Abstract
The Rag1 and Rag2 proteins initiate V(D)J recombination by introducing site-specific DNA double-strand breaks. Cleavage occurs by nicking one DNA strand, followed by a one-step transesterification reaction that forms a DNA hairpin structure. A similar reaction allows Rag transposition, in which the 3′-OH groups produced by Rag cleavage are joined to target DNA. The Rag1 active site DDE triad clearly plays a catalytic role in both cleavage and transposition, but no other residues in Rag1 responsible for transesterification have been identified. Furthermore, although Rag2 is essential for both cleavage and transposition, the nature of its involvement is unknown. Here, we identify basic amino acids in the catalytic core of Rag1 specifically important for transesterification. We also show that some Rag1 mutants with severe defects in hairpin formation nonetheless catalyze substantial levels of transposition. Lastly, we show that a catalytically defective Rag2 mutant is impaired in target capture and displays a novel form of coding flank sensitivity. These findings provide the first identification of components of Rag1 that are specifically required for transesterification and suggest an unexpected role for Rag2 in DNA cleavage and transposition.
INTRODUCTION
Developing lymphocytes rearrange variable (V), diversity (D) and joining (J) gene segments in a combinatorial fashion to generate a diverse repertoire of immunoglobulins and T-cell receptors. This process, called V(D)J recombination (1,2), is initiated when the protein products of the recombination activating genes (Rag)-1 and -2 bind to specific recognition signal sequences (RSS) and introduce double-strand breaks (DSB) between these and the adjacent coding segments. The resulting postcleavage complex then directs the ends to the classical nonhomologous end joining (NHEJ) machinery for repair (3,4). In vitro, these ends can undergo an alternative fate: they can capture an unrelated target DNA and undergo Rag-mediated transposition (5,6). In vivo, however, the Rag proteins are strongly biased towards recombination, with only rare transposition events detected to date (7–11). Deciphering the mechanisms by which the Rag complex carries out these two transesterification reactions should help provide insight into how transposition is suppressed in developing lymphocytes.
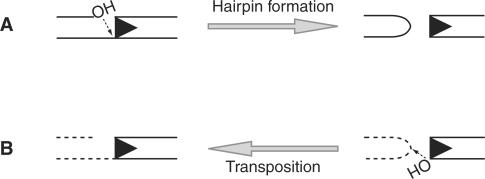
The Rag complex generates DSB in two steps, first introducing a nick between the RSS and the adjacent coding segment (the coding flank) and then using the resulting 3′-hydroxyl group to attack the phosphodiester backbone of the opposite DNA strand. This direct transesterification reaction forms a covalently sealed hairpin structure at the coding flank and a flush DSB at the signal end (Figure 1A) (12). The Rag proteins employ this same chemical mechanism to perform transposition. In this case, after cleavage, the Rag complex uses direct transesterification to insert a segment of DNA terminating in blunt signal ends into a new target location (5,6) (Figure 1B).
Figure 1.
Schematic representation of Rag-mediated transesterification reactions. (A) Hairpin formation is an intramolecular transesterification reaction in which the 3′-hydroxyl liberated by Rag nicking is used to attack the opposite strand of DNA at the heptamer border, resulting in a blunt signal end and a covalently sealed hairpin coding end. The RSS is indicated by a triangle. (B) Transposition is an intermolecular transesterification reaction in which the 3′-hydroxyl of a signal end is used to attack the phosphodiester backbone of the target DNA. Hairpin DNA structures facilitate Rag transposition. Both types of transesterification reaction require the catalytic triad (D600, D708 and E962) as well as the divalent cation Mg2+.
Hairpin formation requires local distortion of the coding flank DNA, in order to allow the attacking nucleophile to align precisely with the phosphodiester bond of the opposite strand. Substrates with unpaired bases at the coding flank facilitate this reaction (13,14). Similarly, transposition is facilitated by certain target DNA structures, including unpaired DNA, triplex DNA and some G-C rich sequences, suggesting that distortion is also required for transposition (5,6,15). The best targets appear to be hairpin tips (16,17), which are thought to adopt a variety of unusual DNA structures determined by the sequence of the terminal nucleotides (18). Indeed, certain hairpin sequences stimulate surprisingly efficient Rag transposition that approaches levels of cleavage (17).
Although there is as yet no structural information about the active site of the Rag complex, Rag1 is considered the catalytic subunit because it contains a number of amino acids required for cleavage (19–22), including a DDE triad responsible for coordinating catalytic metal ions (23–25). Rag2 plays various regulatory roles (3,26–34), but has not been firmly implicated in catalysis. By analogy with other hairpin-forming transposases (35,36), we and others searched for aromatic amino acids in Rag1 that could stabilize an extrahelical, flipped-out base. Through mutational and biochemical analysis, we found an aromatic residue in Rag1, W893, that may be involved in base flipping (20); others have proposed a second candidate, W956 (21). Several other amino acids in Rag1 are critical for hairpin formation and likely play key roles in generating or stabilizing a distorted DNA intermediate (20). One class of hairpin-deficient mutants is unusual in that the defect is conditional, depending on the coding flank sequence: some coding flank sequences prohibit hairpinning, whereas other flanks allow wild-type levels of hairpin formation (37–39). Because unpaired bases in the coding flank rescue this type of hairpinning defect, we and others have suggested that these conditional mutants may have difficulty generating sufficient distortion of the coding flank DNA to allow hairpin formation (13,14,39).
Although these earlier studies provided important clues about the DNA distortion phase of hairpin formation, the transesterification step was not examined. Here, we identify a mutant with a specific defect in transesterification, implicating basic amino acids in the catalytic core of Rag1 in this process. We also show that some Rag1 mutants with severe defects in hairpin formation nonetheless catalyze substantial levels of transposition, and, in several cases, are stimulated by hairpin targets. Lastly, we analyze a catalytically defective Rag2 mutant (the only such mutant identified) and find that it shows a defect in target capture and a novel form of coding flank sensitivity, suggesting an unexpected role for Rag2 in DNA cleavage and transposition.
MATERIALS AND METHODS
Rag proteins
Experiments were performed with recombinant glutathione S-transferase (GST)-tagged mouse core Rag1 (residues 384–1008) and core Rag2 (residues 1–387). 240 μg of Rag1 and 280 μg of Rag2 in the pEBG vector (40) were transiently co-transfected into Chinese hamster ovary (RMP41) cells using 780 μl of Fugene-6 transfection reagent (Roche, Indianapolis, Indiana). Proteins were co-purified as described (39), using GST affinity resin beads (GE, Piscataway, NJ). Multiple protein preparations were made, and the most active preparations were used. Mutant and wild-type Rag proteins shown in this article were prepared at the same time to eliminate variations between different preparations as a confounding factor in interpreting results, and were diluted such that equal amounts of each preparation (∼70 ng of Rag1 and 30 ng of Rag2) were added to reactions, as shown in Supplementary Figure 1.
HMG protein
Recombinant human HMGB1 was transformed into BL21(DE3) Escherichia coli (41). A culture (450 ml) of transformed cells was grown to an OD600 of 0.5; isopropyl-beta-D-thiogalactopyranoside was then added to a final concentration of 1 mM, followed by shaking at 37°C for 4 h. Cells were then resuspended in 20 ml of resuspension buffer (100 mM Tris pH 8.0; 1 mM EDTA; 10% glycerol, w/v) and centrifuged at 10 000 r.p.m. for 15 min at 4°C. Pellet was resuspended in 3 ml of resuspension buffer per gram of pellet. 2-Mercaptoethanol was added to a final concentration of 2 mM, followed by 0.2 vol of 5 mg/ml lysozyme. The mixture was kept on ice for 30 min. Next, Tween-20 was added to a final concentration of 0.1% and NaCl was added to a final concentration of 1 M. The mixture was incubated on ice for 10 min and then centrifuged at 30 000 r.p.m. for 30 min at 4°C. An equal volume of 10 mM imidazole buffer was added to supernatant (0.5 M NaCl; 20% glycerol; 20 mM Tris–Cl, pH 7.9 at 4°C; 10 mM imidazole, pH 8.0; 2 mM 2-mercaptoethanol). Chelating Sepharose (GE, Piscataway, NJ) was prepared with nickel sulphate; slurry was then washed with water, followed by 10 mM imidazole buffer. Slurry was added to lysate, and bound in batch overnight at 4°C. After binding, slurry was washed twice with 10 mM imidazole. HMG was then eluted in five fractions using 60 mM imidazole buffer, dialyzed in dialysis buffer (25 mM Tris, pH 8.0; 1 mM EDTA; 1 mM DTT; 150 mM KCl; 10% glycerol). Finally, HMG was heat-inactivated at 65°C for 30 min.
Oligonucleotide DNA substrates
Oligonucleotides used for cleavage assays were designed according to plasmid pJH290 (20), except that the sequence of the designated two base pairs at the coding flank was varied. For each reaction, both the 12 and 23-RSS are either uncleaved or prenicked and share the same coding flank sequence, whether matched or mismatched. Oligonucleotide-encoded hairpins used for transposition assays were described in (17) and contain the sequence 5′-CTGGGGCTGCAGGxx-xxCCTGCAGCCCCAG, where ‘xx-xx’ represents the tip of the hairpin. For a TG hairpin, ‘xx-xx’ represents TG-CA; for an AC hairpin, ‘xx-xx’ represents AC-GT; for a GC hairpin, ‘xx-xx’ represents GC–GC, etc.
Plasmid DNA substrates
Plasmid targets used for transposition assays were pUC8 and F14C, a pUC8 plasmid with a 106 bp inverted repeat cloned into the EcoRI site (42).
In vitro cleavage and transposition assays
Cleavage reactions and transposition reactions using oligonucleotide substrates were performed as described in (17,20), respectively, except that 70 ng of Rag1 and 30 ng of Rag2 were added to each reaction. Reaction products were detected and quantified using PhosphorImager and ImageQuant software.
Physical analysis of target capture
Target capture assays using radiolabeled hairpin target were performed as described (17), except that 70 ng of Rag1 and 30 ng of Rag2 were added to each reaction. Reactions were performed for 2 h before separation on a native 5% acrylamide gel at 100V, 4°C.
RESULTS
Mutations distinguishing hairpin formation and transposition
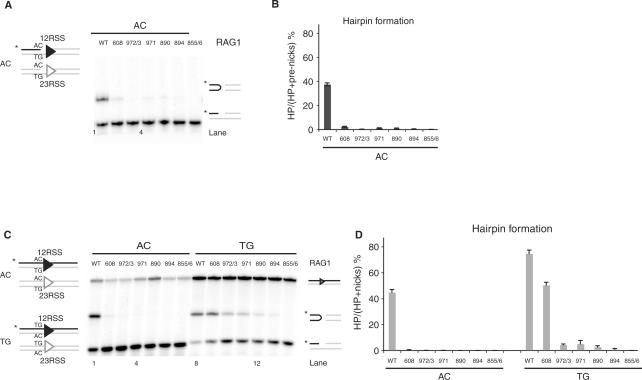
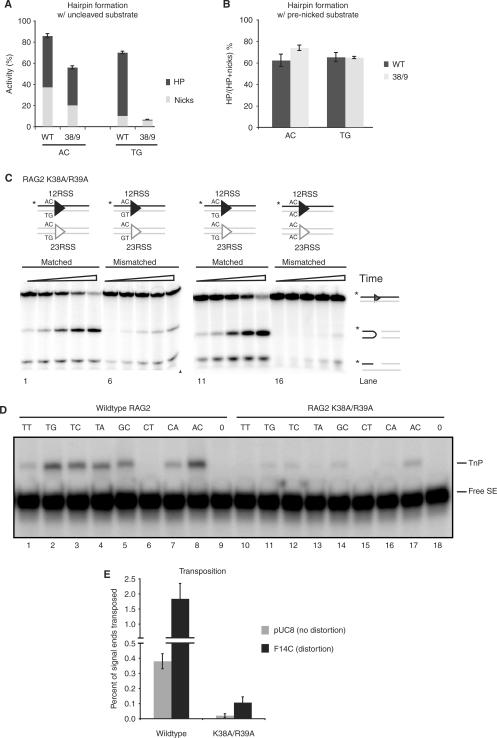
We began by examining a set of Rag1 mutants (K608A, R972A/K973A, F971A, K890A, R894A and R855A/K856A) previously shown to have defects in hairpin formation, as confirmed using prenicked substrates (Figure 2A and B). It was not known whether these defects affect an early step (generation or stabilization of a distorted DNA intermediate) or a later phase of the reaction (transesterification). To determine the reaction step affected by these mutants, we first tested whether the defect was due to coding flank sensitivity. Rag1 mutants mapping to amino acids 606–611 can nick but are unable to form hairpins on substrates with an AC coding flank, although they are capable of hairpinning on a TG coding flank (19,37,39). Only Rag1 K608A showed significant coding flank sensitivity, as evidenced by differential hairpin formation with substrates bearing AC or TG coding flanks (Figure 2C and D). Notably, R855A/K856A showed robust nicking but no detectable hairpinning activity with either TG or AC coding flanks (Figure 2C, lanes 7 and 14) or with substrates bearing a variety of other coding flank sequences (data not shown). Similar results were obtained with the single mutant, R855A (data not shown).
Figure 2.
Rag1 mutants exhibit defective hairpin formation. (A) In vitro cleavage assay using 12- and 23-RSS oligonucleotide substrates that mimic a nick on an AC flank, as depicted in schematic representation on the left. (B) Quantitation of gel in A, showing the percentage of pre-nicks converted to hairpins, was performed using ImageQuant software. Error bars represent SEM of three experiments. (C) In vitro cleavage assay using oligonucleotide substrates with either an AC or TG coding flank on both 12- and 23-RSS, as depicted on the left. Only the strands drawn as black lines were 5′-radiolabeled (*), and thus detectable after denaturing gel electrophoresis. The reaction was performed for 30 min, in the presence of paired 12- and 23-RSS oligonucleotides, purified Rag1 (wild-type or mutants) and Rag2 proteins, HMG and Mg2+. Reaction products were separated on a 12% sequencing gel and visualized by autoradiography. Reaction products are shown on the right (from top to bottom): uncleaved substrates, hairpin and nicks. (D) Quantitation of gel in C, showing the percentage of nicks converted to hairpins, was performed using ImageQuant software. Error bars represent SEM of three experiments.
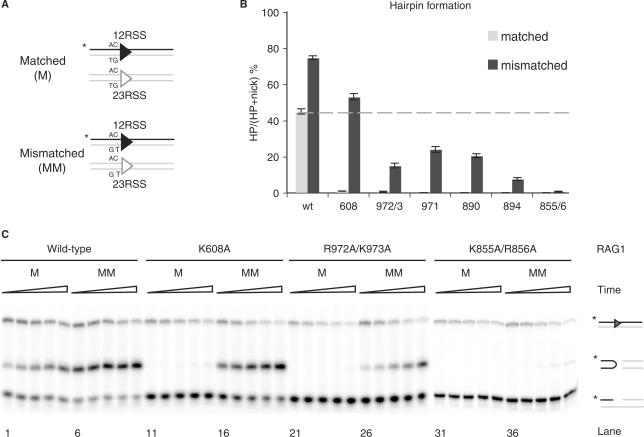
We next investigated the effects of mismatched coding flanks, which can rescue hairpin formation by certain Rag1 mutants [presumably by facilitating DNA distortion at the cleavage site (20,39)] (Figure 3A). In agreement with previous work (13,14,20,39), mismatches enhanced hairpin formation by the wild-type Rag complex and fully rescued hairpin formation by K608A (Figure 3B). Mismatched substrates partially rescued hairpin formation by R972A/K973A, F971A, K890A and R894A (Figure 3B). K855A/R856A shows traces of hairpin products, which could only be detected at long incubation times (Figure 3C), suggesting a specific defect in transesterification.
Figure 3.
Rag1 mutants exhibit varying levels of hairpin formation when provided a mismatched substrate. (A) Schematic representation showing oligonucleotide substrates containing matched (M) or mismatched (MM) base pairs at the coding flank on both 12- and 23-RSS. Only the strands drawn as a black line were 5′-radiolabeled (*), and thus detectable after denaturing gel electrophoresis. (B) Quantitation of in vitro hairpin formation after 30 min incubation time, showing the percentage of nicks converted to hairpins. Reaction products were separated on 12% sequencing gel, visualized by autoradiography and quantified using ImageQuant software. Dashed line represents the wild-type level of hairpin formation at a matched substrate. Error bars show SEM of three experiments. (C) In vitro hairpin formation assay. Representative results from wild-type Rag1, K608A, R972A/K973A and K855A/R856A are shown. Reaction products were separated on a 12% sequencing gel and visualized by autoradiography. Triangles represent increasing incubation times: 5, 10, 20, 40 and 80 min. Reaction products are depicted on the right (from top to bottom): uncleaved substrates, hairpins and nicks.
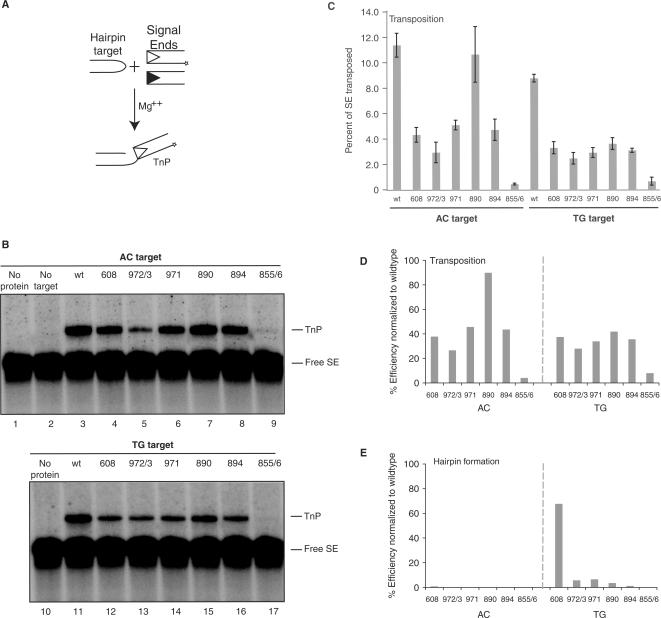
To assess the effects of these mutations on the transesterification step, we tested their ability to catalyze transposition. We used a standard in vitro assay with precleaved signal ends (to bypass hairpin formation) and oligonucleotide hairpin targets chosen to correspond to the substrates with AC or TG flanks shown in Figure 2 (Figure 4A). Several mutants were much more proficient for transposition than hairpin formation (Figure 4B): Rag1 R972A/K973A, F971A, K890A and R894A demonstrated transposition efficiencies of 25–95% of wild-type on an AC hairpin target and 30–40% of wild-type on a TG target (Figure 4C). To compare directly the ability of the mutants to perform hairpin formation and transposition, we normalized the data shown in Figures 2D and 4C, setting wild-type activity at 100% (Figure 4D and E). In this analysis, all Rag1 mutants except for R855A/K856A were much better at transposition than hairpin formation. R855A/K856A, however, was consistently defective for transposition (Figure 4B and C), and single mutant R855A also showed severely defective transposition (data not shown). This defect cannot be explained by its ability to capture target DNA, which is only mildly affected (Figure 5). We conclude that the hairpin formation defects of R972A/K973A, F971A, K890A and R894A do not reflect fundamental problems with the chemistry of transesterification but that R855A/K856A (and perhaps specifically R855A) exhibits a defect in this step.
Figure 4.
Several Rag1 nick-only mutants are able to transpose into different hairpin targets. (A) Schematic representation of in vitro transposition assay. Radiolabeled signal ends (*) are incubated in Ca2+ with Rag proteins and HMG. Annealed hairpin target is then added. Following incubation in Mg2+, transposition products are detected as signal ends covalently attached to the tips of hairpin targets. (B) In vitro transposition products were separated on a 15% sequencing gel, and visualized by autoradiography. (C) Quantitation of in vitro transposition assay shown in (B). Efficiency of transposition was determined as percentage of signal ends transposed by each of the mutants into AC and TG hairpin targets. Error bars represent SEM of five experiments. (D and E) Quantitation of activity of Rag1 mutants R972A/K973A, F971A, K890A, R894A, R855A/K856A as a percentage of wild-type activity in transposition into an AC or TG hairpin target (D) and hairpin formation on an uncleaved substrate with an AC or TG coding flank (E). Free SE, free (not transposed) signal ends; TnP, transposition products.
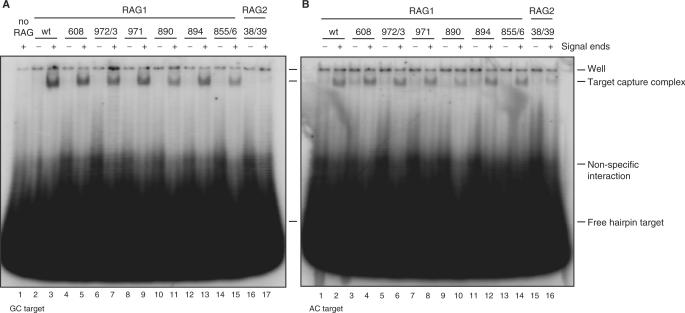
Figure 5.
Signal end complexes formed by Rag1 mutants, but not Rag2 K38A/R39A, are able to capture target. (A) Rag-signal end complexes were formed in vitro and then incubated with radiolabeled GC hairpin target. After 2 h, complexes were separated on a native 5% polyacrylamide gel, which was visualized by autoradiography. (B) Rag-signal end complexes were formed and then incubated with radiolabeled AC hairpin target. After 2 h, complexes were separated on a native 5% polyacrylamide gel, which was visualized by autoradiography. TCC, target capture complex.
Effects of hairpin targets on transposition by the Rag1 mutants
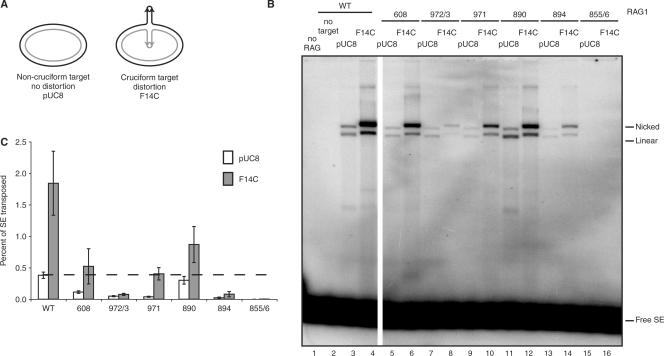
Four Rag1 mutants (R972A/K973A, F971A, K890A and R894A) were quite proficient for transposition into oligonucleotide targets. Given their severe defects in hairpin formation, we wondered whether any of these mutations might specifically affect recognition of hairpin targets, which strongly stimulate transposition by the wild-type Rag complex. To determine this, we used two previously described plasmid targets: pUC8 and pUC8 containing a 106-base pair inverted repeat (F14C) that forms a cruciform structure bearing hairpin tips [Figure 6A; (16,42,43)]. Three mutants (K608A, F971A and K890A), while somewhat impaired for transposition, were consistently and strongly stimulated by F14C (Figure 6B and C). These mutants thus retain the ability to recognize hairpin structures. One mutant (R894A) was modestly stimulated in some experiments. The hairpin target did not stimulate transposition by R972A/K973A, however (Figure 6B, lanes 7 and 8). R855A/K856A was inactive for transposition into both targets (Figure 6B and C, lanes 15 and 16), consistent with a defect in transesterification.
Figure 6.
Rag1 K608A, F971A and K890A are capable of wild-type levels of transposition when provided a cruciform target. (A) Schematic representation of transposition targets. The pUC8 plasmid was used as a nondistorted target control. F14C contains a 106 bp inverted repeat sequence that adopts a cruciform structure. (B) Products of in vitro transposition assay using targets in (A) were separated on a 1% agarose gel and visualized by autoradiography. All lanes were run on the same gel, but a few lanes between WT and 608 were removed for demonstration purposes. (C) Quantitation of percentage of signal ends transposed into either nondistorted pUC8 target or distorted F14C target in (B). Dashed line marks wild-type level of transposition into pUC8 target. Error bars represent SEM of five experiments. Nicked, products of a single signal end insertion into plasmid yield a nicked plasmid transposition product; linear, products of a double signal end insertion into plasmid target yield a linearized plasmid transposition product; free SE, free (un-transposed) signal ends.
A Rag2 mutation affects both nicking and target capture
To better understand the involvement of Rag2 in hairpin formation and transposition, we analyzed the only Rag2 mutation suspected to affect catalysis, K38A/R39A (44). We found, surprisingly, that this mutation causes coding flank sensitivity: it forms hairpins near wild-type levels at an AC flank but is almost inactive with a TG flank (Figure 7A). This is a reversal of the coding flank preferences of Rag1 mutants, which form hairpins efficiently at TG, but not AC, flanks. Even more surprisingly, this conditional defect in cleavage mainly affects the nicking reaction. Total levels of nicking activity on a TG flank (reflected in the amounts of nicks plus hairpins) were only 10% of levels on other substrates (Figure 7A), and a prenicked substrate fully rescued hairpin formation (Figure 7B). Consistent with results in Qiu et al. (44), mismatched base pairs at the coding flank actually inhibited nicking and hairpin formation by K38A/R39A, again in direct contradistinction to the behavior of the Rag1 mutants (Figure 7C); four different pairs of mismatched sequences produced similar results (data not shown). These data indicate that Rag2 plays a previously unsuspected role in cleavage.
Figure 7.
Rag2 K38A/R39A forms hairpins but is deficient for transposition. (A) Quantification of in vitro cleavage reaction to measure hairpin and nick formation was performed using oligonucleotide substrates containing either an AC or TG flank (as in Figure 2A). Reaction products were separated on 12% sequencing gel, visualized by autoradiography and quantified using ImageQuant software. Top part of bar represents the percentage of hairpin products converted from nicks, and bottom part represents the percentage of nicked products. Height of each bar (top + bottom) represents total nicking activity. (B) In vitro cleavage reaction using oligonucleotide substrates containing a nick (as in Figure 2C), with either AC or TG flanks. The reaction was visualized and quantified as in (A). The quantification shows the percentage of nicks converted to hairpins by wild-type and Rag2 K38A/R39A. (C) Cleavage reaction by mutant K38A/R39A, using oligonucleotide substrates containing a matched (M) or mismatched (MM) coding flank. Two different pairs of MM sequences were used as shown schematically above gels. (D) Reaction products from in vitro transposition into multiple targets by Rag2 K38A/R39A. Reaction products were separated on a 15% sequencing gel and visualized as in (A). (E) Quantification of in vitro transposition into nondistorted (pUC8) or distorted (F14C) plasmid targets comparing wild-type and Rag2 K38A/R39A. Reaction products were separated on a 1% native agarose gel, visualized by autoradiography and quantified using ImageQuant software. Graph represents percentage of signal ends transposed into target; error bars show SEM over three experiments. TnP, transposition products; free SE, free (untransposed) signal ends.
Lastly, we investigated the effects of the K38A/R39A mutation on transposition. This mutant showed no detectable transposition of precleaved signal ends into a number of hairpin target sequences and was only weakly active using the most favored targets (Figure 7D). Transposition into plasmid targets was quite weak, although some activity was observed with the F14C cruciform target (Figure 7E). K38A/R39A was severely defective for capture of two hairpin target sequences (Figure 5).
DISCUSSION
Rag1 amino acids critical for transesterification
We previously identified residues in Rag1 involved in hairpin formation and provided evidence that Rag1 possesses a YKEFRK motif that may be functionally similar to the YREK motif found in IS4 family transposases (20). Now, data in the present work have identified amino acids in the Rag complex that are specifically required for transesterification (R855/K856). Transposition by IS4 family transposases, which involves hairpin formation, also requires basic residues: the structure of the Tn5/DNA complex shows that the 5′ phosphate group of thymine 2 of the transferred strand is held in place by specific contacts with R210 and R322 (35). R322 is part of the YREK motif (45), and mutation of this residue diminishes hairpin opening and strand transfer activity (46). We speculate that R855 in Rag1, which, similar to R210 in Tn5, is located N-terminal to and apart from the catalytic E residue, may likewise interact with the phosphate backbone of the DNA, and that the R855A/K856A mutant may fail to correctly position the DNA substrate in the catalytic pocket.
Novel roles for Rag2 in DNA cleavage and transposition
Although it has long been known that Rag2 is required for recombination and transposition, its mechanistic contributions to these reactions have remained unknown. Our analyses of the K38A/R39A Rag2 mutant revealed an unexpected sensitivity to coding flank sequence. [R39A was previously noted to cleave at wild-type levels, but this study was performed using favorable coding flank sequences (47); C.P.L. and D.B.R., unpublished data.] Our data provide strong evidence that Rag2 plays a critical role both in contacting DNA at the RSS-coding flank junction and in site-specific nicking.
The phenotype of Rag2 K38A/R39A differs from that of the coding flank sensitive Rag1 mutants in two interesting respects. K38A/R39A shows diametrically opposite coding flank sequence preferences, and it displays its sequence sensitivity at the nicking step of the reaction rather than the hairpin formation step. The nicking defect could reflect less stable binding of the mutant complexes to particular coding flank sequences. Another (nonexclusive) possibility is that nicking, like hairpin formation, may be promoted by formation of a distorted DNA structure, whose stability is influenced by the coding flank sequence. A precedent for DNA distortion prior to nicking is provided by another hairpin-forming transposase, Tn5: mutation of W323 affects both distortion prior to nicking and the nicking step itself (48). Our observation that unpaired coding flank nucleotides inhibit nicking by K38A/R39A suggests that these unpaired substrates prevent this mutant from creating the proper distortion for nicking. These findings, in conjunction with the previous observation that Rag2 contacts DNA at the coding flank (28,49,50), strongly suggest that the N-terminus of Rag2 contacts the cleavage site and that these contacts are critical for catalysis of nicking. Our data also provide insight into the role of Rag2 in transposition. The severe target binding defect of K38A/R39A indicates Rag2 involvement at an early stage in the recognizing or binding of DNA, and, in particular, participation of the N-terminal region of Rag2 in making critical contacts with target DNA.
Differences between hairpin formation and transposition
We have found that a number of Rag1 mutants with severe defects in hairpin formation remain remarkably competent for transposition. The K890A mutant is particularly striking in that it combines a profound defect in hairpin formation with near-wild-type transposition activity. This suggests that the mutations affect generation of distorted DNA intermediates, while retaining the ability to bind to hairpin targets. In addition, mismatches in the coding flanks facilitated hairpin formation by R894A and R972A/K973A; yet, these mutants failed to show a corresponding increase in transposition activity when presented with hairpin targets, which are normally quite conducive to this reaction. Together, these data suggest that the Rag complex may utilize different combinations of residues to engage unpaired or distorted DNA in the two reactions. It is instructive in this regard to consider studies of phosphorothioate stereoselectivity in Tn10, which suggest that the transesterification steps of hairpin formation and transposition occur with significant conformational differences at the active site (51).
Further analyses of postcleavage and target capture complexes should help define the molecular basis for our observations. Moreover, our identification of mutants that alter the balance between the two alternative transesterification reactions, hairpin formation and transposition, raises the interesting possibility that some Rag mutants could promote increased transposition, perhaps at the expense of efficient V(D)J recombination.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
The authors thank R. Chalmers, J. Bischerour and members of the Roth laboratory, particularly G. Celli, L. Deriano and S. Wong, for critical reading and thoughtful comments. This study was supported by National Institutes of Health (AI36420 to D.B.R.); Irene Diamond Foundation (to D.B.R.). Funding to pay the Open Access publication charges for this article was provided by NIH AI36420.
Conflict of interest statement. None declared.
REFERENCES
- 1.Gellert M. V(D)J recombination: RAG proteins, repair factors, and regulation. Annu. Rev. Biochem. 2002;71:101–132. doi: 10.1146/annurev.biochem.71.090501.150203. [DOI] [PubMed] [Google Scholar]
- 2.Roth DB. Restraining the V(D)J recombinase. Nat. Rev. Immunol. 2003;3:656–666. doi: 10.1038/nri1152. [DOI] [PubMed] [Google Scholar]
- 3.Corneo B, Wendland RL, Deriano L, Cui X, Klein IA, Wong SY, Arnal S, Holub AJ, Weller GR, Pancake BA, et al. Rag mutations reveal robust alternative end joining. Nature. 2007;449:483–486. doi: 10.1038/nature06168. [DOI] [PubMed] [Google Scholar]
- 4.Lee GS, Neiditch MB, Salus SS, Roth DB. RAG proteins shepherd double-strand breaks to a specific pathway, suppressing error-prone repair, but RAG nicking initiates homologous recombination. Cell. 2004;117:171–184. doi: 10.1016/s0092-8674(04)00301-0. [DOI] [PubMed] [Google Scholar]
- 5.Agrawal A, Eastman QM, Schatz DG. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature. 1998;394:744–751. doi: 10.1038/29457. [DOI] [PubMed] [Google Scholar]
- 6.Hiom K, Melek M, Gellert M. DNA transposition by the RAG1 and RAG2 proteins: a possible source of oncogenic translocations. Cell. 1998;94:463–470. doi: 10.1016/s0092-8674(00)81587-1. [DOI] [PubMed] [Google Scholar]
- 7.Chatterji M, Tsai CL, Schatz DG. Mobilization of RAG-generated signal ends by transposition and insertion in vivo. Mol. Cell Biol. 2006;26:1558–1568. doi: 10.1128/MCB.26.4.1558-1568.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reddy YV, Perkins EJ, Ramsden DA. Genomic instability due to V(D)J recombination-associated transposition. Genes Dev. 2006;20:1575–1582. doi: 10.1101/gad.1432706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Messier TL, O’Neill JP, Hou SM, Nicklas JA, Finette BA. In vivo transposition mediated by V(D)J recombinase in human T lymphocytes. EMBO J. 2003;22:1381–1388. doi: 10.1093/emboj/cdg137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clatworthy AE, Valencia MA, Haber JE, Oettinger MA. V(D)J recombination and RAG-mediated transposition in yeast. Mol. Cell. 2003;12:489–499. doi: 10.1016/s1097-2765(03)00305-8. [DOI] [PubMed] [Google Scholar]
- 11.Curry JD, Schulz D, Guidos CJ, Danska JS, Nutter L, Nussenzweig A, Schlissel MS. Chromosomal reinsertion of broken RSS ends during T cell development. J. Exp. Med. 2007;204:2293–2303. doi: 10.1084/jem.20070583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McBlane JF, van Gent DC, Ramsden DA, Romeo C, Cuomo CA, Gellert M, Oettinger MA. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell. 1995;83:387–395. doi: 10.1016/0092-8674(95)90116-7. [DOI] [PubMed] [Google Scholar]
- 13.Ramsden DA, McBlane JF, van Gent DC, Gellert M. Distinct DNA sequence and structure requirements for the two steps of V(D)J recombination signal cleavage. EMBO J. 1996;15:3197–3206. [PMC free article] [PubMed] [Google Scholar]
- 14.Cuomo CA, Mundy CL, Oettinger MA. DNA sequence and structure requirements for cleavage of V(D)J recombination signal sequences. Mol. Cell. Biol. 1996;16:5683–5690. doi: 10.1128/mcb.16.10.5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsai CL, Chatterji M, Schatz DG. DNA mismatches and GC-rich motifs target transposition by the RAG1/RAG2 transposase. Nucleic Acids Res. 2003;31:6180–6190. doi: 10.1093/nar/gkg819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee GS, Neiditch MB, Sinden RR, Roth DB. Targeted transposition by the V(D)J recombinase. Mol. Cell. Biol. 2002;22:2068–2077. doi: 10.1128/MCB.22.7.2068-2077.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Posey JE, Pytlos MJ, Sinden RR, Roth DB. Target DNA structure plays a critical role in RAG transposition. PLoS Biol. 2006;4:e350. doi: 10.1371/journal.pbio.0040350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kabotyanski EB, Zhu C, Kallick DA, Roth DB. Hairpin opening by single-strand specific nucleases. Nucleic Acids Res. 1995;23:3872–3881. doi: 10.1093/nar/23.19.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huye LE, Purugganan MM, Jiang MM, Roth DB. Mutational analysis of all conserved basic amino acids in RAG-1 reveals catalytic, step arrest, and joining-deficient mutants in the V(D)J recombinase. Mol. Cell. Biol. 2002;22:3460–3473. doi: 10.1128/MCB.22.10.3460-3473.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu CP, Sandoval H, Brandt VL, Rice PA, Roth DB. Amino acid residues in Rag1 crucial for DNA hairpin formation. Nat. Struct. Mol. Biol. 2006;13:1010–1015. doi: 10.1038/nsmb1154. [DOI] [PubMed] [Google Scholar]
- 21.Grundy GJ, Hesse JE, Gellert M. Requirements for DNA hairpin formation by RAG1/2. Proc. Natl Acad. Sci. USA. 2007;104:3078–3083. doi: 10.1073/pnas.0611293104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li W, Chang FC, Desiderio S. Rag-1 mutations associated with B-cell-negative scid dissociate the nicking and transesterification steps of V(D)J recombination. Mol. Cell. Biol. 2001;21:3935–3946. doi: 10.1128/MCB.21.12.3935-3946.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Landree MA, Wibbenmeyer JA, Roth DB. Mutational analysis of RAG-1 and RAG-2 identifies three active site amino acids in RAG-1 critical for both cleavage steps of V(D)J recombination. Genes Dev. 1999;13:3059–3069. doi: 10.1101/gad.13.23.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fugmann SD, Villey IJ, Ptaszek LM, Schatz DG. Identification of two catalytic residues in RAG1 that define a single active site within the RAG1/RAG2 protein complex. Mol. Cell. 2000;5:97–107. doi: 10.1016/s1097-2765(00)80406-2. [DOI] [PubMed] [Google Scholar]
- 25.Kim DR, Dai Y, Mundy CL, Yang W, Oettinger MA. Mutations of acidic residues in RAG1 define the active site of the V(D)J recombinase. Genes Dev. 1999;13:3070–3080. doi: 10.1101/gad.13.23.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hiom K, Gellert M. A stable RAG1-RAG2-DNA complex that is active in V(D)J cleavage. Cell. 1997;88:65–72. doi: 10.1016/s0092-8674(00)81859-0. [DOI] [PubMed] [Google Scholar]
- 27.Akamatsu Y, Oettinger MA. Distinct roles of RAG1 and RAG2 in binding the V(D)J recombination signal sequences. Mol. Cell. Biol. 1998;18:4670–4678. doi: 10.1128/mcb.18.8.4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Swanson PC, Desiderio S. RAG-2 promotes heptamer occupancy by RAG-1 in the assembly of a V(D)J initiation complex. Mol. Cell. Biol. 1999;19:3674–3683. doi: 10.1128/mcb.19.5.3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sekiguchi JA, Whitlow S, Alt FW. Increased accumulation of hybrid V(D)J joins in cells expressing truncated versus full-length RAGs. Mol. Cell. 2001;8:1383–1390. doi: 10.1016/s1097-2765(01)00423-3. [DOI] [PubMed] [Google Scholar]
- 30.Akamatsu Y, Monroe R, Dudley DD, Elkin SK, Gartner F, Talukder SR, Takahama Y, Alt FW, Bassing CH, Oettinger MA. Deletion of the RAG2 C terminus leads to impaired lymphoid development in mice. Proc. Natl Acad. Sci. USA. 2003;100:1209–1214. doi: 10.1073/pnas.0237043100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Elkin SK, Matthews AG, Oettinger MA. The C-terminal portion of RAG2 protects against transposition in vitro. Embo J. 2003;22:1931–1938. doi: 10.1093/emboj/cdg184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsai CL, Schatz DG. Regulation of RAG1/RAG2-mediated transposition by GTP and the C-terminal region of RAG2. Embo J. 2003;22:1922–1930. doi: 10.1093/emboj/cdg185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.West KL, Singha NC, De Ioannes P, Lacomis L, Erdjument-Bromage H, Tempst P, Cortes P. A direct interaction between the RAG2 C terminus and the core histones is required for efficient V(D)J recombination. Immunity. 2005;23:203–212. doi: 10.1016/j.immuni.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 34.Liu Y, Subrahmanyam R, Chakraborty T, Sen R, Desiderio S. A plant homeodomain in RAG-2 that binds hypermethylated lysine 4 of histone H3 is necessary for efficient antigen-receptor-gene rearrangement. Immunity. 2007;27:561–571. doi: 10.1016/j.immuni.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davies DR, Goryshin IY, Reznikoff WS, Rayment I. Three-dimensional structure of the Tn5 synaptic complex transposition intermediate. Science. 2000;289:77–85. doi: 10.1126/science.289.5476.77. [DOI] [PubMed] [Google Scholar]
- 36.Zhou L, Mitra R, Atkinson PW, Hickman AB, Dyda F, Craig NL. Transposition of hAT elements links transposable elements and V(D)J recombination. Nature. 2004;432:995–1001. doi: 10.1038/nature03157. [DOI] [PubMed] [Google Scholar]
- 37.Sadofsky MJ, Hesse JE, van Gent DC, Gellert M. RAG-1 mutations that affect the target specificity of V(D)J recombination: a possible direct role of RAG-1 in site recognition. Genes Dev. 1995;9:2193–2199. doi: 10.1101/gad.9.17.2193. [DOI] [PubMed] [Google Scholar]
- 38.Roman CAJ, Baltimore D. Genetic evidence that the RAG1 protein directly participates in V(D)J recombination through substrate recognition. Proc. Natl Acad. Sci. USA. 1996;93:2333–2338. doi: 10.1073/pnas.93.6.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kale SB, Landree MA, Roth DB. Conditional RAG-1 mutants block the hairpin formation step of V(D)J recombination. Mol. Cell. Biol. 2001;21:459–466. doi: 10.1128/MCB.21.2.459-466.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spanopoulou E, Cortes P, Shih C, Huang C.-M, Silver DP, Svec P, Baltimore D. Localization, interaction, and RNA binding properties of the V(D)J recombination-activating proteins RAG1 and RAG2. Immunity. 1995;3:715–726. doi: 10.1016/1074-7613(95)90061-6. [DOI] [PubMed] [Google Scholar]
- 41.Huye LE, Roth DB. Differential requirements for cis and trans V(D)J cleavage: effects of substrate length. Nucleic Acids Res. 2000;28:4903–4911. doi: 10.1093/nar/28.24.4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sinden RR, Zheng GX, Brankamp RG, Allen KN. On the deletion of inverted repeated DNA in Escherichia coli: effects of length, thermal stability, and cruciform formation in vivo. Genetics. 1991;129:991–1005. doi: 10.1093/genetics/129.4.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oussatcheva EA, Shlyakhtenko LS, Glass R, Sinden RR, Lyubchenko YL, Potaman VN. Structure of branched DNA molecules: gel retardation and atomic force microscopy studies. J. Mol. Biol. 1999;292:75–86. doi: 10.1006/jmbi.1999.3043. [DOI] [PubMed] [Google Scholar]
- 44.Qiu JX, Kale SB, Yarnell Schultz H, Roth DB. Separation-of-function mutants reveal critical roles for RAG2 in both the cleavage and joining steps of V(D)J recombination. Mol. Cell. 2001;7:77–87. doi: 10.1016/s1097-2765(01)00156-3. [DOI] [PubMed] [Google Scholar]
- 45.Rezsohazy R, Hallet B, Delcour J, Mahillon J. The IS4 family of insertion sequences: evidence for a conserved transposase motif. Mol. Microbiol. 1993;9:1283–1295. doi: 10.1111/j.1365-2958.1993.tb01258.x. [DOI] [PubMed] [Google Scholar]
- 46.Naumann TA, Reznikoff WS. Tn5 transposase active site mutants. J. Biol. Chem. 2002;277:17623–17629. doi: 10.1074/jbc.M200742200. [DOI] [PubMed] [Google Scholar]
- 47.Fugmann SD, Schatz DG. Identification of basic residues in RAG2 critical for DNA binding by the RAG1-RAG2 complex. Mol. Cell. 2001;8:899–910. doi: 10.1016/s1097-2765(01)00352-5. [DOI] [PubMed] [Google Scholar]
- 48.Bischerour J, Chalmers R. Base-flipping dynamics in a DNA hairpin processing reaction. Nucleic Acids Res. 2007;35:2584–2595. doi: 10.1093/nar/gkm186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eastman QM, Villey IJ, Schatz DG. Detection of RAG protein-V(D)J recombination signal interactions near the site of DNA cleavage by UV cross-linking. Mol. Cell. Biol. 1999;19:3788–3797. doi: 10.1128/mcb.19.5.3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kirch SA, Rathbun G, Oettinger MA. Dual role of RAG2 in V(D)J recombination: catalysis and regulation of ordered Ig gene assembly. EMBO J. 1998;17:4881–4886. doi: 10.1093/emboj/17.16.4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kennedy AK, Haniford DB, Mizuuchi K. Single active site catalysis of the successive phosphoryl transfer steps by DNA transposases: insights from phosphorothioate stereoselectivity. Cell. 2000;101:295–305. doi: 10.1016/s0092-8674(00)80839-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.