouse origin. The MLE-15 cell line was used since it is one of the few distal epithelial cell lines that express both Nkx2.1 and SP-C. However, changes in levels of expression of TF and SP-C were evaluated in primary rat AEC (Figures 7C and 8) because they have been better characterized as a model of AEC transdifferentiation in vitro than mouse AEC. Ongoing studies, however, suggest that mouse and rat AEC behave similarly when cultured on appropriate substrata, forming high-resistance monolayers and demonstrating reciprocal changes in phenotype-specific markers over time. Two-hybrid assay demonstrated a significant increase in luciferase activity of the reporter construct pFR-luc (Figure 3) after cotransfection of pM3/Nkx2.1 and pVP16/Foxp2. Some activation of the luciferase reporter was observed after co-transfection of pM3/Nkx2.1 and pVP16 compared with pM3 or pVP16 individually. A similar increase was observed in a previous study investigating interactions between Foxa2 and Nkx2.1 (36) using the same pM3/Nkx2.1 construct, suggesting that Nkx2.1 alone or as part of the fusion protein may have the ability to bind to VP16. Nevertheless, this effect is significantly augmented by co-transfection of pVP16/Foxp2 consistent with interaction between Foxp2 and Nkx2.1. This interaction was further confirmed by GST pull-down assay. In this instance, Foxp2 interacted exclusively with the Nkx2.1 homeodomain, which is frequently characteristic of interactions between Fox TF and homeodomain proteins, although other co-factors have been shown to variably interact with N-terminal, C-terminal, or homeodomain of Nkx2.1 (29, 31, 49).
In summary, we demonstrate that, in addition to its ability to directly repress SP-C through binding to its cognate binding site in the SP-C promoter (11), Foxp2 interacts with the Nkx2.1 homeodomain to inhibit Nkx2.1-DNA binding and transcriptional stimulation of the SP-C promoter through a DNA-independent mechanism. As mentioned above, Foxp2 has been shown to repress basal transcriptional activity of the SP-C promoter by approximately 40% (11). Foxp2 has two inherent transcriptional repression subdomains, supporting its ability to repress gene expression by direct interaction with target promoters. Foxp2–DNA complex formation by EMSA and more robust occupancy of the SP-C promoter by FoxP2 on D8 than D1 indicate that Foxp2 confers transcriptional repression in part through direct interaction with its cognate binding site in a DNA-dependent manner. In addition, Foxp2 inhibits transcriptional activation of SP-C by Nkx2.1 by preventing interaction of Nkx2.1 with its cognate DNA binding site. Since Foxp2 itself does not bind to the Nkx2.1 oligonucleotide (Figure 6, lane 4), these findings suggest that interaction of Foxp2 with the Nkx2.1 homeodomain provokes its dissociation from DNA and down-regulation of SP-C. We speculate that both DNA-dependent and -independent mechanisms function coordinately to regulate SP-C, and that the relative levels of expression of FoxP2 and Nkx2.1 determine which mechanism predominates in a particular cellular context. Specifically, when levels of FoxP2 are high relative to Nkx2.1 (e.g., in AT1 cells), DNA-dependent mechanisms predominate due to greater binding of FoxP2 to its cognate site as shown by ChIP. When levels of FoxP2 are low relative to Nkx2.1 (e.g., in AT2 cells), SP-C is activated by Nkx2.1. During transdifferentiation from AT2 to AT1 cell phenotype (e.g., following lung injury), when levels of FoxP2 and Nkx2.1 are intermediate, FoxP2 prevents binding of Nkx2.1 to DNA through interactions with the homeodomain, leading to down-regulation of SP-C in a DNA-independent manner. These results suggest a mechanism whereby reciprocal expression of FoxP2 and Nkx2.1, and differential interactions between them, regulate expression of SP-C in a cell-specific fashion to ensure suppression of AT2 cell-associated genes during transition to the AT1 cell phenotype.
Acknowledgments
The authors note with appreciation the expert technical assistance of Juan Ramon Alvarez and Monica Flores.
This work was supported by the Hastings Foundation and National Institutes of Health Research Grants DE-10742 (to D.K.A.), DE-14183 (to D.K.A.), HL-38578 (to Z.B.), HL-38621 (to E.D.C.), HL-56590 (to P.M.), HL-62659 (to Z.B.), HL-64365 (to E.D.C.), HL-73471 (to P.M.), HL-071589 (to E.E.M.), and HL-075334 (to C.L.). E.E.M. is supported by the American Heart Association. E.D.C. is Hastings Professor and Kenneth T. Norris Jr. Chair of Medicine. B.F. is J. Harold and Edna L. LaBriola Chair in Genetic Orthopaedic Research. Z.B. is Ralph Edgington Chair in Medicine.
Originally Published in Press as DOI: 10.1165/rcmb.2007-0350OC on January 31, 2008
Conflict of Interest Statement: None of the authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Crapo JD, Barry BE, Gehr P, Bachofen M, Weibel ER. Cell number and cell characteristics of the normal human lung. Am Rev Respir Dis 1982;126:332–337. [DOI] [PubMed] [Google Scholar]
- 2.Evans MJ, Cabral LJ, Stephens RJ, Freeman G. Renewal of alveolar epithelium in the rat following exposure to NO2. Am J Pathol 1973;70:175–198. [PMC free article] [PubMed] [Google Scholar]
- 3.Adamson IY, Bowden DH. The type 2 cell as progenitor of alveolar epithelial regeneration. A cytodynamic study in mice after exposure to oxygen. Lab Invest 1974;30:35–42. [PubMed] [Google Scholar]
- 4.Adamson IY, Bowden DH. Derivation of type 1 epithelium from type 2 cells in the developing rat lung. Lab Invest 1975;32:736–745. [PubMed] [Google Scholar]
- 5.Diglio CA, Kikkawa Y. The type II epithelial cells of the lung: IV. Adaption and behavior of isolated type II cells in culture. Lab Invest 1977;37:622–631. [PubMed] [Google Scholar]
- 6.Danto SI, Zabski SM, Crandall ED. Reactivity of alveolar epithelial cells in primary culture with type I cell monoclonal antibodies. Am J Respir Cell Mol Biol 1992;6:296–306. [DOI] [PubMed] [Google Scholar]
- 7.Brody JS, Williams MC. Pulmonary alveolar epithelial cell differentiation. Annu Rev Physiol 1992;54:351–371. [DOI] [PubMed] [Google Scholar]
- 8.Cheek JM, Evans MJ, Crandall ED. Type I cell-like morphology in tight alveolar epithelial monolayers. Exp Cell Res 1989;184:375–387. [DOI] [PubMed] [Google Scholar]
- 9.Carlsson P, Mahlapuu M. Forkhead transcription factors: key players in development and metabolism. Dev Biol 2002;250:1–23. [DOI] [PubMed] [Google Scholar]
- 10.Mazet F, Yu JK, Liberles DA, Holland LZ, Shimeld SM. Phylogenetic relationships of the Fox (Forkhead) gene family in the Bilateria. Gene 2003;316:79–89. [DOI] [PubMed] [Google Scholar]
- 11.Shu W, Yang H, Zhang L, Lu MM, Morrisey EE. Characterization of a new subfamily of winged-helix/forkhead (Fox) genes that are expressed in the lung and act as transcriptional repressors. J Biol Chem 2001;276:27488–27497. [DOI] [PubMed] [Google Scholar]
- 12.Perrone L, Pasca di Magliano M, Zannini M, Di Lauro R. The thyroid transcription factor 2 (TTF-2) is a promoter-specific DNA-binding independent transcriptional repressor. Biochem Biophys Res Commun 2000;275:203–208. [DOI] [PubMed] [Google Scholar]
- 13.Whitsett JA, Tichelaar JW. Forkhead transcription factor HFH-4 and respiratory epithelial cell differentiation. Am J Respir Cell Mol Biol 1999;21:153–154. [DOI] [PubMed] [Google Scholar]
- 14.Ikeda K, Shaw-White JR, Wert SE, Whitsett JA. Hepatocyte nuclear factor 3 activates transcription of thyroid transcription factor 1 in respiratory epithelial cells. Mol Cell Biol 1996;16:3626–3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stroud JC, Wu Y, Bates DL, Han A, Nowick K, Paabo S, Tong H, Chen L. Structure of the forkhead domain of FOXP2 bound to DNA. Structure 2006;14:159–166. [DOI] [PubMed] [Google Scholar]
- 16.Banham AH, Beasley N, Campo E, Fernandez PL, Fidler C, Gatter K, Jones M, Mason DY, Prime JE, Trougouboff P, et al. The FOXP1 winged helix transcription factor is a novel candidate tumor suppressor gene on chromosome 3p. Cancer Res 2001;61:8820–8829. [PubMed] [Google Scholar]
- 17.Vargha-Khadem F, Gadian DG, Copp A, Mishkin M. FOXP2 and the neuroanatomy of speech and language. Nat Rev Neurosci 2005;6:131–138. [DOI] [PubMed] [Google Scholar]
- 18.Lai CS, Fisher SE, Hurst JA, Vargha-Khadem F, Monaco AP. A forkhead-domain gene is mutated in a severe speech and language disorder. Nature 2001;413:519–523. [DOI] [PubMed] [Google Scholar]
- 19.Bettelli E, Dastrange M, Oukka M. Foxp3 interacts with nuclear factor of activated T cells and NF-kappa B to repress cytokine gene expression and effector functions of T helper cells. Proc Natl Acad Sci USA 2005;102:5138–5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu MM, Li S, Yang H, Morrisey EE. Foxp4: A novel member of the Foxp subfamily of winged-helix genes co-expressed with Foxp1 and Foxp2 in pulmonary and gut tissues. Gene Expr Patterns 2002;2:223–228. [DOI] [PubMed] [Google Scholar]
- 21.Li S, Weidenfeld J, Morrisey EE. Transcriptional and DNA binding activity of the Foxp1/2/4 family is modulated by heterotypic and homotypic protein interactions. Mol Cell Biol 2004;24:809–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bruno MD, Bohinski RJ, Huelsman KM, Whitsett JA, Korfhagen TR. Lung cell-specific expression of the murine surfactant protein A (SP-A) gene is mediated by interactions between the SP-A promoter and thyroid transcription factor-1. J Biol Chem 1995;270:6531–6536. [DOI] [PubMed] [Google Scholar]
- 23.Li C, Cai J, Pan Q, Minoo P. Two functionally distinct forms of NKX2.1 protein are expressed in the pulmonary epithelium. Biochem Biophys Res Commun 2000;270:462–468. [DOI] [PubMed] [Google Scholar]
- 24.Morotti RA, Gutierrez MC, Askin F, Profitt SA, Wert SE, Whitsett JA, Greco MA. Expression of thyroid transcription factor-1 in congenital cystic adenomatoid malformation of the lung. Pediatr Dev Pathol 2000;3:455–461. [DOI] [PubMed] [Google Scholar]
- 25.Zhou L, Lim L, Costa RH, Whitsett JA. Thyroid transcription factor-1, hepatocyte nuclear factor-3beta, surfactant protein B, C, and Clara cell secretory protein in developing mouse lung. J Histochem Cytochem 1996;44:1183–1193. [DOI] [PubMed] [Google Scholar]
- 26.Stahlman MT, Gray ME, Whitsett JA. Expression of thyroid transcription factor-1(TTF-1) in fetal and neonatal human lung. J Histochem Cytochem 1996;44:673–678. [DOI] [PubMed] [Google Scholar]
- 27.Kelly SE, Bachurski CJ, Burhans MS, Glasser SW. Transcription of the lung-specific surfactant protein C gene is mediated by thyroid transcription factor 1. J Biol Chem 1996;271:6881–6888. [DOI] [PubMed] [Google Scholar]
- 28.Glasser SW, Burhans MS, Eszterhas SK, Bruno MD, Korfhagen TR. Human SP-C gene sequences that confer lung epithelium-specific expression in transgenic mice. Am J Physiol 2000;278:L933–L945. [DOI] [PubMed] [Google Scholar]
- 29.Liu C, Glasser SW, Wan H, Whitsett JA. GATA-6 and thyroid transcription factor-1 directly interact and regulate surfactant protein-C gene expression. J Biol Chem 2002;277:4519–4525. [DOI] [PubMed] [Google Scholar]
- 30.Bachurski CJ, Yang GH, Currier TA, Gronostajski RM, Hong D. Nuclear factor I/thyroid transcription factor 1 interactions modulate surfactant protein C transcription. Mol Cell Biol 2003;23:9014–9024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park KS, Whitsett JA, Di Palma T, Hong JH, Yaffe MB, Zannini M. Taz interacts with TTF-1 and regulates expression of surfactant protein-c. J Biol Chem 2004;279:17384–17390. [DOI] [PubMed] [Google Scholar]
- 32.Lin S, Perl AK, Shannon JM. Erm/thyroid transcription factor 1 interactions modulate surfactant protein C transcription. J Biol Chem 2006;281:16716–16726. [DOI] [PubMed] [Google Scholar]
- 33.Mann RS, Chan SK. Extra specificity from extradenticle: The partnership between HOX and PBX/EXD homeodomain proteins. Trends Genet 1996;12:258–262. [DOI] [PubMed] [Google Scholar]
- 34.Foucher I, Montesinos ML, Volovitch M, Prochiantz A, Trembleau A. Joint regulation of the MAP1B promoter by HNF3beta/Foxa2 and Engrailed is the result of a highly conserved mechanism for direct interaction of homeoproteins and fox transcription factors. Development 2003;130:1867–1876. [DOI] [PubMed] [Google Scholar]
- 35.Nakano T, Murata T, Matsuo I, Aizawa S. OTX2 directly interacts with LIM1 and HNF-3beta. Biochem Biophys Res Commun 2000;267:64–70. [DOI] [PubMed] [Google Scholar]
- 36.Minoo P, Hu L, Xing Y, Zhu NL, Chen H, Li M, Borok Z, Li C. Physical and functional interactions between homeodomain NKX2.1 and winged helix/forkhead FOXA1 in lung epithelial cells. Mol Cell Biol 2007;27:2155–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Borok Z, Danto SI, Zabski SM, Crandall ED. Defined medium for primary culture de novo of adult rat alveolar epithelial cells. In Vitro Cell Dev Biol 1994;30A:99–104. [DOI] [PubMed] [Google Scholar]
- 38.Wikenheiser KA, Vorbroker DK, Rice WR, Clark JC, Bachurski CJ, Oie HK, Whitsett JA. Production of immortalized distal respiratory epithelial cell lines from surfactant protein C/simian virus 40 large tumor antigen transgenic mice. Proc Natl Acad Sci USA 1993;90:11029–11033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou B, Ann DK, Li X, Kim KJ, Lin H, Minoo P, Crandall ED, Borok Z. Hypertonic induction of aquaporin-5: novel role of hypoxia-inducible factor-1alpha. Am J Physiol 2007;292:C1280–C1290. [DOI] [PubMed] [Google Scholar]
- 40.Crighton D, Woiwode A, Zhang C, Mandavia N, Morton JP, Warnock LJ, Milner J, White RJ, Johnson DL. P53 represses RNA polymerase III transcription by targeting TBP and inhibiting promoter occupancy by TFIIIB. EMBO J 2003;22:2810–2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shu W, Lu MM, Zhang Y, Tucker PW, Zhou D, Morrisey EE. Foxp2 and Foxp1 cooperatively regulate lung and esophagus development. Development 2007;134:1991–2000. [DOI] [PubMed] [Google Scholar]
- 42.Zhong Q, Zhou B, Minno P, Ann DK, Crandall ED, Borok Z. Role of forkhead homologue in rhabdomysosarcoma (FKHR) in regulation of alveolar epithelial cell (AEC) transdifferentiation in vitro. Am J Respir Crit Care Med 2007;175:A340. [Google Scholar]
- 43.Marshak S, Benshushan E, Shoshkes M, Havin L, Cerasi E, Melloul D. Functional conservation of regulatory elements in the pdx-1 gene: PDX-1 and hepatocyte nuclear factor 3beta transcription factors mediate beta-cell-specific expression. Mol Cell Biol 2000;20:7583–7590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jonsson H, Peng SL. Forkhead transcription factors in immunology. Cell Mol Life Sci 2005;62:397–409. [DOI] [PubMed] [Google Scholar]
- 45.Foucher I, Volovitch M, Frain M, Kim JJ, Souberbielle JC, Gan L, Unterman TG, Prochiantz A, Trembleau A. Hoxa5 overexpression correlates with IGFBP1 upregulation and postnatal dwarfism: evidence for an interaction between Hoxa5 and Forkhead box transcription factors. Development 2002;129:4065–4074. [DOI] [PubMed] [Google Scholar]
- 46.Kim JJ, Taylor HS, Akbas GE, Foucher I, Trembleau A, Jaffe RC, Fazleabas AT, Unterman TG. Regulation of insulin-like growth factor binding protein-1 promoter activity by FKHR and HOXA10 in primate endometrial cells. Biol Reprod 2003;68:24–30. [DOI] [PubMed] [Google Scholar]
- 47.Sawaya PL, Luse DS. Two members of the HNF-3 family have opposite effects on a lung transcriptional element; HNF-3 alpha stimulates and HNF-3 beta inhibits activity of region I from the Clara cell secretory protein (CCSP) promoter. J Biol Chem 1994;269:22211–22216. [PubMed] [Google Scholar]
- 48.Watada H, Mirmira RG, Kalamaras J, German MS. Intramolecular control of transcriptional activity by the NK2-specific domain in NK-2 homeodomain proteins. Proc Natl Acad Sci USA 2000;97:9443–9448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yan C, Naltner A, Martin M, Naltner M, Fangman JM, Gurel O. Transcriptional stimulation of the surfactant protein B gene by STAT3 in respiratory epithelial cells. J Biol Chem 2002;277:10967–10972. [DOI] [PubMed] [Google Scholar]
- 50.Li C, Zhu NL, Tan RC, Ballard PL, Derynck R, Minoo P. Transforming growth factor-beta inhibits pulmonary surfactant protein B gene transcription through SMAD3 interactions with NKX2.1 and HNF-3 transcription factors. J Biol Chem 2002;277:38399–38408. [DOI] [PubMed] [Google Scholar]