Abstract
Sre1, the fission yeast sterol regulatory element binding protein, is an endoplasmic reticulum membrane-bound transcription factor that responds to changes in oxygen-dependent sterol synthesis as an indirect measure of oxygen availability. Under low oxygen, Sre1 is proteolytically cleaved and the released N-terminal transcription factor (Sre1N) activates gene expression essential for hypoxic growth. Here, we describe an oxygen-dependent mechanism for regulation of Sre1 that is independent of sterol-regulated proteolysis. Using yeast expressing only Sre1N, we show that Sre1N turnover is regulated by oxygen. Ofd1, an uncharacterized prolyl 4-hydroxylase-like 2-oxoglutarate-Fe(II) dioxygenase, accelerates Sre1N degradation in the presence of oxygen. However, unlike the prolyl 4-hydroxylases that regulate mammalian hypoxia-inducible factor, Ofd1 uses multiple domains to regulate Sre1N degradation by oxygen; the Ofd1 N-terminal dioxygenase domain is required for oxygen sensing and the Ofd1 C-terminal domain accelerates Sre1N degradation. Our data support a model whereby the Ofd1 N-terminal dioxygenase domain is an oxygen sensor that regulates the activity of the C-terminal degradation domain.
Keywords: degradation, hydroxylase, oxygen, SREBP, yeast
Introduction
Most eukaryotic cells, from unicellular organisms to cells in mammalian tissues, experience fluctuating environmental oxygen concentrations. To adapt to these changes in cellular oxygen supply, cells must detect changes in oxygen concentration and elicit a physiological response to allow the organism or tissue to adapt. In mammals and yeast, transcription factors have been identified that activate a programme of gene expression to promote cell growth and survival in response to a decrease in oxygen (Emerling and Chandel, 2005).
In mammals, the central regulator of hypoxic gene expression is the heterodimeric transcription factor hypoxia-inducible factor (HIF), which consists of one of three α-subunits (HIF-1α, HIF-2α, and HIF-3α) and one β-subunit (HIF-1β) (for reviews see Schofield and Ratcliffe, 2005; Gordan and Simon, 2007). Both the degradation and activity of the HIF-α subunit are regulated by oxygen-dependent post-translational hydroxyl modifications performed by enzymes belonging to the 2-oxoglutarate (OG)-Fe(II) dioxygenase family (Ozer and Bruick, 2007). Under normoxic conditions, HIF-α is hydroxylated on two proline residues by a family of three conserved 2-OG-Fe(II)-dependent prolyl 4-hydroxylases, named PHD1–3 (Dann and Bruick, 2005). Hydroxylated HIF-1α is recognized by the von Hippel–Lindau (VHL) protein-containing E3 ligase, ubiquitinylated, and degraded by proteasome (Kaelin, 2005). Transcriptional activation by HIF-1 is further controlled by FIH, an oxygen-dependent, 2-OG-Fe(II)-dependent asparaginyl hydroxylase that hydroxylates HIF-1α and inhibits recruitment of the coactivators p300/CBP, preventing full activation of HIF-1 target genes (Hirota and Semenza, 2005). Hypoxia inhibits these 2-OG-Fe(II) dioxygenases, leading to the accumulation of the HIF-α subunit and expression of genes essential for adaptation to hypoxia.
In fission yeast, the Sre1 transcription factor is a principal regulator of low oxygen gene expression (Todd et al, 2006). Yeast Sre1 is the orthologue of mammalian sterol regulatory element binding protein (SREBP), a basic helix–loop–helix leucine zipper, endoplasmic reticulum membrane-bound transcription factor that regulates expression of genes involved in cholesterol and lipid synthesis (Hughes et al, 2005; Espenshade and Hughes, 2007). When the cellular concentration of sterols is high, SREBP is retained in the endoplasmic reticulum in an inactive form. When sterol levels decrease, SREBP and its binding partner SREBP cleavage-activating protein (SCAP) are transported to the Golgi where SREBP is proteolytically cleaved, allowing the N-terminal transcription factor to enter the nucleus and activate genes required for cholesterol synthesis and uptake (Goldstein et al, 2006).
Our recent studies in fission yeast revealed that Sre1 is proteolytically activated in response to low oxygen (Hughes et al, 2005). Upon shifting to low-oxygen conditions, oxygen-dependent sterol synthesis is inhibited and Sre1 is proteolytically cleaved, releasing the N-terminal transcription factor domain (Sre1N) (Hughes et al, 2005, 2007). Sre1N upregulates expression of genes required for low-oxygen growth (Hughes et al, 2005). Microarray studies revealed that Sre1 controls the expression of 68% of the genes that are upregulated >2-fold under anaerobic conditions, indicating that Sre1 is an important regulator of low-oxygen gene expression (Todd et al, 2006).
Sre1 activation occurs in response to either chemical inhibition of sterol synthesis or when sterol synthesis is inhibited by low-oxygen conditions (Hughes et al, 2005). Interestingly, accumulation of Sre1N is more rapid and robust when sterol synthesis is inhibited by low oxygen than by chemical inhibitors, despite equivalent decreases in sterol synthesis (Hughes et al, 2005). These data suggest that while sterol depletion activates Sre1 proteolysis, oxygen depletion may additionally regulate Sre1N by a sterol-independent mechanism.
In this study, we investigated the mechanism of oxygen-dependent, sterol-independent regulation of Sre1N. Using an engineered yeast strain that expresses only Sre1N, we show that Sre1N is stabilized under low oxygen. We demonstrate that the 2-OG-Fe(II) dioxygenase Ofd1 mediates this oxygen-dependent response by accelerating the turnover of Sre1N in the presence of oxygen. However, in contrast to the HIF 2-OG-Fe(II) dioxygenases, Ofd1 uses separate domains to sense oxygen and regulate Sre1N turnover. The Ofd1 N-terminal dioxygenase domain is required for oxygen sensing whereas the C-terminal domain accelerates Sre1N degradation. Our data suggest that the Ofd1 N-terminal dioxygenase domain controls the activity of its C-terminal degradation domain in response to oxygen availability by an autoregulatory mechanism.
Results
Oxygen regulates Sre1N independently of proteolysis
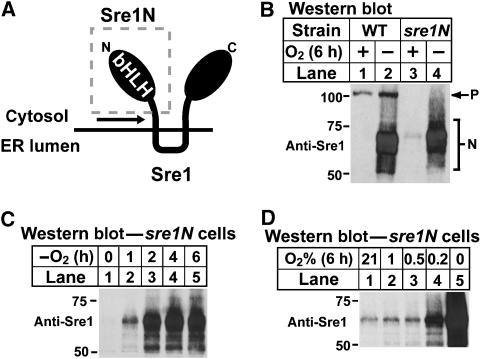
To examine whether oxygen regulates Sre1 by a sterol-independent mechanism, we generated a yeast strain that expressed only the truncated nuclear form of Sre1 by inserting a stop codon before the first predicted transmembrane domain of Sre1 at the endogenous sre1+ locus (Figure 1A). This strain, designated sre1N, expresses the soluble nuclear form of Sre1 (Sre1N, amino acids (aa) 1–440) but lacks the full-length precursor form of Sre1. Sre1N is sufficient to rescue the low-oxygen sensitivity of sre1Δ and scp1Δ yeast (Hughes et al, 2005). To determine whether oxygen regulates Sre1N, wild-type and sre1N cells were grown in the presence or absence of oxygen, and Sre1N was assayed by western blotting. Sre1N migrates as a broad smear on SDS–PAGE due to hyperphosphorylation of the cleaved transcription factor. As previously reported (Hughes et al, 2005), in wild-type cells Sre1 was cleaved in response to low oxygen (Figure 1B, lanes 1 and 2). Surprisingly, in sre1N cells, Sre1N increased dramatically under low-oxygen conditions (Figure 1B, lanes 3 and 4). The increase in Sre1N in the absence of oxygen was independent of proteolysis as this strain does not express the precursor form of Sre1.
Figure 1.
Sre1N accumulates under low oxygen. (A) Diagram of Sre1. A stop codon was inserted after aa 440 directly before the first predicted transmembrane domain to generate strain sre1N. bHLH denotes the N-terminal transcription factor domain of Sre1. (B) Wild-type and sre1N cells were grown in the presence or absence of oxygen for 6 h. P and N denote the precursor and nuclear forms, respectively. (C) sre1N cells were grown for increasing times in the absence of oxygen. (D) sre1N cells were grown at the indicated oxygen concentration for 6 h. Whole-cell lysates were subjected to western blot analysis with an antibody directed against the N-terminus of Sre1 (anti-Sre1 IgG). For all figures, molecular mass markers (kDa) are shown.
To examine the kinetics of Sre1N accumulation under low oxygen, we grew sre1N cells in the absence of oxygen for increasing periods of time. Sre1N induction occurred within 1 h and increased to 4–6 h in the absence of oxygen (Figure 1C). To determine the oxygen concentration at which Sre1N accumulated, we grew sre1N cells at decreasing oxygen concentrations (Figure 1D). Sre1N increased in the absence of oxygen and at 0.2% oxygen, but not 0.5 or 1% oxygen, suggesting that regulation of Sre1N by oxygen occurred between 0.2 and 0.5% oxygen. Taken together, these results indicate that oxygen regulates Sre1N by a mechanism that is independent of Sre1 precursor proteolysis.
Sre1N is stabilized under low oxygen
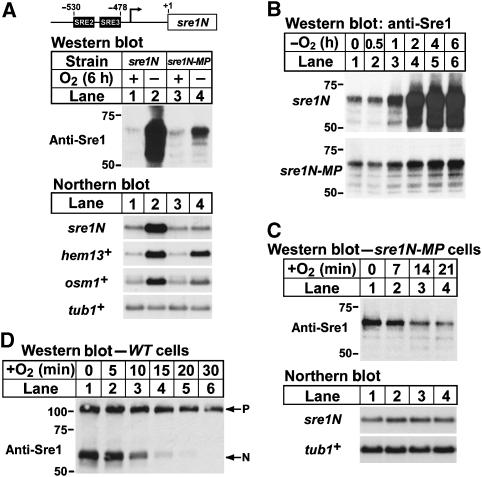
Sre1N activates its own transcription in a positive feedback loop and maximal upregulation of Sre1N in the absence of oxygen requires this positive feedback regulation (Hughes et al, 2005; Todd et al, 2006). Therefore, the increase in Sre1N under low oxygen could be due to oxygen-dependent regulation of sre1N transcription, translation, or Sre1N degradation. To determine if the low-oxygen induction of Sre1N required an increase in sre1N transcription, we mutated the two Sre1N binding sites (SRE2 and SRE3) in the sre1N promoter required for positive feedback regulation and designated this strain sre1N-MP (mutant promoter) (Figure 2A) (Todd et al, 2006). sre1N cells grown under low oxygen for 6 h showed a dramatic induction of both Sre1N protein and mRNA (Figure 2A, lanes 1 and 2). Mutating the SRE2 and SRE3 sequence elements in the sre1N promoter blocked the oxygen-dependent induction of sre1N mRNA (Figure 2A, lanes 3 and 4). Despite the loss of sre1N transcriptional regulation, Sre1N still increased in the absence of oxygen (Figure 2A, lanes 3 and 4). Consistent with the induction of Sre1N in sre1N-MP cells, expression of the Sre1 target genes hem13+ and osm1+ increased in sre1N-MP cells in the absence of oxygen (Todd et al, 2006). This oxygen-dependent induction of Sre1N in sre1N-MP cells occurred between 0.5 and 1 h and was maximal 4–6 h after a shift to low-oxygen conditions (Figure 2B). The kinetics of Sre1N induction were identical in sre1N and sre1N-MP cells (Figure 2B), but the amplitude of the induction differed due to the loss of positive feedback regulation in sre1N-MP cells. These results suggest that the low-oxygen induction of Sre1N is due to a post-transcriptional mechanism and that positive feedback at the sre1N promoter is required for maximal Sre1N expression under low oxygen.
Figure 2.
Sre1N is stabilized under low oxygen. (A) Yeast cells expressing Sre1N from either the wild-type (sre1N) or SRE2+3 mutated sre1N promoter (sre1N-MP) were grown in the presence or absence of oxygen for 6 h. (B) sre1N or sre1N-MP cells were grown for increasing times in the absence of oxygen. (C) sre1N-MP cells were grown in the absence of oxygen for 6 h. At t=0, cells were shifted to the presence of oxygen and samples were collected at the indicated times. (D) Wild-type cells were grown for 6 h in the absence of oxygen. At t=0, cultures were shifted to the presence of oxygen and samples were collected at the indicated times. Whole-cell extracts were subjected to western blot analysis using anti-Sre1 IgG and total RNA (5 μg) was subjected to northern analysis with the indicated 32P-labelled probes. α-Tubulin (tub1+) mRNA served as a loading control.
Next, we investigated whether oxygen regulates Sre1N at the level of protein translation or degradation. Polysome profiling experiments indicated that sre1+ mRNA is not subject to translational control by oxygen (Sehgal et al, 2008), suggesting that oxygen may regulate Sre1N at the level of protein turnover. To determine whether oxygen affects Sre1N stability, we examined Sre1N in sre1N-MP cells where sre1N mRNA is not regulated by oxygen. sre1N-MP cells were grown in the absence of oxygen to increase Sre1N. Cells were then shifted to the presence of oxygen at t=0 and samples were harvested at the indicated times. No cycloheximide was added to inhibit protein translation. sre1N mRNA was constant, whereas Sre1N decreased rapidly in the presence of oxygen (Figure 2C). Together with the results in Figure 2B, these data suggest that oxygen regulates Sre1N degradation.
To confirm that Sre1N generated from the Sre1 precursor by proteolysis is also rapidly degraded in the presence of oxygen, wild-type cells were grown in the absence of oxygen to induce Sre1 cleavage. Cells were then shifted to the presence of oxygen and samples were collected after increasing times. Reintroduction of oxygen inhibits Sre1 proteolysis (data not shown), thereby preventing production of Sre1N and allowing us to monitor the turnover of Sre1N. On oxygen addition, Sre1N decreased rapidly, indicating that Sre1N is unstable in the presence of oxygen (Figure 2D). Collectively, these results indicate that Sre1N is a short-lived protein whose stability is regulated by oxygen.
Sre1N requires the proteasome for degradation
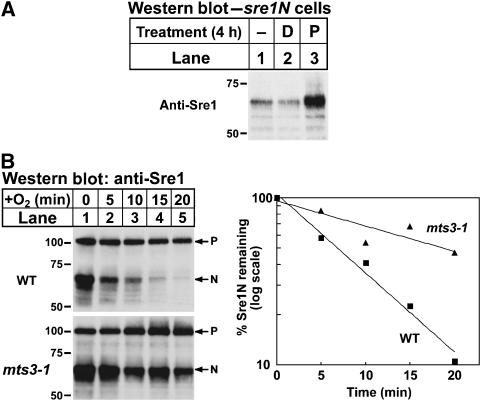
As Sre1N is rapidly turned over in the presence of oxygen, next we tested whether the proteasome is required for Sre1N degradation. sre1N cells were grown in the presence or absence of a proteasome inhibitor and Sre1N was assayed by western blotting. Treatment with the proteasome inhibitor increased Sre1N, whereas treatment with DMSO (the vehicle for the proteasome inhibitor) had no effect (Figure 3A). To test genetically if Sre1N turnover required the proteasome, we assayed Sre1N degradation in wild-type or mts3-1 cells containing a temperature-sensitive proteasome mutation (Gordon et al, 1996). Wild-type and mts3-1 cells expressing full-length Sre1 were cultured in the absence of oxygen for 4 h at 30°C to activate Sre1 cleavage and accumulate Sre1N. Cells were then incubated in the absence of oxygen at the non-permissive temperature (35.5°C) for 2 h to inhibit proteasome function in mts3-1 cells. Finally, cycloheximide was added, cells were shifted to the presence of oxygen to inhibit Sre1 proteolysis at 35.5°C, and whole-cell lysates were collected after increasing times. Sre1N was more stable in mts3-1 cells and displayed a half-life of 20 min compared with 6 min in wild-type cells (Figure 3B). Together, these results indicate that Sre1N degradation requires the proteasome.
Figure 3.
Sre1N is degraded by the proteasome. (A) sre1N cells were grown ±2% DMSO (D) or 200 μM proteasome inhibitor II (P) for 4 h. (B) Wild-type and mts3-1 cells were grown in the absence of oxygen at 30°C for 4 h. The hypoxic workstation was shifted to 35.5°C for an additional 2 h. At t=0, cycloheximide (100 μg/ml) was added and cells were shifted to the presence of oxygen at 35.5°C. Samples were collected at the indicated times. The percent of Sre1N remaining at each time point relative to t=0 is quantified to the right. Whole-cell lysates were subjected to western blot analysis using anti-Sre1 IgG. P denotes the precursor form of Sre1 and N denotes the nuclear form.
Ofd1 is a 2-OG-Fe(II) dioxygenase that negatively regulates Sre1N
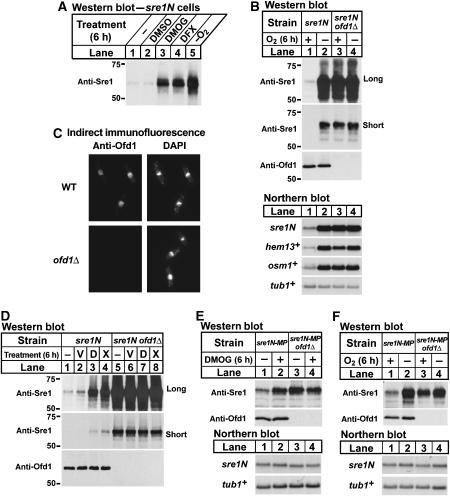
Degradation of the mammalian hypoxic transcription factor HIF-1α is controlled by a family of 2-OG-Fe(II)-dependent dioxygenases. These oxygen-dependent prolyl hydroxylases are inhibited under low oxygen, leading to an increase in HIF-1α protein and transcription of genes required for hypoxic adaptation (Ozer and Bruick, 2007). To test whether Sre1N is regulated by a 2-OG-Fe(II) dioxygenase, we cultured sre1N cells in the presence of specific (dimethyloxalylglycine (DMOG)) or nonspecific (deferoxamine (DFX)) inhibitors of 2-OG-Fe(II) dioxygenases. Treatment with DMOG or DFX or growing cells under low oxygen increased Sre1N, whereas treatment with DMSO (the vehicle for DMOG) had no effect (Figure 4A). These data suggest that Sre1N is negatively regulated by a 2-OG-Fe(II) dioxygenase.
Figure 4.
Ofd1 is a 2-OG-Fe(II) dioxygenase that negatively regulates Sre1N. (A) sre1N cells were grown for 6 h ±0.5% DMSO, 10 mM DMOG, or 200 μM DFX, or for 6 h in the absence of oxygen. (B) sre1N and sre1N ofd1Δ cells were grown in the presence or absence of oxygen for 6 h. (C) Wild-type and ofd1Δ cells were analysed by indirect immunofluorescence using affinity-purified anti-Ofd1 antibody and 4′,6-diamidino-2-phenylindole (DAPI) to stain DNA. (D) sre1N and sre1N ofd1Δ cells were grown for 6 h in the presence or absence of 0.5% DMSO (V), 10 mM DMOG (D), or 200 μM DFX (X). (E) sre1N-MP and sre1N-MP ofd1Δ cells were grown in the presence of 0.5% DMSO or 10 mM DMOG for 6 h. (F) sre1N-MP and sre1N-MP ofd1Δ cells were grown in the presence or absence of oxygen for 6 h. Whole-cell extracts were subjected to western blot analysis using anti-Sre1 IgG or anti-Ofd1 IgG as indicated. (B, D) Both long and short exposures to film are shown. Total RNA (5 μg) was subjected to northern analysis with the indicated 32P-labelled probes. α-Tubulin (tub1+) mRNA served as a loading control.
Transcription factors often control expression of their own regulators to elicit feedback control (Stiehl et al, 2006). Two Sre1-dependent genes identified by microarray analysis belong to the 2-OG-Fe(II) dioxygenase class of enzymes and these genes emerged as candidate regulators of Sre1N (Aravind and Koonin, 2001; Todd et al, 2006). We refer to these genes as ofd1+ (SPBC6B1.08c) and ofd2+ (SPAP8A3.02c), for 2-oxoglutarate and Fe(II) dioxygenase 1 and 2. We confirmed that ofd1+ and ofd2+ mRNAs increased under low oxygen and that this increase required Sre1 (Supplementary Figure S1A). In addition, Ofd1 protein increased under low oxygen (Supplementary Figure S1B).
To determine whether Ofd1 or Ofd2 regulates Sre1N, we cultured sre1N, sre1N ofd1Δ, and sre1N ofd2Δ cells in the presence or absence of oxygen and assayed Sre1N by western blotting. As expected, low oxygen increased Sre1N in sre1N cells (Figure 4B, lanes 1 and 2). Surprisingly, deletion of ofd1+ dramatically increased Sre1N in the presence of oxygen (Figure 4B, compare lanes 1 and 3). Growth of sre1N ofd1Δ cells under low oxygen did not further increase Sre1N, suggesting that the oxygen-dependent regulation of Sre1N required ofd1+ (Figure 4B, lanes 3 and 4, short exposure). Expression of Sre1N target genes, including sre1N itself, mirrored levels of Sre1N (Figure 4B, lower panels). Deletion of ofd2+ had no effect on regulation of Sre1N levels by oxygen (Supplementary Figure S2). These results indicate that Ofd1 is a negative regulator of Sre1N in the presence of oxygen.
To determine the intracellular localization of endogenous Ofd1, we performed indirect immunofluorescence using an affinity-purified Ofd1 antibody. Ofd1 was concentrated in the nucleus with some cytosolic staining (Figure 4C). Staining was specific as ofd1Δ cells showed no signal. Recent global protein localization studies confirmed this result and revealed that an Ofd1–YFP fusion protein localized primarily to the nucleus (Matsuyama et al, 2006). These data indicate that Sre1N and Ofd1 localize to the same subcellular compartment.
To investigate whether the 2-OG-Fe(II) dioxygenase inhibitors act through Ofd1 to increase Sre1N, we cultured sre1N and sre1N ofd1Δ cells in the presence of DMOG or DFX. As expected, Sre1N increased in response to the inhibitors DMOG and DFX in cells expressing Ofd1 (Figure 4D, lanes 1–4). However, these inhibitors had no effect on Sre1N in ofd1Δ cells (Figure 4D, lanes 5–8, short exposure). These results suggest that DMOG and DFX act through Ofd1 to regulate Sre1N.
To determine if Ofd1 regulates Sre1N by a post-transcriptional mechanism, we examined the function of Ofd1 in sre1N-MP cells lacking positive feedback regulation of sre1N transcription. We cultured sre1N-MP and sre1N-MP ofd1Δ cells with DMOG (Figure 4E) or under low oxygen (Figure 4F). sre1N mRNA was unchanged, whereas Sre1N increased after treatment with DMOG or growth under low oxygen (Figure 4E and F, lanes 1 and 2). Importantly, deletion of ofd1+ increased Sre1N in sre1N-MP cells without a change in sre1N mRNA (Figure 4E and F, compare lanes 1 and 3). Consistent with the results in Figure 4B and D, regulation by DMOG and low oxygen was abrogated in cells lacking Ofd1 (Figure 4E and F). Collectively, these results demonstrate that Ofd1 is a negative regulator of Sre1N in the presence of oxygen and that regulation of Sre1N by low oxygen and DMOG requires Ofd1.
Ofd1 accelerates Sre1N degradation
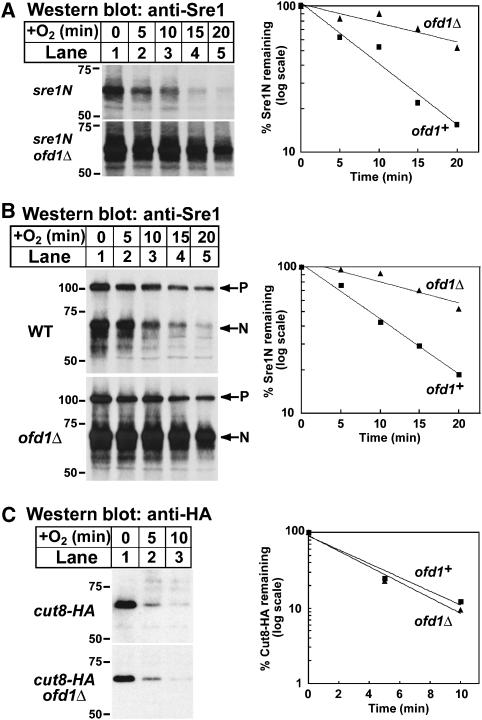
Based on our data that oxygen regulates Sre1N turnover and the role of the HIF 2-OG-Fe(II) dioxygenases in protein stability, we tested whether Ofd1 regulates the turnover of Sre1N. To assay the stability of Sre1N in the presence of oxygen, we grew sre1N and sre1N ofd1Δ cells in the absence of oxygen to accumulate equivalent amounts of Sre1N. Cycloheximide was added to inhibit protein translation, cells were shifted to the presence of oxygen, and whole-cell lysates were collected after increasing times. Sre1N half-life increased in sre1N ofd1Δ cells (t1/2=23 min) compared with sre1N cells (t1/2=7 min), indicating that Ofd1 accelerated Sre1N turnover (Figure 5A). To test whether Ofd1 regulated the physiological degradation of Sre1N, we assayed the turnover of Sre1N generated from the Sre1 precursor by proteolysis. In Figure 5B, wild-type and ofd1Δ cells expressing full-length Sre1 were cultured in the absence of oxygen to activate Sre1 cleavage and accumulate Sre1N. Cycloheximide was added, cells were shifted to the presence of oxygen to inhibit Sre1 proteolysis, and whole-cell lysates were collected after increasing times. Consistent with the results in sre1N cells, Sre1N half-life increased from 8 min in wild-type cells to 21 min in ofd1Δ cells. The stabilization of Sre1N was specific insomuch as turnover of another short-lived nuclear protein Cut8-HA was unchanged in ofd1Δ cells (Figure 5C) (Takeda and Yanagida, 2005). Taken together, these data demonstrate that Ofd1 accelerates the degradation of Sre1N.
Figure 5.
Ofd1 accelerates Sre1N turnover in the presence of oxygen. (A) sre1N and sre1N ofd1Δ cells were grown in the absence of oxygen for 6 h. At t=0, cycloheximide (100 μg/ml) was added and cells were shifted to the presence of oxygen. Samples were collected at the indicated times. (B) Wild-type and ofd1Δ cells were grown in the absence of oxygen for 6 h. At t=0, cycloheximide (100 μg/ml) was added and cells were shifted to the presence of oxygen. Samples were collected at the indicated times. P denotes the precursor form of Sre1 and N denotes the nuclear form. (C) Wild-type and ofd1Δ cells expressing Cut8-HA were grown in the absence of oxygen for 6 h. At t=0, cycloheximide (100 μg/ml) was added and cells were shifted to the presence of oxygen. Samples were collected at the indicated times. (A–C) Whole-cell extracts were subjected to western blot analysis using anti-Sre1 IgG or anti-HA IgG as indicated. The percent of Sre1N or Cut8-HA remaining at each time point relative to t=0 is quantified to the right; ofd1+ (squares) and ofd1Δ (triangles).
Ofd1 dioxygenase domain is required for oxygen regulation but not Sre1N degradation
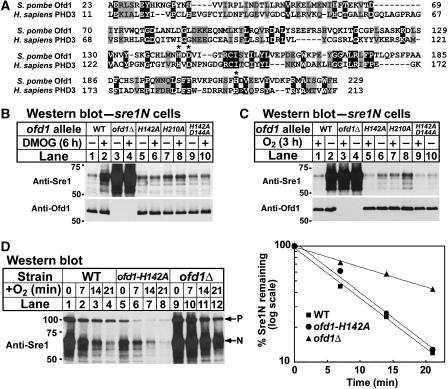
Ofd1 belongs to a family of conserved uncharacterized 2-OG-Fe(II) dioxygenase enzymes and a Conserved Database Domain search revealed that Ofd1 is most similar to prolyl 4-hydroxylase-like 2-OG-Fe(II) dioxygenases (Aravind and Koonin, 2001). 2-OG-Fe(II) dioxygenases contain three iron-coordinating residues contained in an HXD/E…H motif (Clifton et al, 2006). Alignment of the prolyl 4-hydroxylase domain of Schizosaccharomyces pombe Ofd1 and the human HIF prolyl 4-hydroxylase PHD3 revealed that these three iron-coordinating residues are conserved in S. pombe Ofd1 (Figure 6A). Mutation of any one of these residues abolished the activity of the HIF prolyl hydroxylase (Bruick and McKnight, 2001). To test if the prolyl 4-hydroxylase domain (hereafter referred to as the dioxygenase domain) is required for Ofd1 activity, we generated sre1N strains containing mutations of the Ofd1 iron-coordinating residues at the endogenous ofd1+ locus and assayed Sre1N in cells treated with DMOG (Figure 6B) or grown under low oxygen (Figure 6C). If the enzymatic activity of the Ofd1 dioxygenase domain is required for accelerating Sre1N degradation in the presence of oxygen, Ofd1 iron-coordinating mutants should have increased Sre1N, similar to ofd1Δ cells (Figure 4B). Surprisingly, mutation of the iron-coordinating residues had no effect on Sre1N in untreated cells, suggesting that dioxygenase activity is not required for Sre1N degradation (Figure 6B and C). However, unlike cells expressing wild-type Ofd1, treatment of Ofd1 iron-coordinating mutants with DMOG (Figure 6B) or under low oxygen (Figure 6C) failed to increase Sre1N, suggesting that the function of the dioxygenase domain had been disrupted. Thus, iron-coordinating mutations in the dioxygenase domain of Ofd1 did not block Sre1N degradation, but rather created a constitutively active Ofd1 that was no longer inhibited by DMOG or low oxygen. Consistent with this conclusion, the Ofd1 iron-coordinating mutants accelerated Sre1N turnover as rapidly as wild-type Ofd1 (Figure 6D). Collectively, these results indicate that the dioxygenase domain of Ofd1 is required for DMOG and oxygen regulation of Sre1N stability, but not for accelerating Sre1N degradation.
Figure 6.
Prolyl 4-hydroxylase domain of Ofd1 is required for oxygen sensing, but not Sre1N degradation. (A) Sequence alignment of the prolyl 4-hydroxylase domains of S. pombe Ofd1 (aa 23–229) and human HIF PHD3 (NP_071356) (aa 11–213). Identical residues are shaded in black and similar residues are shaded in grey. Residues predicted to be required for iron coordination are indicated by an asterisk. (B) sre1N, sre1N ofd1Δ, sre1N ofd1-H142A, sre1N ofd1-H210A, or sre1N ofd1-H142A;D144A cells were grown in 0.5% DMSO or 10 mM DMOG for 6 h. (C) sre1N, sre1N ofd1Δ, sre1N ofd1-H142A, sre1N ofd1-H210A, or sre1N ofd1-H142A;D144A cells were grown in the presence or absence of oxygen for 3 h. (D) sre1N, sre1N ofd1-H142A, or sre1N ofd1Δ cells were grown in the absence of oxygen for 6 h. At t=0, cycloheximide (100 μg/ml) was added and cells were shifted to the presence of oxygen. Samples were collected at the indicated times. P denotes the precursor form of Sre1 and N denotes the nuclear form. The percent of Sre1N remaining at each time point relative to t=0 is quantified to the right. Whole-cell extracts were subjected to western blot analysis using anti-Sre1 IgG or anti-Ofd1 IgG as indicated.
Ofd1 C-terminus accelerates Sre1N degradation
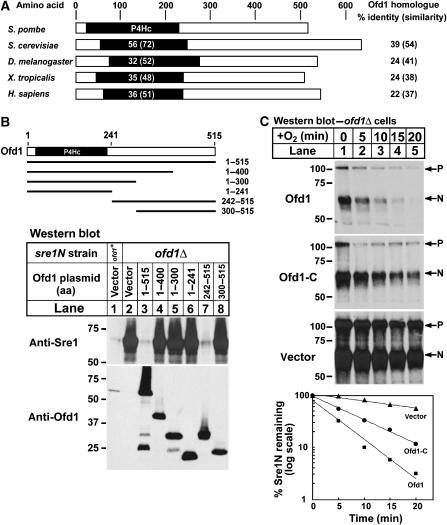
Given that the dioxygenase domain of Ofd1 was not required for accelerating Sre1N degradation, another domain of Ofd1 must perform this function. Sequence analysis and alignments of the eukaryotic Ofd1 homologues indicated that Ofd1 homologues are ∼500 aa and contain a prolyl 4-hydroxylase domain at the N-terminus (Figure 7A). Notably, the domain architecture of Ofd1 differs from the HIF prolyl hydroxylase family, placing Ofd1 in a distinct family of 2-OG-Fe(II) dioxygenases (Aravind and Koonin, 2001). To determine which region of Ofd1 is required for degrading Sre1N, Ofd1 truncations were expressed from a plasmid in sre1N or sre1N ofd1Δ cells in the presence of oxygen. Expression of full-length Ofd1 (aa 1–515) decreased Sre1N levels, but Ofd1 with a C-terminal deletion (Ofd1 aa 1–400) failed to decrease Sre1N (Figure 7B, lanes 1–4). Interestingly, expression of the Ofd1 C-terminus (aa 242–515) alone decreased Sre1N levels (Figure 7B, lane 7). An intact C-terminal domain was required as expression of Ofd1 (aa 300–515) failed to decrease Sre1N (Figure 7B, lane 8). Thus, the N-terminal dioxygenase domain is not required for Sre1N degradation.
Figure 7.
The C-terminus of Ofd1 is required for accelerating Sre1N degradation. (A) Domain structure of Ofd1 homologues from S. pombe, S. cerevisiae Tpa1p, D. melanogaster CG31120-PA, X. tropicalis LOC549076, and H. sapiens OGFOD1. The black box denotes the prolyl-4-hydroxylase (P4Hc) domain. The percent identity (similarity) of the P4Hc domain of each homologue to the P4Hc domain of S. pombe Ofd1 is shown in the black box. The percent identity (similarity) to S. pombe Ofd1 across the entire length of each homologue is shown to the right. (B) The indicated amino acids of Ofd1 were expressed from a plasmid using the thiamine-repressible Nmt* promoter in sre1N (lane 1) or sre1N ofd1Δ cells (lanes 2–8). Cells were grown in the presence of oxygen in minimal medium lacking thiamine to induce expression of Ofd1. (C) ofd1Δ cells containing an Nmt* plasmid expressing full-length Ofd1, the C-terminus of Ofd1 (Ofd1-C, aa 242–515), or empty vector were grown overnight in minimal medium lacking thiamine to induce expression of Ofd1. Cells were shifted to rich medium and grown in the absence of oxygen for 6 h. At t=0, cycloheximide (100 μg/ml) was added and cells were shifted to the presence of oxygen. Samples were collected at the indicated times. The percent of Sre1N remaining at each time point relative to t=0 is quantified below. Whole-cell lysates were subjected to western blot analysis using anti-Sre1 IgG or anti-Ofd1 IgG.
To test if the Ofd1 C-terminus is sufficient to accelerate Sre1N degradation, ofd1Δ cells containing plasmids expressing full-length Ofd1, the Ofd1 C-terminal domain (aa 242–515), or empty vector were assayed for Sre1N degradation. Cells were grown in the absence of oxygen for 6 h to accumulate Sre1N. Cycloheximide was added, cells were shifted to the presence of oxygen, and whole-cell lysates were collected after increasing times. Cells lacking Ofd1 degraded Sre1N slowly (Figure 7C, vector; t1/2=23 min). Expression of the Ofd1 C-terminal domain accelerated Sre1N turnover (Figure 7C, t1/2=7 min), matching the kinetics in wild-type sre1N cells (Figure 5A), and overexpression of full-length Ofd1 further accelerated Sre1N degradation (Figure 7C, t1/2=4 min). These results demonstrate that the Ofd1 C-terminal domain (aa 242–515) is both necessary and sufficient to accelerate Sre1N degradation.
Dual regulation of Sre1 by oxygen
Our data are consistent with a model in which oxygen acts through two distinct mechanisms to control Sre1 in fission yeast. First, Sre1N must be proteolytically released from the membrane (Figure 8A, control point 1). In the presence of oxygen, this cleavage is inhibited by oxygen-dependent sterol synthesis. When sterol synthesis decreases under low oxygen, Sre1N is proteolytically released from the membrane (Hughes et al, 2005, 2007). Second, the 2-OG-Fe(II) dioxygenase Ofd1 accelerates the degradation of Sre1N in the presence of oxygen (Figure 8A, control point 2). Under low oxygen, Sre1N degradation is inhibited and Sre1N accumulates. The rapid increase in Sre1N observed in response to low oxygen is due to regulation of Sre1 at both control points.
Figure 8.
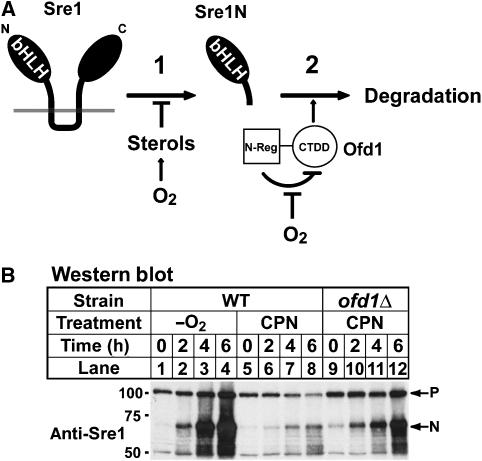
Oxygen regulates Sre1 by two distinct mechanisms. (A) Model for regulation of Sre1 by oxygen. Sre1 proteolysis is controlled by sterol synthesis, which requires oxygen (1). Under low oxygen, sterol synthesis is inhibited and Sre1 proteolysis is upregulated, leading to increased Sre1N. In addition, degradation of Sre1N is regulated by oxygen (2). In the absence of oxygen, the Ofd1 N-Reg inhibits the Ofd1 CTDD, leading to increased Sre1N stability. In the presence of oxygen, N-Reg inhibition is blocked and Ofd1 CTDD accelerates Sre1N degradation. (B) Wild-type or ofd1Δ cells were grown in the absence of oxygen or treated with 200 μM compactin (CPN) for the indicated times. Whole-cell lysates were subjected to western blot analysis using anti-Sre1 IgG.
Previously, we observed that the kinetics and amplitude of Sre1 activation in response to low oxygen were more robust than Sre1 activation in response to chemical inhibition of sterol synthesis (Figure 8B, lanes 1–4 versus lanes 5–8) (Hughes et al, 2005). Based on the proposed model, the blunted response in chemically treated cells may be due to the continued degradation of Sre1N by Ofd1 in the presence of oxygen. Accordingly, the model predicts that Sre1 activation by chemical inhibition in ofd1Δ cells in the presence of oxygen should mirror Sre1 activation in wild-type cells under low oxygen. To test this prediction, wild-type and ofd1Δ cells were grown in the presence of the sterol synthesis inhibitor compactin or in the absence of oxygen for increasing times. Activation of Sre1 in ofd1Δ cells treated with compactin (Figure 8B, lanes 9–12) was more rapid and robust than in wild-type cells treated with compactin (Figure 8B, lanes 5–8) and was comparable to wild-type cells grown in the absence of oxygen (Figure 8B, lanes 1–4). These results confirm a prediction of the model proposed in Figure 8A and demonstrate that oxygen acts through two distinct mechanisms to regulate Sre1.
Discussion
Inhibition of sterol synthesis by low oxygen stimulates proteolysis of the Sre1 membrane-bound transcription factor (Hughes et al, 2005). In this study, we describe a second mechanism by which oxygen regulates Sre1 and identify Sre1N as a target of a new family of 2-OG-Fe(II) dioxygenase. Using a yeast strain expressing the N-terminal transcription factor domain of Sre1, we found that Sre1N accumulates under low oxygen and is rapidly degraded by the proteasome in the presence of oxygen (Figures 1, 2 and 3). The prolyl 4-hydroxylase-like 2-OG-Fe(II) dioxygenase Ofd1 mediates this oxygen-dependent regulation by accelerating the degradation of Sre1N in the presence of oxygen (Figures 4 and 5). However, in contrast to the prolyl 4-hydroxylases that regulate HIF-α activity, Ofd1 uses two distinct domains to regulate Sre1N degradation: the Ofd1 N-terminal dioxygenase domain is required for oxygen regulation (Figure 6) and the Ofd1 C-terminus (aa 242–515) accelerates Sre1N degradation (Figure 7).
Based on these findings, we propose a mechanism whereby the Ofd1 C-terminal degradation domain (CTDD) (aa 242–515) accelerates Sre1N degradation and the N-terminal dioxygenase domain is an oxygen-sensing regulatory domain (N-Reg) that controls the activity of the CTDD (Figure 8A). In the absence of oxygen and dioxygenase activity, the N-Reg domain inhibits the CTDD, leading to increased Sre1N stability. Conversely, in the presence of oxygen and dioxygenase activity, this inhibition is lost, leading to increased Sre1N degradation through the Ofd1 CTDD. In this way, Ofd1 acts as both an oxygen sensor and an effector to regulate Sre1N levels in response to changes in environmental oxygen.
The identification of the 2-OG-Fe(II) dioxygenase Ofd1 as a regulator of Sre1N stability leads to a model in which oxygen acts at two control points to regulate Sre1N. Oxygen-dependent sterol synthesis regulates Sre1 cleavage and requires fission yeast SCAP, Scp1 (Figure 8A, control point 1) (Hughes et al, 2005, 2007). Additionally, oxygen acts through Ofd1 to regulate Sre1N stability (Figure 8A, control point 2). This dual regulation of Sre1 facilitates rapid activation of hypoxic gene expression and adaptation of cells to changes in oxygen supply. Additionally, Sre1N induces expression of sre1+ mRNA, increasing the levels of Sre1 precursor protein (Todd et al, 2006). The positive feedback loop results in a dramatic upregulation of Sre1N in the absence of oxygen (Figure 8B, lanes 1–4) (Hughes et al, 2005). Conversely, when oxygen increases, sterol synthesis resumes, Sre1 proteolysis is blocked, and Sre1N is rapidly degraded, turning off hypoxia-specific transcription (Figure 5B).
Ofd1 accelerates Sre1N degradation, but the mechanism of Ofd1 action is unknown. The presence of a conserved prolyl 4-hydroxylase domain in Ofd1 suggests that Ofd1 may act as a protein hydroxylase. In mammals, the HIF PHDs hydroxylate HIF-α on two proline residues and target the α-subunit for ubiquitinylation and degradation by the proteasome (Dann and Bruick, 2005). However, in contrast to the HIF hydroxylases, the Ofd1 N-Reg dioxygenase domain is not required for Sre1N degradation. Instead, the Ofd1 CTDD is sufficient to accelerate Sre1N degradation. Therefore, in fission yeast, hydroxylation of Sre1N is not a prerequisite for degradation. Consistent with this, sequence database searches revealed that fission yeast lacks a homologue of VHL, the E3 ligase that recognizes the hydroxyproline signal in HIF-α. Whether regulation of Ofd1 by the N-Reg dioxygenase domain requires hydroxylation of a protein substrate remains to be determined. In addition, although Sre1N is degraded by the proteasome (Figure 3), understanding how the Ofd1 CTDD accelerates Sre1N turnover requires additional studies.
Several lines of evidence suggest that the Ofd1 homologues define a functionally distinct family of 2-OG-Fe(II) dioxygenases that have a broad role in eukaryotic biology: (1) Ofd1 uses separate domains to respond to oxygen availability and accelerate Sre1N degradation, suggesting that Ofd1 has autoregulatory properties. (2) There is no sequence conservation outside of the prolyl 4-hydroxylase domain between Ofd1 and the collagen prolyl hydroxylases, the HIF PHDs, or the Dictyostelium Skp1 prolyl hydroxylase (van der wel et al, 2005). (3) Sequence database searches identified homologues of Ofd1 in a wide range of eukaryotes from fungi to humans (Figure 7A) (Aravind and Koonin, 2001). (4) A homologue of Ofd1 is present in Saccharomyces cerevisiae, which does not contain a conserved Sre1 pathway, indicating that Ofd1 likely functions in other pathways. The S. cerevisiae Ofd1 homologue, Tpa1p, has been implicated in the control of translation termination, mRNA poly(A) tail length, and mRNA stability (Keeling et al, 2006), but the requirement for the dioxygenase domain has not been investigated. Future studies will focus on understanding how this unique 2-OG-Fe(II) dioxygenase regulates the CTDD to control degradation of Sre1N in response to oxygen.
Materials and methods
Materials
Yeast extract was obtained from Becton Dickinson and Co.; amino acids were from Sigma; horseradish peroxidase-conjugated affinity-purified donkey anti-rabbit and anti-mouse immunoglobulin G (IgG) were from Jackson ImmunoResearch; anti-HA 12CA5 mouse monoclonal antibody was from Roche; oligonucleotides were from Integrated DNA Technologies; DFX was from Sigma; compactin (mevastatin) and proteasome inhibitor II was from Calbiochem; and DMOG was from Frontier Scientific Inc.
Yeast strains and culture
Wild-type haploid S. pombe KGY425 (h−, his3-D1, leu1-32, ura4-D18, ade6-M210) and KGY461 (h+, his3-D1, leu1-32, ura4-D18, ade6-M210) were obtained from American Type Culture Collection (Burke and Gould, 1994). S. pombe strain sre1Δ has been described previously (Hughes et al, 2005). S. pombe strains ofd1Δ, ofd2Δ, and cut8-3xHA were generated from wild-type haploid yeast (KGY425 or KGY461) by homologous recombination using established techniques (Bahler et al, 1998). S. pombe strain sre1N contains a precise deletion of the sequence coding for Sre1 aa 441–900 and does not contain a selectable marker. To generate sre1N, we first created an sre1Δ∷ura4+ strain lacking the entire open reading frame (ORF) by homologous recombination. A PCR product containing base pairs 1–1320 of the sre1+ ORF followed by a TAA stop codon was amplified from an sre1N plasmid template using oligonucleotides containing 79 bp of homology to sequences directly upstream (sre1N forward) or downstream (sre1N reverse) of the sre1+ ORF. This PCR product was transformed into the sre1Δ∷ura4+ strain and transformants were selected on minimal medium containing 5-fluoroorotic acid (5-FOA) to counter-select for expression of the ura4+ gene product (Boeke et al, 1987). Transformants were screened by PCR and confirmed by sequencing.
Strain sre1N-MP (MP, mutant promoter) had the Sre1 DNA binding sequences SRE2 and SRE3 mutated in the sre1N promoter to prevent positive feedback regulation by Sre1N. The sequence of SRE2 was changed from 5′-ATCACCCCAT-3′ to 5′-ATATACCATA-3′ and the sequence of SRE3 was changed from 5′-GTCAGTCCAC-3′ to 5′-GTATATCATA-3′. To generate sre1N-MP, base pairs −550 to −450 upstream of the sre1+ ORF were replaced with ura4+ by homologous recombination. This strain was subsequently transformed with an 850 bp PCR product containing sequences directly upstream of the sre1+ ORF amplified using oligonucleotides SRE2+3F and SRE2+3R from an SRE2*SRE3* mutant plasmid template previously described (Todd et al, 2006). Transformants were selected on minimal medium containing 5-FOA to counter-select for expression of the ura4+ gene product (Boeke et al, 1987). Transformants were screened by PCR and confirmed by sequencing.
Strains containing mutations in Ofd1 iron-coordinating residues at the endogenous ofd1+ locus were generated by the pop-in/pop-out replacement method (Rothstein, 1991). Briefly, Ofd1 iron-coordinating mutant integrating plasmids were linearized with AscI, transformed into wild-type strain KGY425, and cells were grown on minimal medium lacking uracil. Integration at the ofd1+ locus was confirmed by Southern blotting. Strains containing integrated Ofd1 mutant plasmid were plated on minimal medium containing 5-FOA to select for strains that excised the ura4+ integrated marker. Mutations in Ofd1 iron-coordinating residues were confirmed by Southern blotting and DNA sequencing. Oligonucleotides used to generate the above strains are described in Supplementary Table S1. Standard genetic techniques were used to generate S. pombe strains not described above (Moreno et al, 1991).
Yeast strains were grown to exponential phase at 30°C in yeast extract plus supplements (225 μg/ml each of histidine, leucine, adenine, lysine, and uracil) or in Edinburgh minimal medium where indicated using standard techniques (Hughes et al, 2005). Anaerobic growth conditions were maintained using an In vivo2 400 workstation (Biotrace Inc.) as described previously (Hughes et al, 2005; Todd et al, 2006).
Plasmids
Ofd1 iron-coordinating mutant integrating plasmids were created by splice overlap PCR and contained the ofd1+ ORF plus 500 bp of flanking sequence at each end, an AscI site after base pair 200 of the ofd1+ ORF, and the ura4+ HindIII fragment from pREP4X inserted into pBluescript II SK(+) (Forsburg, 1993). DNA was amplified from genomic DNA or plasmid templates containing Ofd1 iron-coordinating mutations (H142A: CAT to GCT; H210A: CAT to GCT; H142A D144A: CAT to GCT and GAT to GCC) previously introduced by QuikChange PCR mutagenesis (Stratagene). Plasmids containing N- or C-terminal deletions of Ofd1 expressed from the thiamine-repressible Nmt* promoter were generated by insertion of ofd1+ PCR fragments into XhoI–SmaI sites of pREP41X (Forsburg, 1993).
Antibody preparation
Polyclonal antiserum against full-length Ofd1 was generated by immunizing rabbits with bacterially expressed antigen using a standard protocol (Covance). A bacterial expression plasmid for Ofd1 was created by cloning PCR products generated using an S. pombe Lindner 972 genomic DNA template into the expression vector pGEX-KG (Guan and Dixon, 1991). Antigen containing an N-terminal glutathione-S-transferase (GST) fusion to Ofd1 was purified from Escherichia coli using glutathione agarose (Sigma) and then cleaved by thrombin (Novagen) to remove the GST tag. Ofd1-specific antibodies were isolated from rabbit serum by affinity chromatography using the AminoLink Plus Immobilization Kit (Pierce) according to the manufacturer's instructions.
Indirect immunofluorescence
Immunofluorescent staining was performed as previously described with modification (Hagan and Hyams, 1988). Cells were fixed by the addition of 1/9 volume of 37% formaldehyde. After 1 min, 1/9 volume of 2% glutaraldehyde was added. Cultures were incubated for 1 h at 30°C, then centrifuged, resuspended, and washed in PEM (0.1 M PIPES, pH 6.9, 1 mM EGTA, 1 mM MgSO4). Cells were resuspended in PEMS (PEM with 1.2 M sorbitol) at 5 × 107 cells/ml. Zymolyase 100T (Seikagaku) was added to 0.5 mg/ml and incubated at room temperature for 1 h to remove the cell wall. Cells were pelleted and resuspended in 1% Triton X-100 in PEMS for 10 min and washed in PEM. Unreacted aldehydes were quenched by washing once with a fresh solution of PEM containing 1 mg/ml sodium borohydride (Sigma). Cells were washed in PEM, resuspended in PEMBAL (PEM with 1% bovine serum albumin (fatty acid and globulin free), 0.1% sodium azide, and 0.1 M L-lysine hydrochloride), and incubated at room temperature for 30 min. Cells were incubated with affinity-purified Ofd1 (4 μg/ml) overnight at room temperature and washed three times in PEMBAL. Cells were then incubated with 20 μg/ml goat anti-rabbit Alexa 488 antibody (Molecular Probes) at room temperature for 4 h. Cells were washed three times in PEMBAL and once in phosphate-buffered saline (PBS). Then, the cells were resuspended in PBS with 0.2 μg/ml DAPI (Molecular Probes). Cells were washed in PBS and mounted on coverslips coated with poly-L-lysine (1 mg/ml).
Northern and western blotting
Total RNA isolation and northern blot analysis were performed as described previously (Hughes et al, 2005). PCR fragments for ofd1+, ofd2+, sre1N, and tub1+ used for northern probe synthesis were generated using gene-specific primer pairs (Supplementary Table S1). Probes for hem13+ and osm1+ have been described previously (Todd et al, 2006). Blots were exposed to film and/or a PharosFX Plus Molecular Imager (Bio-Rad) and quantified using Quantity One Software (Bio-Rad) where indicated. Whole-cell yeast extract preparation and western blot analysis using anti-Sre1 IgG and horseradish peroxidase-conjugated anti-rabbit IgG were performed as described previously (Hughes et al, 2005). Anti-Ofd1 IgG serum was used at 1:10 000 dilution.
Protein alignments
The homologues of Ofd1 were identified and the percent identity/similarity was determined using the position-specific iterated basic local alignment search tool (PSI-BLAST) from the NCBI database with two iterations (Altschul et al, 1997, 2005). P4Hc domains were identified using the Conserved Domain Database available from NCBI (Marchler-Bauer et al, 2007). The amino acids corresponding to the P4Hc domain of each orthologue used for sequence alignment were S. pombe Ofd1 (NP_596087) (aa 23–229), S. cerevisiae Tpa1p (NP_010969) (aa 53–246), Drosophila CG31120-PA (NP_733061) (aa 78–274), X. tropicalis LOC549076 (NP_001016322) (aa 49–239), and human OGFOD1 (NP_060703) (aa 61–238). Sequence alignment of the prolyl 4-hydroxylase domains of S. pombe Ofd1 and human HIF PHD3 was determined by PSI-BLAST using three iterations.
Measurement of protein half-life
For protein half-life analysis, indicated yeast strains were grown in the absence of oxygen to exponential phase and then shifted to aerobic conditions in the presence of cycloheximide (100 μg/ml) (Sigma). Samples were collected and whole-cell lysates (40 μg) were subjected to western blot analysis. Blots were developed using the HyGLO HRP Detection Kit (Denville Scientific), exposed to film, and imaged using the Versadoc Imaging System (Bio-Rad). Quantification was performed using Quantity One Software (Bio-Rad). An exponential trend line was used to calculate the line slope (k) and the protein half-life was calculated using the equation t1/2=(ln 2)/k (Belle et al, 2006).
Supplementary Material
Supplementary Information
Acknowledgments
We thank members of the Espenshade Lab for experimental suggestions and critical evaluation of this manuscript. We kindly thank the laboratory of Colin Gordon (Medical Research Council, UK) for providing the mts3-1 strain. This work was supported by a grant from the National Institutes of Health (HL-077588) (to PJE). BTH is the recipient of an American Heart Association Predoctoral Fellowship and PJE is the recipient of a Burroughs Wellcome Fund Career Award in the Biomedical Sciences.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Wootton JC, Gertz EM, Agarwala R, Morgulis A, Schaffer AA, Yu YK (2005) Protein database searches using compositionally adjusted substitution matrices. FEBS J 272: 5101–5109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Koonin EV (2001) The DNA-repair protein AlkB, EGL-9, and leprecan define new families of 2-oxoglutarate- and iron-dependent dioxygenases. Genome Biol 2: RESEARCH0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A III, Steever AB, Wach A, Philippsen P, Pringle JR (1998) Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14: 943–951 [DOI] [PubMed] [Google Scholar]
- Belle A, Tanay A, Bitincka L, Shamir R, O'Shea EK (2006) Quantification of protein half-lives in the budding yeast proteome. Proc Natl Acad Sci USA 103: 13004–13009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke JD, Trueheart J, Natsoulis G, Fink GR (1987) 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol 154: 164–175 [DOI] [PubMed] [Google Scholar]
- Bruick RK, McKnight SL (2001) A conserved family of prolyl-4-hydroxylases that modify HIF. Science 294: 1337–1340 [DOI] [PubMed] [Google Scholar]
- Burke JD, Gould KL (1994) Molecular cloning and characterization of the Schizosaccharomyces pombe his3 gene for use as a selectable marker. Mol Gen Genet 242: 169–176 [DOI] [PubMed] [Google Scholar]
- Clifton IJ, McDonough MA, Ehrismann D, Kershaw NJ, Granatino N, Schofield CJ (2006) Structural studies on 2-oxoglutarate oxygenases and related double-stranded beta-helix fold proteins. J Inorg Biochem 100: 644–669 [DOI] [PubMed] [Google Scholar]
- Dann CE III, Bruick RK (2005) Dioxygenases as O2-dependent regulators of the hypoxic response pathway. Biochem Biophys Res Commun 338: 639–647. [DOI] [PubMed] [Google Scholar]
- Emerling BM, Chandel NS (2005) Oxygen sensing: getting pumped by sterols. Sci STKE 2005: pe30. [DOI] [PubMed] [Google Scholar]
- Espenshade PJ, Hughes AL (2007) Regulation of sterol synthesis in eukaryotes. Annu Rev Genet 41: 401–427 [DOI] [PubMed] [Google Scholar]
- Forsburg SL (1993) Comparison of Schizosaccharomyces pombe expression systems. Nucleic Acids Res 21: 2955–2956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein JL, DeBose-Boyd RA, Brown MS (2006) Protein sensors for membrane sterols. Cell 124: 35–46 [DOI] [PubMed] [Google Scholar]
- Gordan JD, Simon MC (2007) Hypoxia-inducible factors: central regulators of the tumor phenotype. Curr Opin Genet Dev 17: 71–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon C, McGurk G, Wallace M, Hastie ND (1996) A conditional lethal mutant in the fission yeast 26 S protease subunit mts3+ is defective in metaphase to anaphase transition. J Biol Chem 271: 5704–5711 [DOI] [PubMed] [Google Scholar]
- Guan KL, Dixon JE (1991) Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal Biochem 192: 262–267 [DOI] [PubMed] [Google Scholar]
- Hagan IM, Hyams JS (1988) The use of cell division cycle mutants to investigate the control of microtubule distribution in the fission yeast Schizosaccharomyces pombe. J Cell Sci 89 (Part 3): 343–357 [DOI] [PubMed] [Google Scholar]
- Hirota K, Semenza GL (2005) Regulation of hypoxia-inducible factor 1 by prolyl and asparaginyl hydroxylases. Biochem Biophys Res Commun 338: 610–616 [DOI] [PubMed] [Google Scholar]
- Hughes AL, Lee CS, Bien CM, Espenshade PJ (2007) 4-Methyl sterols regulate fission yeast SREBP-Scap under low oxygen and cell stress. J Biol Chem 282: 24388–24396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Todd BL, Espenshade PJ (2005) SREBP pathway responds to sterols and functions as an oxygen sensor in fission yeast. Cell 120: 831–842 [DOI] [PubMed] [Google Scholar]
- Kaelin WG (2005) Proline hydroxylation and gene expression. Annu Rev Biochem 74: 115–128 [DOI] [PubMed] [Google Scholar]
- Keeling KM, Salas-Marco J, Osherovich LZ, Bedwell DM (2006) Tpa1p is part of an mRNP complex that influences translation termination, mRNA deadenylation, and mRNA turnover in Saccharomyces cerevisiae. Mol Cell Biol 26: 5237–5248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Anderson JB, Derbyshire MK, DeWeese-Scott C, Gonzales NR, Gwadz M, Hao L, He S, Hurwitz DI, Jackson JD, Ke Z, Krylov D, Lanczycki CJ, Liebert CA, Liu C, Lu F, Lu S, Marchler GH, Mullokandov M, Song JS, Thanki N, Yamashita RA, Yin JJ, Zhang D, Bryant SH (2007) CDD: a conserved domain database for interactive domain family analysis. Nucleic Acids Res 35: D237–D240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama A, Arai R, Yashiroda Y, Shirai A, Kamata A, Sekido S, Kobayashi Y, Hashimoto A, Hamamoto M, Hiraoka Y, Horinouchi S, Yoshida M (2006) ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol 24: 841–847 [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P (1991) Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol 194: 795–823 [DOI] [PubMed] [Google Scholar]
- Ozer A, Bruick RK (2007) Non-heme dioxygenases: cellular sensors and regulators jelly rolled into one? Nat Chem Biol 3: 144–153 [DOI] [PubMed] [Google Scholar]
- Rothstein R (1991) Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol 194: 281–301 [DOI] [PubMed] [Google Scholar]
- Schofield CJ, Ratcliffe PJ (2005) Signalling hypoxia by HIF hydroxylases. Biochem Biophys Res Commun 338: 617–626 [DOI] [PubMed] [Google Scholar]
- Sehgal A, Hughes BT, Espenshade PJ (2008) Oxygen-dependent, alternative promoter controls translation of tco1+ in fission yeast. Nucleic Acids Res 36: 2024–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiehl DP, Wirthner R, Koditz J, Spielmann P, Camenisch G, Wenger RH (2006) Increased prolyl 4-hydroxylase domain proteins compensate for decreased oxygen levels. Evidence for an autoregulatory oxygen-sensing system. J Biol Chem 281: 23482–23491 [DOI] [PubMed] [Google Scholar]
- Takeda K, Yanagida M (2005) Regulation of nuclear proteasome by Rhp6/Ubc2 through ubiquitination and destruction of the sensor and anchor Cut8. Cell 122: 393–405 [DOI] [PubMed] [Google Scholar]
- Todd BL, Stewart EV, Burg JS, Hughes AL, Espenshade PJ (2006) Sterol regulatory element binding protein is a principal regulator of anaerobic gene expression in fission yeast. Mol Cell Biol 26: 2817–2831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der wel H, Ercan A, West CM (2005) The Skp1 prolyl hydroxylase from Dictyostelium is related to the hypoxia-inducible factor-alpha class of animal prolyl 4-hydroxylases. J Biol Chem 280: 14645–14655 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information